Abstract
Purpose
Phage display technology can be used to identify peptide sequences which bind rapidly and specifically to tumors responding to sunitinib therapy. These peptides may help to address problems with current methods of assessing tumor response to therapy which can be slow and have limited usage.
Experimental Design
The peptide of interest was isolated after four rounds of biopanning in MDA-MB-231 and MCF-7 xenografted tumors. The peptide’s binding location was investigated with immunohistochemistry. Its in vivo ability to bind to breast tumors responding to therapy was determined by treating nude mice, xenografted with various tumor cell lines, with sunitinib and using near infrared imaging to assess the ability of the peptide conjugated to Alexafluor-750 to bind tumors.
Results
EGEVGLG was the dominant sequence isolated from biopanning. This peptide showed increased binding relative to control groups in two cancer cell lines (MDA-MB-435 and MCF-7 human breast) responding to sunitinib treatment while no elevated binding occurred in vitro when samples were incubated with tumor cells that are unresponsive to sunitinib treatment (B16 melanoma and BxPC3 pancreatic). Mice xenografted with tumors that are responsive to sunitinib therapy showed increased peptide binding when compared to untreated control. Mice bearing tumors unresponsive to sunitinib therapy showed no increased peptide binding between treated and untreated groups.
Conclusion
The use of recombinant peptides to assess the pharmacodynamic response of cancer holds promise in minimizing the duration of ineffective treatment regimens in patients, potentially providing a more rapid and less invasive assessment of cancer response to systemic therapy.
Keywords: Biomarkers and intervention studies, Noninvasive imaging in animal models, Molecular diagnosis and prognosis
INTRODUCTION
Current efforts to detect primary and metastatic tumor response to treatment largely depend on measuring changes in tumor size. Measuring tumor volume is a lengthy process that can take weeks or months before noticeable changes can be observed. Biopsies are not always possible with certain inaccessible metastatic tumor sites like lung, brain, or liver. Sampling error can factor into this process which could lead to an inaccurate measurement of tumor response to therapy. Thus, there is a strong need for new methods to rapidly and noninvasively differentiate between tumors that are responding to treatment (1).
The ability to quickly predict the outcome of an individual’s treatment would allow physicians to effectively optimize a treatment regimen to target a patient’s specific malignancy. Current technologies such as glucose analogue fluorine-18 fluorodeoxyglucose (FDG) PET/CT scans show the potential of this form of assessment (2),(3). However, many factors limit the effectiveness of this form of FDG/PET, such as the inability to detect tumors of small size (4), increased FDG update during endogenous wound repair (5), and inability to detect slow-growing cancers (6).
Recently, recombinant peptides were identified that are capable of recognizing cancer response to therapy rapidly and noninvasively (7). By implanting tumors in nude mice and treating them with ionizing radiation, biopanning of the T7 phage-displayed peptide library in lung and brain cancers permitted for selection of peptides that bind to receptors activated in response to radiation therapy. Screening of phage-displayed peptide libraries has been established as a way to discover peptide ligands that bind to tumor vasculature, cancer cells, or specific molecular targets (8–10). Phage display technology allows for the insertion of random DNA sequences into the bacteriophage genome which encodes the phage capsid proteins. This leads to new peptide expression on the phage surface that binds to cell surface molecules (11, 12).
In the present study, we have discovered a novel set of recombinant peptides selected from phage displayed peptide libraries that are capable of detecting cancer response to the tyrosine kinase inhibitor (TKI) sunitinib (SU11248; Sutent). These peptides can be labeled with internal emitters to provide a means to monitor and predict cancer response to sunitinib treatment in tumor bearing mice. Elevated binding levels of the peptide in the tumor region occur days before the tumor begins displaying a response to therapy. This suggests it may be used as a rapid and non-invasive means of predicting treatment efficacy.
MATERIALS AND METHODS
Sunitinib Synthesis
Sunitinib was synthesized in the Vanderbilt Institute of Chemical Biology using the five-step method previously described (13, 14). Sunitinib was administered in vivo using intraperitoneal injection at either the subtherapeutic dose of 4 mg/kg or the therapeutic dose of 40 mg/kg (11). All protocols in animal experiments were reviewed and approved by the Vanderbilt University Institutional Animal Care & Use Committee.
Biopanning Phage-displayed Libraries
In vivo biopanning was done as described (7, 11) with a T7 phage-based random peptide library. The phage-displayed peptide library represents 1×108 independent clones of phages expressing random nonamer peptides that are displayed on T7 phages as fusion proteins with the amino terminus of 10A capsid protein. Mice bearing either MDA-MB-231 or MCF-7 breast tumors were subject to treatments of vehicle control (PBS) or 40 mg/kg sunitinib given intraperitoneally for 3 consecutive days. Treatment began 30 days after tumor implantation. The phage libraries were administered 4 hours after the last treatment. Phages were recovered after being in circulation for 16 hours by harvesting the tumors in the mice. Certain peptide candidates were eliminated by negative selection, since they would remain bound to other organs in the mice. The first round of biopanning was done with mice implanted with MDA-MB-231 tumors. Phages were recovered from excised tumors and were subjected to three more rounds of selection with either MDA-MB-231 or MCF-7 breast cancer cell lines.
