Abstract
Amelogenins are the major proteins involved in tooth enamel formation. In the present study we have cloned and sequenced four novel amelogenins from three amphibian species in order to analyze similarities and differences between mammalian and non-mammalian amelogenins. The newly sequenced amphibian amelogenin sequences were from a Red-eyed tree frog (Litoria chloris) and a Mexican axolotl (Ambystoma mexicanum). We identified two amelogenin isoforms in the Eastern Red-backed Salamander (Plethodon cinereus). Sequence comparisons confirmed that non-mammalian amelogenins are overall shorter than their mammalian counterparts, contain less proline and less glutamine, and feature shorter polyproline tripeptide repeat stretches than mammalian amelogenins. We propose that unique sequence parameters of mammalian amelogenins might be a pre-requisite for complex mammalian enamel prism architecture.
Introduction
Amelogenin is the major protein component (90%) of the mammalian enamel protein matrix (1–2). A series of genetic, antisense, knockout, and crystal growth studies of the recent decade have established amelogenin’s pivotal role in the control of enamel crystal growth and enamel formation (3–6). While amelogenins are not the only proteins in the developing enamel matrix, they have nevertheless been attributed a major role in the growth of elongated enamel crystals (3–6). In previous studies from our laboratory, we have characterized the structure of the amelogenin-rich enamel protein matrix (4, 7, 8) and its functional changes related to enamel crystal growth (4). Our studies have established a functional relevance for the structured amelogenin matrix to control enamel crystal growth (4) and a close relationship between crystal nucleation and changes in matrix configuration during initial crystal formation (8).
In the recent years, our knowledge of the enamel protein composition and function of non-mammalian vertebrates has seen significant progress. Amelogenin sequences from reptilian and amphibian teeth have been published (9–10), and biochemical and immunohistochemical studies have enhanced our knowledge of enamel protein homologies between different vertebrate species (12–16). Immunohistochemical findings have confirmed earlier reports on a predominance of enamelins in shark enameloid, compared to a predominance of amelogenins in reptilian and mammalian enamel (16). Based on molecular phylogenetic studies, the amelogenin signal peptide in exon 2 has been linked to a similar region of the SPARC, and suggesting that exon 2 was duplicated to amelogenin approximately 630 MYA (11). According to homology analyses, intermediaries in the molecular evolution of amelogenins from SPARC were SPARCL1, enamelin, and ameloblastin (17).
In the present study we have generated sequence data for four novel amphibian amelogenins to analyze and compare key parameters of mammalian and non-mammalian amelogenins. In addition, we have documented prismatic mammalian enamel with non-prismatic reptilian and amphibian microarchitecture.
Materials and Methods
Source and isolation of the genomic DNA
For the present analysis, three amphibian species were chosen, a Red-eyed tree frog, (Litoria chloris); a Mexican axolotl (Ambystoma mexicanum) and an Eastern Red-backed Salamander (Plethodon cinereus). Amphibians were euthanized according to approved guidelines by the UIC animal care committee. The genomic DNA was isolated using GenElute™ Mammalian Genomic DNA Miniprep (Sigma-Aldrich Co., St. Louis, Mo) following the manufacturer’s instruction. The isolated genomic DNA was kept at −80°C for future use.
RNA isolation
RNA isolation was performed as previously reported (10). Briefly, amphibian jaws were removed and immediately frozen in liquid nitrogen and homogenized. The homogenized teeth tissue in TRI AGENT® reagent (Sigma) was mixed with 0.2 ml of chloroform and shaken vigorously for 30 sec. The mixture was centrifuged at 12,000× g for 20 min at 4 °C after 10 min incubation at room temperature. The aqueous phase was mixed with equal volume of isopropanol, and then centrifuged at 12,000× g for 20 min at 4 °C. The pellet was washed with 70% ethanol and dissolved in DEPC-treated H2O. The isolated RNA was kept at −80 °C for future use.