Peptide Design and Imaging Specifications
The isolated peptide, EGEVGLG, was synthesized with two lysine residues for biotin and imaging agent conjugation and three glycine linkers to separate the targeting peptide from the biotin and the imaging agent. Biotinylated-KKGGGEGEVGLG synthetic peptide was purchased from Genemed Synthesis Inc (San Antonio, TX) with biotin attached to the N-terminus lysine residue. This peptide was conjugated with streptavidin (Pierce, Rockford, IL) for 2 hours to create a 1:4 molar ratio of peptide:streptavidin. Conjugated peptide was then incubated with Alexafluor 594 or Alexafluor 750 dye (Invitrogen) for an additional hour, which bound to the remaining lysine residue of the synthesized peptide. Labeled complexes of biotinylated peptide-streptavidin-Alexafluor conjugates were then used in vitro and in vivo for imaging purposes.
Co-culture Assay
Four coverslips, each containing 1×104 human umbilical vein endothelial cells (HUVECs), in the sixth passage (Lonza, Walkersville, MD), were placed on the bottom layers of co-culture plates (Fisher, Wilkes Barre, PA). Cells were grown for one day in the plate before 3×105 of either MDA-MB-231, B16, or BxPC3, cells were added to the superior layer of the plate. HUVECs were allowed to interact for an additional day before 0.5 μg/mL of sunitinib was added into the bottom dish. The cells were incubated for 1 hour before they were harvested. Coverslips were blocked for 30 minutes with 5% BSA and 1% Streptavidin and incubated for 1 hour with a Streptavidin-Peptide-Alexafluor 594 complex (Invitrogen, Eugene, OR). The HUVEC nuclei were stained with DAPI and images of nuclei and peptide binding were taken using a Zeiss Axiophot fluorescent microscope at 40X magnification.
In a second assay, 3×105 MDA-MB-231 cells were plated on cover slips and co-cultured with HUVECs. HUVECs were also layered in co-culture plates. Both culture types were either treated with 0.5 μg/mL sunitinib or left untreated, incubated with the EGEVGLG peptide and imaged as before. Positive and negative controls of sunitinib treated and untreated MDA/HUVEC co-cultures with the peptide incubated on HUVECs were used.
Background fluorescence of sunitinib was determined by co-culturing cells as before, treating with sunitinib and imaging for fluorescence. These values were subtracted from all treated groups to normalize them against background fluroscence. Quantification of peptide and cell co-localization was performed using Metamorph Offline software.
Tumor Models
B16 murine melanoma, BxPC3 human pancreatic adenocarcinoma, MDA-MB-231 human breast cancer, and MCF-7 human breast cancer cell lines were purchased from American Type Culture Collection (ATCC; Manassas, VA). MDA-MB-435 cells transfected with GFP (MDA-MB-435-GFP) was a gift from G. Mundy (Vanderbilt University, Nashville, TN). Heterotopic tumor models were developed by subcutaneously inoculating cell suspensions (6×106 cells or adjusted for different cell types) into nude mice. Estradiol pellets (Innovative Resarch of America, Sarasota, FL) were implanted subcutaneously in nude mice two days before they were xenografted with the estrogen receptor/progesterone receptor (ER/PR) positive MCF-7 tumor cell line. Nude mice had tumors implanted into their right hind limbs and were used for experiments when the tumor size reached approximately 300 mm3 in volume (30 days after implantation).
Immunohistochemistry
Paraffin embedded tumor samples were taken from mice which had received 4 mg/kg sunitinib, 40 mg/kg sunitinib, or vehicle control. Samples were stained using an antibody for the von Willebrand Factor (vWF) (DakoCytomation, Carpinteria, CA) at a 1:100 dilution from the original stock solution of 3.1 g/L and incubated overnight. A final three washes with PBS for 5 minutes per wash was done before placing a coverslip over the sample. Images were taken using a fluorescent microscope at 20X magnification.
Tumor Growth Study
Mice were implanted with MDA-MB-435-GFP tumors into their right hind limbs. Treatment was started when tumor sizes reached approximately 300 mm3 in volume. Treatment conditions included sunitinib at 4 mg/kg or 40 mg/kg, and vehicle control. Treatments were given through intraperitoneal injection daily for 5 days. Tumor size was measured every other day using calipers. Fold-increase in tumor volume (compared to tumor size on the first day of treatment) was calculated to show tumor responsiveness to treatment. Intensity of GFP was also measured on the days when tumor volume was measured with calipers.
Near Infrared Imaging
Labeled complexes of biotinylated peptide-streptavidin-Alexafluor conjugates were injected into circulation using tail vein injection in tumor bearing mice being treated with vehicle control or sunitinib 4 hours after the final sunitinib treatment. Near infrared images were taken using the IVIS imaging system with an ICG filter setting at various time points after the injection. Radiance (photons/s/cm2) was measured in the region of interest (ROI) by using the LivingImage software.
Statistical Analyses
Student’s t-test was used to perform group comparisons. Linear correlations of peptide binding and tumor response to treatment were developed by use of the correlation coefficient of tumor growth and radiance datasets (SigmaPlot). The same linear correlation was performed with GFP radiance and tumor growth.
RESULTS
Selection of Phage-displayed Peptides
The primary goal of this study was to identify novel recombinant peptides capable of differentiating responsive mouse models of cancer from those that are unresponsive to sunitnib therapy. Due to the spatial separation of organs, in vivo selection allows for the differentiation between peptides that bind specifically to responsive tumors versus those binding to unresponsive tumors and other normal tissues (7). The phages that bound to responsive tumors were enriched through a total of four serial rounds of biopanning. The first round of biopanning was performed in MDA-MB-231 tumors, with three subsequent rounds of selection done in either MDA-MB-231 or MCF-7 tumors. This helped select for peptides that bound multiple types of tumors.
The peptides discovered after biopanning in the MCF-7 or MDA-MB-231 tumors are displayed on Table 1. Thirty-six phage plaques were amplified by use of PCR and were sequenced. The relative abundance of each of these sequences is listed in Table 1. The peptide EGEVGLG was found to be the predominant phage-encoded peptide isolated from both the MDA-MB-231 and MCF-7 tumor screens and was selected for subsequent experiments.
Table 1.
| Sequence | From MDA-MB-231 | From MCF-7 |
|---|---|---|
| EGEVGLG | 58% | 67% |
| MRRSVGS | 14% | 12% |
| SSAVL | 8% | 18% |
| VLI | 8% | 0% |
| SAGSVAL | 6% | 0% |
| FGVR | 3% | 1% |
| GFWEGGL | 3% | 0% |
EGEVGLG Peptide Recognizes Sunitinib-treated Endothelium
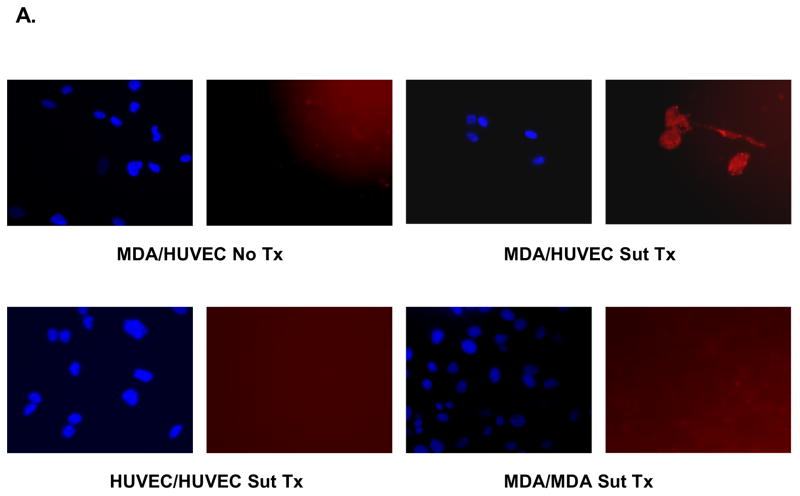
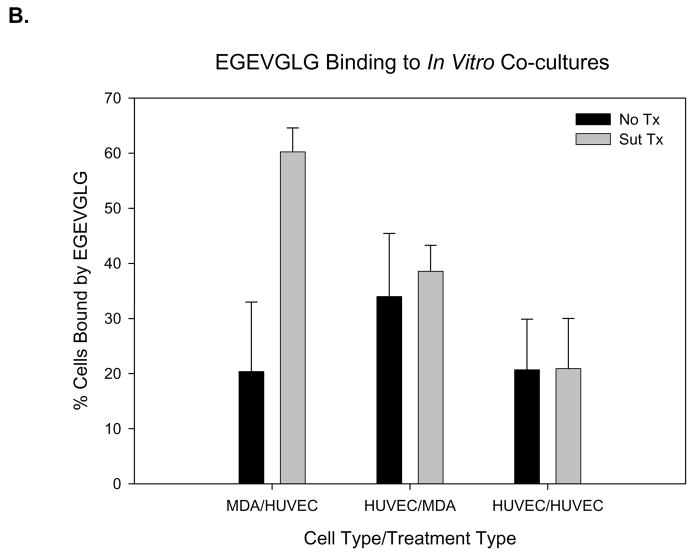
We have previously shown that screened phage peptides bind to tumor vasculature (7). As a preliminary assessment of the differentiating capability of the EGEVGLG peptide, an in vitro assay was performed. Using co-culture plates, MDA-MB-231 cells were co-cultured with HUVECs and treated with 0.5 μg/mL sunitinib or vehicle control for one hour. HUVECs were then incubated for another hour with the fluorescent labeled EGEVGLG peptide. The HUVECs were then imaged for peptide binding. We found (Fig. 1A–B) a significant increase in the amount of EGEVGLG peptide binding in the sunitinib-treated HUVECs when compared to the untreated control group (P < 0.05).
Fig. 1.