cDNA synthesis, cloning and sequencing
The sequencing strategy was as described by Wang et al. (10, 18). Briefly, primers were selected based on sequence analysis of amelogenin genes from selected species with the aid of Lasergene software (DNASTAR Inc., Madison, WI). The consensus sequences among different species were chosen as primers for amplification of novel amelogenin genes. Reverse transcriptase reaction was performed using SuperSript™ II Reverse Transcriptase (Invitrogen, Carlsbad, CA). The PCR products were purified with QIAquick® Gel Extraction Kit (Qiagen Inc., Valencia, CA), ligated to pGEM®-T Easy vector, and transformed into JM109 competent cells (Promega, Madison, WI). Transformants were picked up, and cultivated in LB medium containing ampicillin at a final concentration of 50μg/ml. Recombinant plasmids were isolated with Wizard® Plus SV Minipreps DNA Purification System (Promega), identified by enzyme digestion with EcoR I, and sequenced using the ABI 377 sequencer (Northwoods DNA Inc, Becida, MN). Three colonies were selected and sequenced 2 times from both orientations with either T7 or SP6 primers.
Sequence analysis
The four newly discovered amelogenin amino acid sequences were deduced from the nucleotide sequence, and sequence analyses were performed to identify its features. Sequences were manually aligned to represent optimum interspecies homology.
Scanning electron microscopy
For scanning electron microscopy, frog, squamate, and mammalian teeth from the du Brul collection at the University of Illinois were longitudinally sectioned and etched using EDTA (ethylenediaminetetraacetic acid). Etched enamel surfaces were then analyzed using a Joel JSM-6320F at the UIC RRC laboratory.
Results
Higher complexity of tooth enamel microstructure in mammals versus amphibians/reptilians
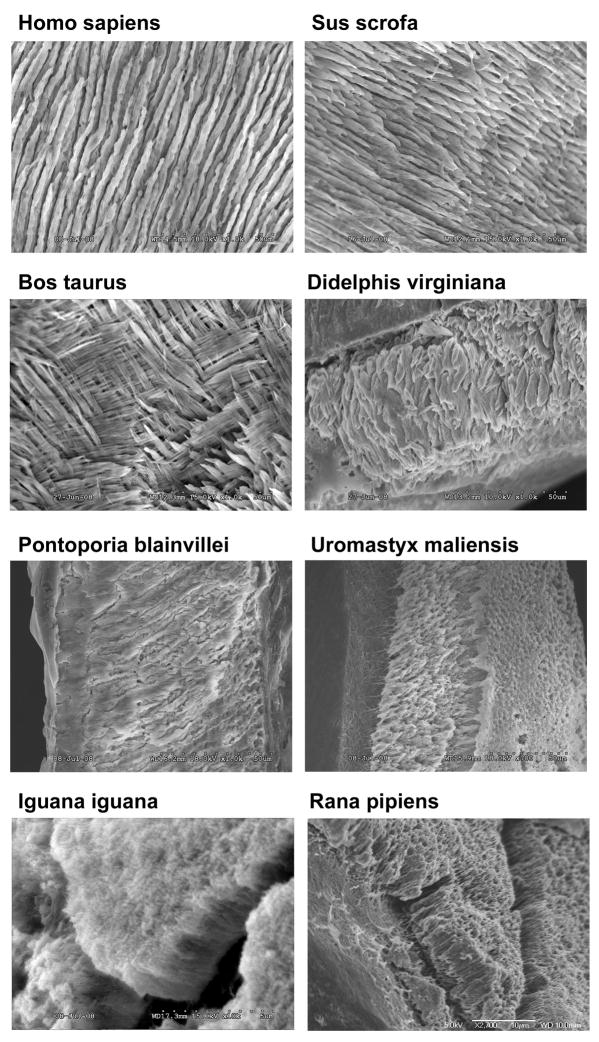
Scanning electron micrographs illustrated prismatic organization of enamel microstructure in all mammals investigated (human, Homo sapiens; pig, Sus scrofa; steer, Bos Taurus; Virginia opossum, Didelphis virginiana; and La Plata river dolphin, Pontoporia blainvillei), while there was no prismatic enamel structure in most squamates and amphibians (e.g. Green Iguana, Iguana iguana; Leopard frog, Rana pipiens). The Spiny-tailed lizard (Uromastyx maliensis) was unique among squamates as its enamel is prismatic (Fig. 1).
Figure 1. Enamel prisms and the non-mammalian/mammalian transition.