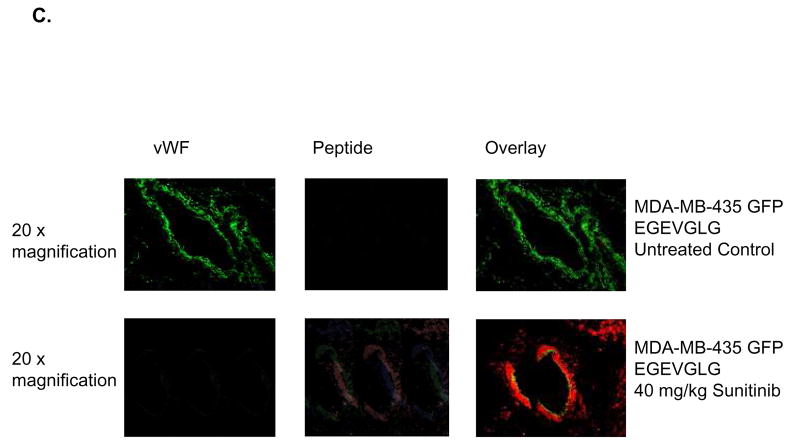
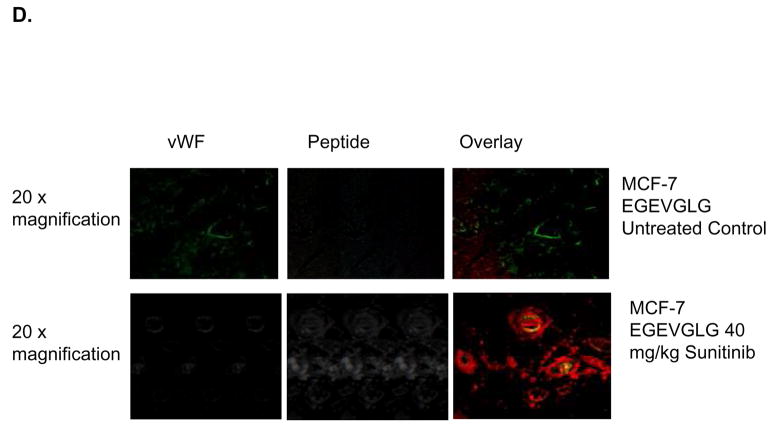
EGEVGLG peptide differentiates treated from untreated endothelial cells. A and B, HUVECs were co-cultured for 1 day with MDA-MB-231 cells to simulate tumor/tumor vasculature interaction. After treatment with 0.5 μg/mL sunitinib for 1 hour and incubation with fluorescently labeled EGEVGLG peptide for another hour, HUVECs were imaged for binding of peptide. The experiment was repeated with HUVEC/MDA co-cultures with the peptide incubated on the MDA cells and with HUVEC/HUVEC “co-cultures”. C and D Immunohistochemistry of heterotopic tumor samples from breast cancer xenografted mice. Tumor samples were taken from mice that were treated with either vehicle control (top panel) or sunitinib (bottom panel) for 3 days. C, MDA-MB-435 and D, MCF-7 samples were taken from samples at 96 or 48 hours, respectively, after the final treatment. Samples were stained for tumor vasculature using vWF antibody (left panel) and were incubated with peptide for 1 hour and imaged (middle panel); these images were overlayed (right panel) to identify areas of colocalization.
Investigation of the mechanism of EGEVGLG binding was performed with a second co-culture assay for differences in binding of EGEVGLG between sunitinib treated and untreated groups (Fig 1A–B). The groups were (using the nomenclature upper level/lower level of coculture plate) MDA/HUVEC, HUVEC/MDA, and HUVEC/HUVEC. This showed a statistically significant increase in binding of EGEVGLG in only the co-culture of MDA/HUVEC with the peptide incubated on the HUVEC (P < 0.05). There was no significant increase in binding in treated versus untreated in both the HUVEC/HUVEC and HUVEC/MDA with the peptide incubated with MDA (P > 0.05). There was also a baseline of fluorescence of sunitinib that was accounted for by incubating cells with the drug and imaging for fluorescence. This baseline level was subtracted from all sunitinib treated groups (Fig. 1A–B).
To further investigate the location of peptide binding, tumor samples were resected from mice that were treated with either 40 mg/kg sunitinib or vehicle control once daily for 5 days. MDA-MB-435 and MCF-7 tumor samples were taken at 96 and 48 hours, respectively, after the final treatment. Immunohistochemistry was performed and samples were stained for tumor vasculature and incubated with peptide for 1 hour (Fig. 1C–D). MDA-MB-435 and MCF-7 samples treated with sunitinib (Fig. 1C–D, bottom panels) showed EGEVGLG peptide binding that colocalized with tumor vasculature stained with von Willebrand Factor (vWF) while vehicle-treated groups showed minimal peptide binding (Fig. 1C–D, top panels). These results indicate that the EGEVGLG peptide binds to tumor vasculature after sunitinib treatment in an ex vivo model.
Sunitinib Treatment Elicits Tumor Growth Delay in MDA-MB-435-GFP Tumors
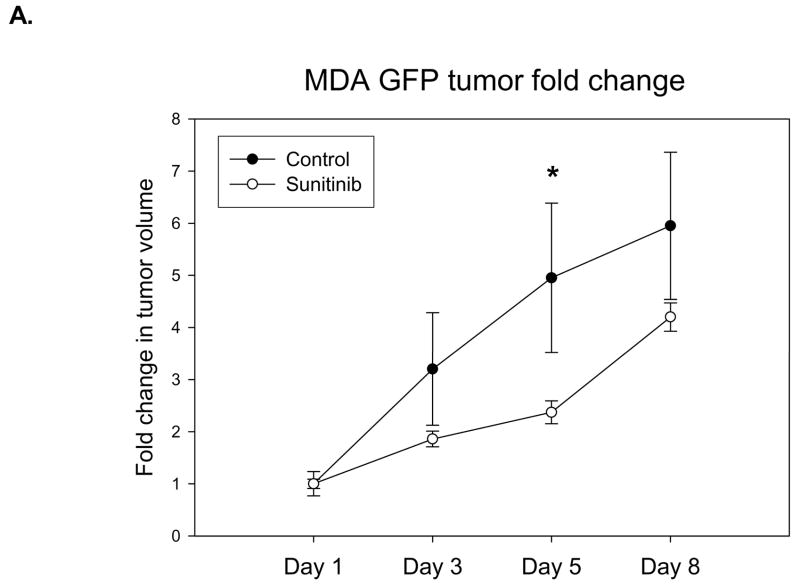
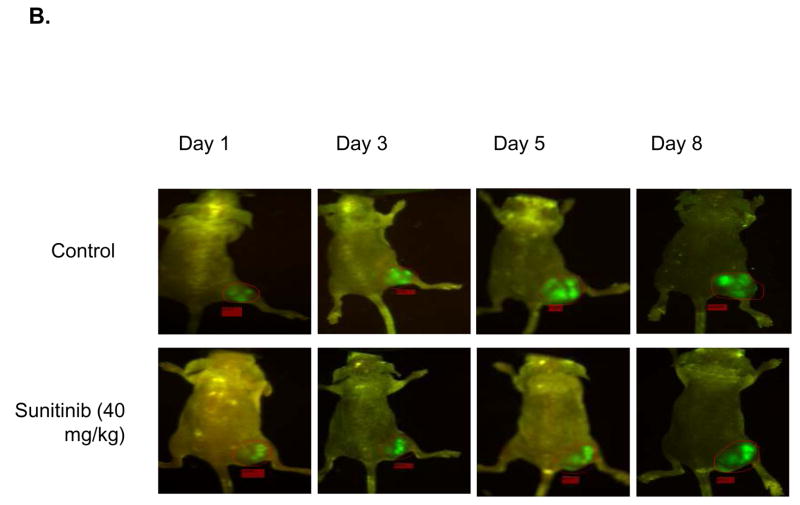
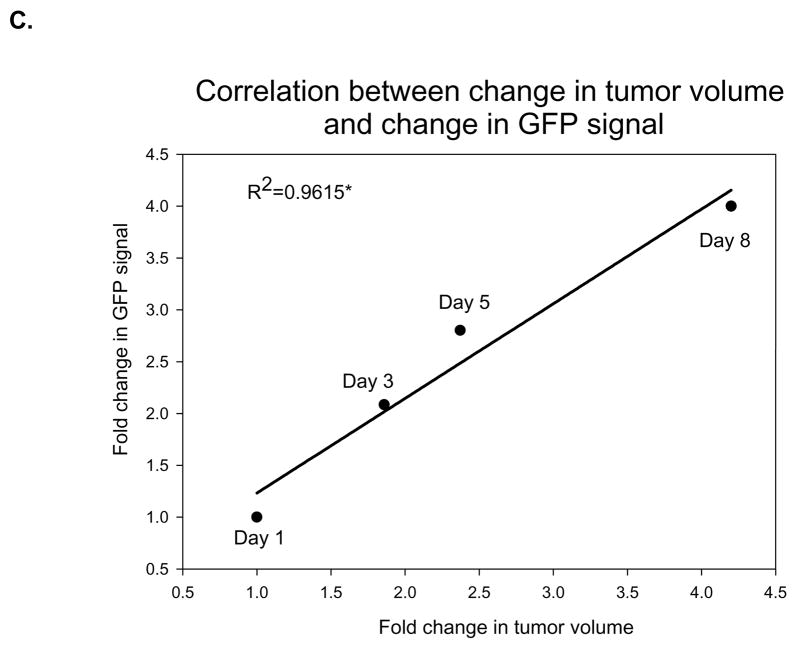
To develop a model that could address metastatic disease changes, a stable cell line of MDA-MB-435 cancer cells transfected with GFP was implanted into the right hind limbs of nude mice. The mice were then given daily treatments of sunitinib at 40 mg/kg while their tumors volumes were measured by caliper. At the same time, images were taken that captured GFP radiance from the tumors. It was observed that the treated mice had smaller average tumor volume than the control group (Fig. 2A). A significant difference in volume was observed the fifth day after the end of treatment (P < 0.05). Imaging reveals an increase in GFP radiance for both control and treated groups over time (Fig. 2B). We found a significant correlation (P < 0.05) between fold-change in tumor volume and fold-change in GFP signal in the sunitinib-treated group over time (Fig. 2C).
Fig. 2.
Nude mice implanted with MDA-MB-435-GFP show correlation between tumor growth delay and GFP radiance. Nude mice (n=5 per group) were injected with MDA-MB-435-GFP tumor cells and were given vehicle control or 40 mg/kg of sunitinib once daily for five days. A, Tumor volumes were measured by caliper throughout the experiment and there was significant growth delay by 5 days (*P < 0.05) B, Images of GFP radiance were taken the same day as the caliper measurements. C, A significant correlation (*P < 0.05) is observed when the fold-change in tumor volume over time is compared to the fold-change in GFP signal over time in the group treated with 40 mg/kg of sunitinib.