Scanning electron micrographs resolve long and parallel enamel prisms in omnivores (human, Homo sapiens, and pig, Sus scrofa). Note the pronounced plywood structure in ruminants (steer, Bos taurus) and marsupials (Virginia opossum, Didelphis virginiana). The enamel layer of dolphins (La Plata river dolphin, Pontoporia blainvillei) is fairly thin for eutherians and consists mostly of radial enamel. The Spiny-tailed lizard (Uromastyx maliensis) is unique among squamates as its enamel is prismatic. In most squamates (e.g. Green Iguana, Iguana iguana) and amphibians (e.g. Leopard frog, Rana pipiens) the enamel is devoid of prisms.
Four novel amphibian amelogenin genes confirm conserved elements of tetrapod amelogenins
For this study, RNA was isolated from the tooth-bearing elements of three amphibian species, a Red-eyed tree frog, Litoria chloris; a Mexican axolotl Ambystoma mexicanum; and an Eastern Red-backed Salamander Plethodon cinereus. Based on RNA extracts, four novel amphibian amelogenin cDNAs were cloned, sequenced, and deposited in Genbank (Red-eyed tree frog, Litoria chloris, accession number DQ069788; Mexican axolotl Ambystoma mexicanum, accession number DQ069791; and two amelogenin isoforms from the Eastern Red-backed Salamander Plethodon cinereus, accession numbers DQ069789 and DQ069789). Translated amino acid sequences are listed an aligned in Fig. 2. In this alignment, sequence elements encoded by exons 2, 3, and 5 were highly conserved, while portions of the exon 6 encoded domain were highly variable among species (Fig. 2). Nevertheless, also in the exon 6 encoded regions, unique polyproline tripeptide repeats were fairly conserved (Fig. 2).
Figure 2. Conserved regions and evolutionary “hotspots” among tetrapod amelogenins.
Note the highly conserved sequence elements in encoded by exons 2, 3, and 5 (marked in red and green). In contrast, the region encoded by exon 6 reveals significant differences among species. Conserved areas of polyproline tripeptide repeats are labeled in gray. This alignment features four newly discovered amphibian amelogenins (Red-eyed tree frog, Litoria chloris, accession number DQ069788; Mexican axolotl Ambystoma mexicanum, accession number DQ069791; Eastern Red-backed Salamander Plethodon cinereus, accession numbers DQ069789 and DQ069789). The Leopard frog (Rana pipiens) amelogenin sequence was reported earlier by our group (#AY695795).
Mammalian amelogenins are longer, contain more proline and glutamine, and have more proline-tripeptide repeats that their amphibian/reptilian counterparts
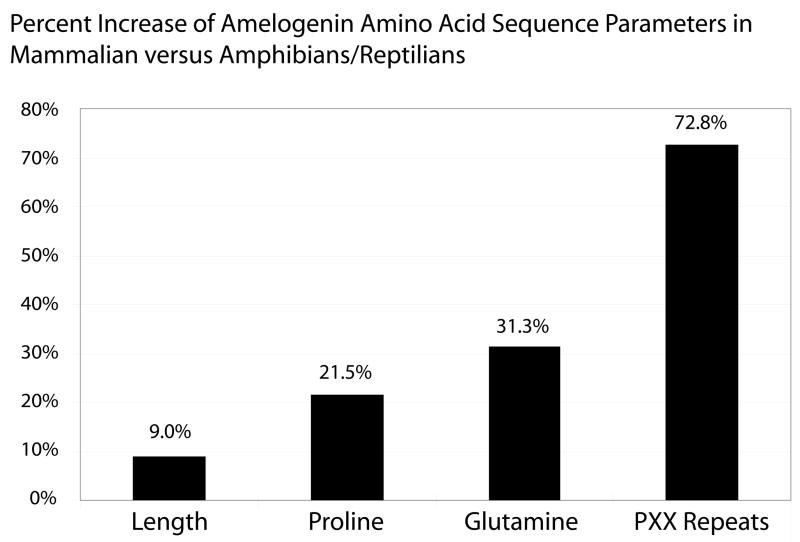
In order to determine differences between mammalian and non-mammalian amelogenin proteins, a number of key parameters such as length, amino acid composition, and the number of proline repeats were analyzed based on amelogenin sequences presented in Fig. 2. Comparing mammalian and non-mammalian amelogenins, the overall amelogenin length increased by 9%, the proline content increased by 21.5%, the glutamine content increased by 31.3%, and the number of proline-tripeptide repeats increased by 72.8% (Fig. 3).