EGEVGLG Peptide Detects Treatment Response in Tumor Bearing Mice
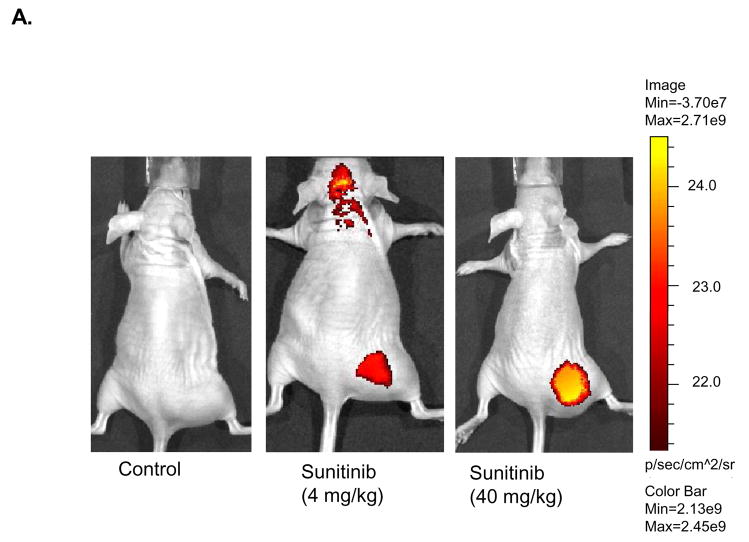
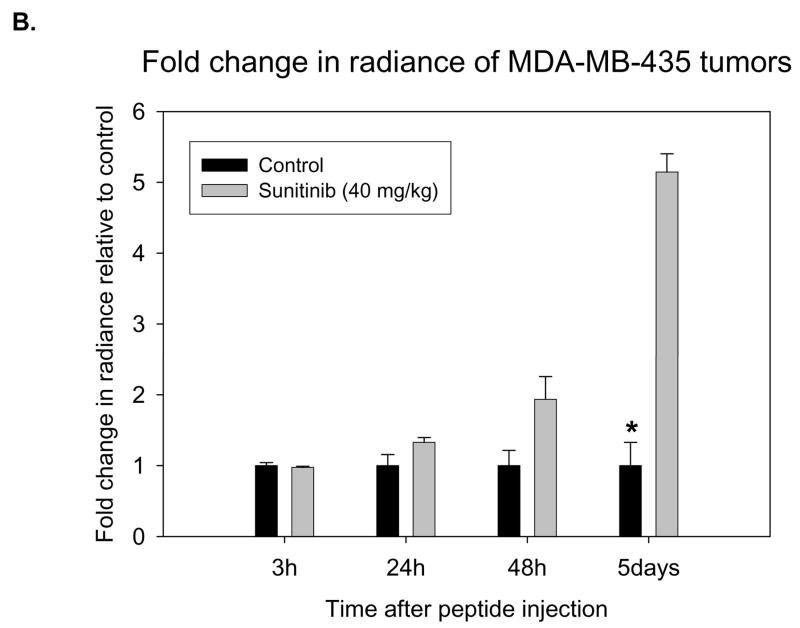
Having shown the potential of EGEVGLG to bind selectively to treated tumor sections ex vivo, the next step was to test the peptide’s binding specificity in vivo. Nude mice were implanted with MDA-MB-435-GFP tumors. Tumor-bearing mice were treated with once daily doses of sunitinib for five days. Four hours after the final sunitinib treatment, the mice were injected with EGEVGLG peptide-dye complex. Images of the mice 48 hours after peptide injection (Fig. 3A) showed a graded increase in tumor radiance as dose of drug increased from vehicle control (left panel) to 4 mg/kg sunitinib (middle panel) to 40 mg/kg sunitinib (right panel). Radiance of images of the mice was quantified and the peptide radiance from the tumor’s region of interest was quantified relative to the control (Fig. 3B). The average radiance of the treated group was noticeably higher than the control group beginning 24 hours after peptide injection, and this difference became statistically significant (P < 0.05) on the fifth day after peptide injection.
Fig. 3.
Nude mice implanted with MDA-MB-435-GFP tumor cells show increased peptide binding relative to control. Nude mice (n=5 per group) were injected with MDA-MB-435-GFP tumor cells and were given vehicle control, 4 mg/kg, or 40 mg/kg of sunitinib once daily for five consecutive days. Peptide-dye complexes were injected 4 hours after the fifth day of treatment. A, Mice were imaged 48 hours after initial peptide injection. B, Radiance of groups treated with control or 40 mg/kg were quantified and significant difference was observed on day 5 post-treatment (*P < 0.05).
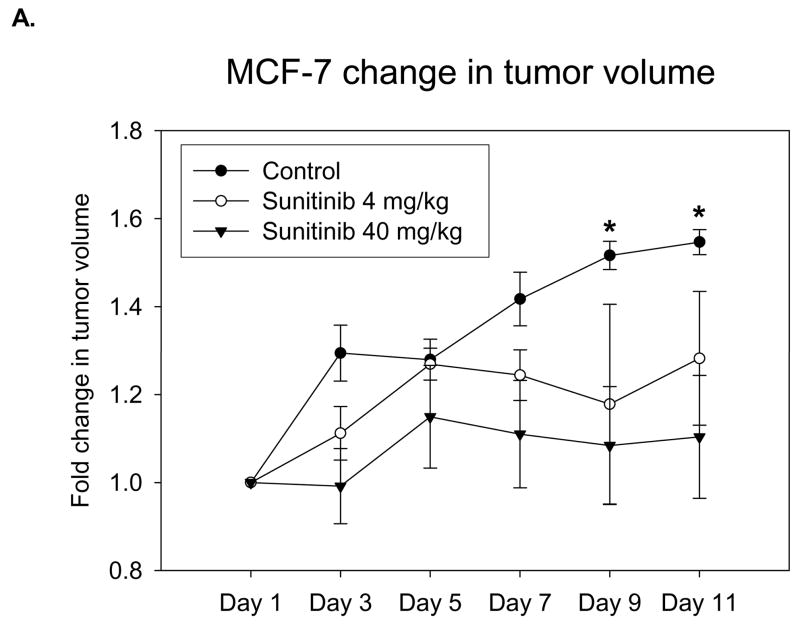
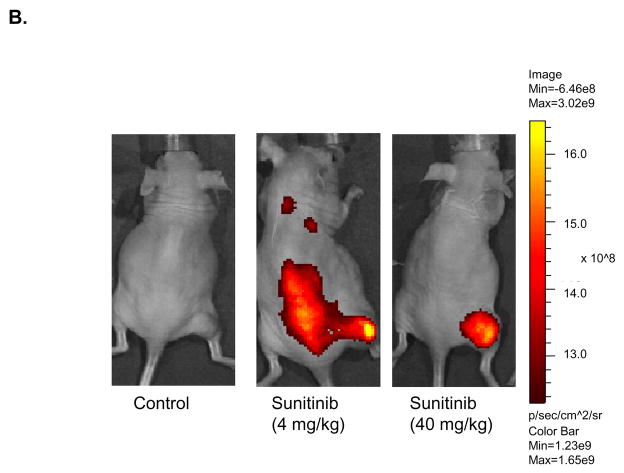
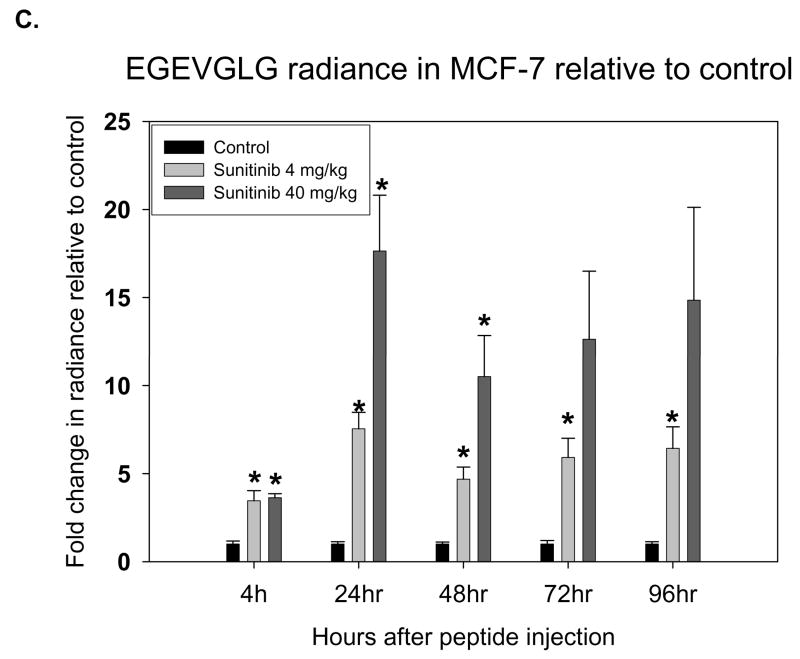
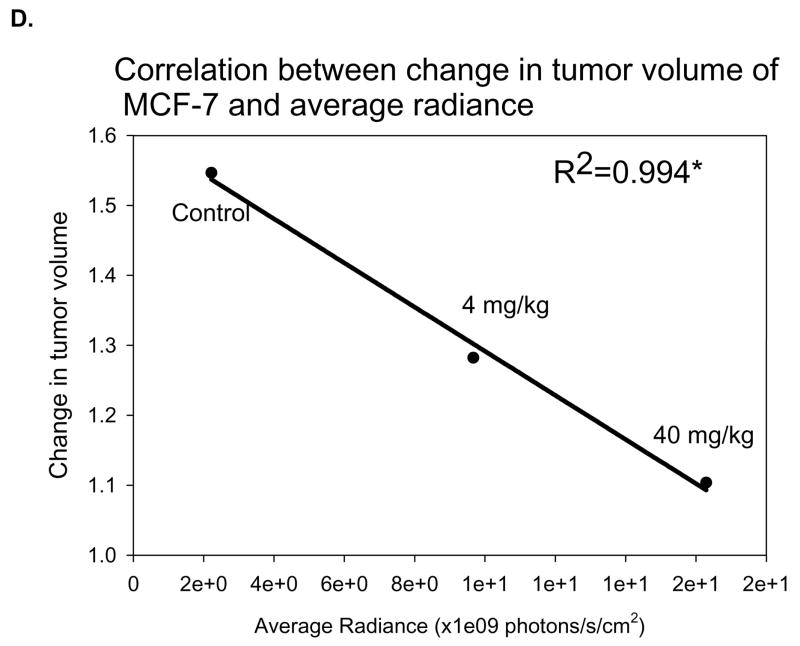
MCF-7 was the second in vivo tumor model used to assess the differential binding ability of the EGEVGLG peptide. Like the aforementioned experiment, MCF-7 was implanted into the hind limbs of nude mice treated daily with sunitinib. Groups treated with 4 or 40 mg/kg of sunitinib had average tumor volumes that were smaller than the control (Fig. 4A). These differences became significant on days 9 and 11 after treatment. Images of the mice taken at 48 hours after initial injection of the peptide-dye complex (Fig. 4B) showed a marked increase in peptide binding in the 4 mg/kg and 40 mg/kg treated groups (middle and right panels, respectively) when compared to the control group (left panel). The fold-change in tumor radiance was higher in both treated groups and significantly higher (P < 0.05) for the 40 mg/kg treatment group starting at 24 hours and was maintained even at 96 hours after peptide injection (Fig. 4C). Fig. 4D shows a significant correlation between the average peptide radiance in the three experimental groups at 24 hours and their average tumor volumes on day 11 (P < 0.05). These imaging studies with the EGEVGLG peptide showed that this peptide preferentially binds to sunitinib-treated responsive tumors when compared to untreated controls, as the intensity of peptide binding correlates with a decrease in tumor size and decrease in GFP activity.