Figure 3. Differences between mammalian and non-mammalian amelogenin sequence parameters.
A numerical comparison of key parameters of the translated amelogenin protein sequence between mammalian and non-mammalian species revealed that the overall amelogenin length was increased by 9%, the proline content increased by 21.5%, the glutamine content increased by 31.3%, and the number of proline-tripeptide repeats increased by 72.8%.
Discussion
Already studies by Bonass et al. pointed toward a functional significance of the amelogenin center domain as an evolutionary “hotspot” by identifying seven tandem repeats of a section of nucleotides with the consensus sequence CTGCAGCCC (19). Further support for the importance of the central amelogenin domain for enamel crystal growth has been provided by transgenic studies documenting that LRAP (a small amelogenin-derived peptide containing most of the A- and B-domain) fails to rescue an amelogenin null phenotype (20). Recent studies have linked the emergence of genes with new functions to gene duplication and alternative splicing (21, 22) providing theoretical support for a novel evolutionary mechanism by which mammalian amelogenin may have evolved through tandem exon duplication and substitution alternative splicing. According to evolutionary studies, the rapid evolution of the central amelogenin domain is primarily accomplished by insertions of PXX or PXQ tripeptide motifs (19), with both proline and glutamine causing structural rigidity of the newly added tripeptide complexes (23). These studies suggest that amelogenin evolution is associated with significant alterations in the physico-chemical properties of the amelogenin molecule.
Here we have confirmed that amphibian amelogenins are overall shorter than their mammalian counterparts (9), contain less proline and less glutamine, and most significantly, feature shorter polyproline tripeptide repeat stretches than mammalian amelogenins. Moreover, there is ample evidence for a simple, prism-less enamel microarchitecture in most amphibians and reptilians, while mammalian enamel is often organized into prisms that frequently form a plywood structure (24). While direct evidence for a relationship between amelogenin gene structure and enamel prism architecture has not yet been established, we suggest that the increased length and changed composition of mammalian amelogenins provides a basis for an organized protein matrix that might promote increased enamel crystal length and prismatic architecture. Especially the presence of polyproline repeat motifs would provide a potential explanation for the increased rigidity of mammalian amelogenin protein assemblies and thus a mechanism for an orderly growth of long and parallel apatite crystals in complex prism patterns. In addition, the dolphin with its thin and radial enamel and its short polyproline amelogenin repeat motifs might provide one example of a “link” species, in which the lack of typical mammalian amelogenin characteristics are associated with a reduction in mammalian enamel features.
References
- 1.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980a;255:9760–8. [PubMed] [Google Scholar]
- 2.Fincham AG, Lau EC, Simmer J, Zeichner-David M. Amelogenin biochemistry form and function. In: Slavkin H, Price P, editors. Chemistry and Biology of Mineralized Tissues. Amsterdam: Excerpta Media; 1992. pp. 187–201. [Google Scholar]
- 3.Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–5. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 4.Diekwisch T, David S, Bringas P, Jr, Santos V, Slavkin HC. Antisense inhibition of AMEL translation demonstrates supramolecular controls for enamel HAP crystal growth during embryonic mouse molar development. Development. 1993;117:471–82. doi: 10.1242/dev.117.2.471. [DOI] [PubMed] [Google Scholar]
- 5.Gibson CW, Yuan Z-A, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 6.Iijima M, Moriwaki Y, Wen HB, Fincham AB, Moradian-Oldak J. Elongated Growth of Octacalcium phosphate crystals in Recombinant amelogenin gels under controlled ionic flow. J Dent Res. 2002;81:69–73. doi: 10.1177/002203450208100115. [DOI] [PubMed] [Google Scholar]
- 7.Diekwisch TGH, Berman BJ, Gentner S, Slavkin HC. Initial enamel crystals are not spatially associated with mineralized dentine. Cell Tissue Res. 1995;279:149–67. doi: 10.1007/BF00300701. [DOI] [PubMed] [Google Scholar]
- 8.Diekwisch TG. Subunit compartments of secretory stage enamel matrix. Connect Tissue Res. 1998;38:101–111. doi: 10.3109/03008209809017026. discussion 139–45. [DOI] [PubMed] [Google Scholar]
- 9.Toyosawa S, O’hUigin C, Figueroa F, Tichy H, Klein J. Identification and characterization of amelogenin genes in monotremes, reptiles, and amphibians. Proc Natl Acad Sci U S A. 1998;95:13056–61. doi: 10.1073/pnas.95.22.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Ito Y, Luan X, Yamane A, Diekwisch TG. Amelogenin sequence and enamel biomineralization in Rana pipiens. J Exp Zoolog B Mol Dev Evol. 2005;304:177–186. doi: 10.1002/jez.b.21035. [DOI] [PubMed] [Google Scholar]
- 11.Delgado S, Casane D, Bonnaud L, Laurin M, Sire JY, Girondot M. Molecular evidence for precambrian origin of amelogenin, the major protein of vertebrate enamel. Mol Biol Evol. 2001;18:2146–2153. doi: 10.1093/oxfordjournals.molbev.a003760. [DOI] [PubMed] [Google Scholar]
- 12.Slavkin HC, Diekwisch T. Evolution in tooth developmental biology: of morphology and molecules. Anat Rec. 1996;245:131–50. doi: 10.1002/(SICI)1097-0185(199606)245:2<131::AID-AR3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Slavkin HC, Diekwisch TGH. Molecular strategies of tooth enamel formation are highly conserved during vertebrate evolution. Ciba Found Symp. 1997;205:73–80. doi: 10.1002/9780470515303.ch6. discussion 81–84. [DOI] [PubMed] [Google Scholar]
- 14.Kogaya Y. Immunohistochemical localisation of amelogenin-like proteins and type I collagen and histochemical demonstration of sulphated glycoconjugates in developing enameloid and enamel matrices of the larval urodele (Triturus pyrrhogaster) teeth. J Anat. 1999;195:455–64. doi: 10.1046/j.1469-7580.1999.19530455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishiyama M, Mikami M, Shimokawa H, Oida S. Amelogenin protein in tooth germs of the snake Elaphe quadrivirgata, immunohistochemistry, cloning and cDNA sequence. Arch Histol Cytol. 1998;61:467–74. doi: 10.1679/aohc.61.467. [DOI] [PubMed] [Google Scholar]
- 16.Satchell PG, Anderton X, Ryu OH, Luan X, Ortega AJ, Opamen R, Berman BJ, Witherspoon DE, Gutmann JL, Yamane A, Zeichner-David M, Simmer JP, Shuler CF, Diekwisch TGH. Conservation and variation in enamel protein distribution during tooth development across vertebrates. Mol Dev Evol. 2002;294:91–106. doi: 10.1002/jez.10148. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 2003;100:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Fan JL, Ito Y, Luan X, Diekwisch TG. Identification and characterization of a squamate reptilian amelogenin gene: Iguana iguana. J Exp Zoolog B Mol Dev Evol. 2006;306:393–406. doi: 10.1002/jez.b.21096. [DOI] [PubMed] [Google Scholar]
- 19.Bonass WA, Kirkham J, Brookes SJ, Shore RC, Robinson C. Isolation and characterization of an alternatively-spliced rat amelogenin cDNA: LRAP – a highly conserved, functional alternatively-spliced amelogenin? Biochimica et Biochphysica Acta. 1994;1219:690–692. doi: 10.1016/0167-4781(94)90228-3. [DOI] [PubMed] [Google Scholar]
- 20.Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, Hall B, D’Angelo M, Decker S, Piddington R, Abrams WR, Kulkarni AB, Gibson CW. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- 21.Kondrashov FA, Koonin EV. Origin of alternative splicing by tandem exon duplication. Human Mol Genetics. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- 22.Sankoff D. Gene and genome duplication. Curr Opin Genet Dev. 2001;11:681–684. doi: 10.1016/s0959-437x(00)00253-7. [DOI] [PubMed] [Google Scholar]
- 23.Anishetty S, Pennathur G, Anishetty R. Tripeptide analysis of protein structures. BMC Struct Biol. 2002;2:9. doi: 10.1186/1472-6807-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander PM. Presmless enamel in amniotes: terminology, function, and evolution. In: Teaford MF, Smith MM, Ferguson MWJ, editors. Development, Function and Evolution of Teeth. 2000. pp. 92–106. [Google Scholar]