Fig. 4.
Nude mice implanted with MCF-7 show increased peptide binding relative to control. Nude mice (n=4 per group) were injected with MCF-7 tumor cells and were given vehicle control, 4 mg/kg or 40 mg/kg of sunitinib once daily for five consecutive days. A, Tumor volumes were measured by caliper daily (*P < 0.05). B, Peptide-dye complexes were injected 4 hours after the fifth day of treatment, and the mice were imaged at various intervals. Shown are representative images at 48 hours after peptide injection. C, Radiance was quantified from the images taken at time intervals after peptide injection (*P < 0.05) D, Radiance of tumor at 24 hours after peptide injection was correlated with a change in tumor volume on day 11 (*P < 0.05).
EGEVGLG Differentiates between Responsive and Unresponsive Tumors
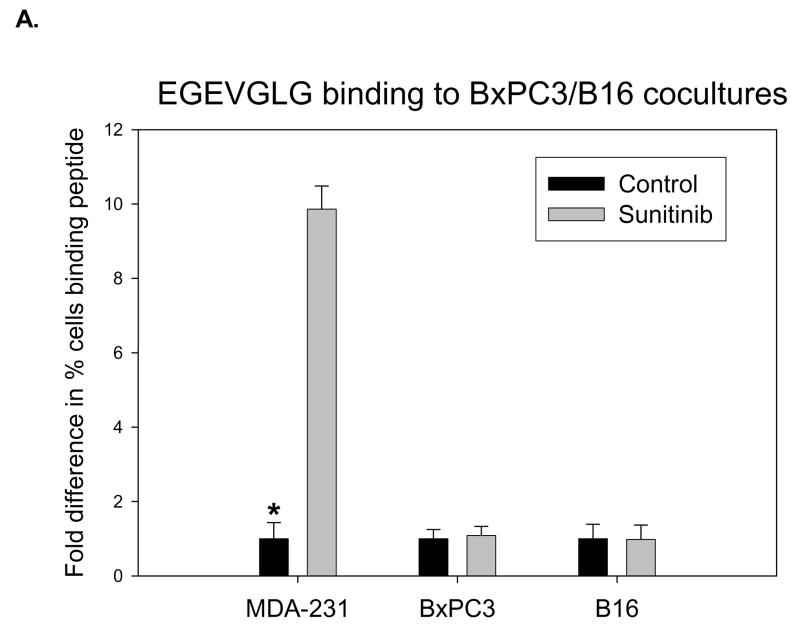
The previously described co-culture experiments were performed using B16 melanoma and BxPC3 pancreatic tumor cell lines which have been shown in previous studies (7) to be unresponsive to sunitinib therapy. HUVECs were co-cultured with MDA-MB-231, B16, or BxPC3 cells to simulate tumor vasculature. After treatment with 0.5 μg/mL sunitinib for one hour and incubation with fluorescently labeled EGEVGLG peptide for another hour, HUVECs were imaged for peptide binding. MDA-MB-231 showed a significant differential binding (over 9 times, P < 0.05) in the sunitinib-treated group relative to the untreated control (Fig. 5A). No differential binding was observed in either the B16 or BxPC3 cells lines between the sunitinib-treated and untreated control groups (Fig. 5A).
Fig. 5.
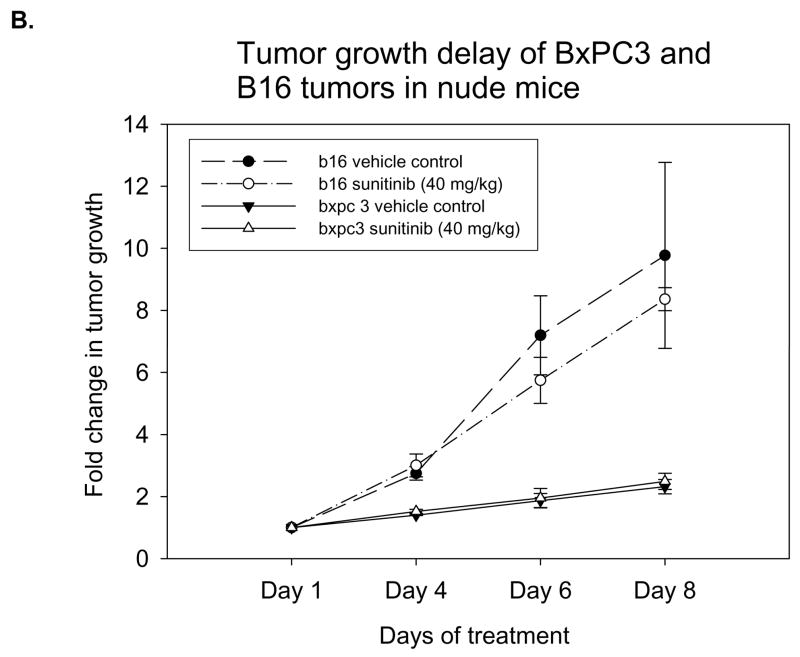
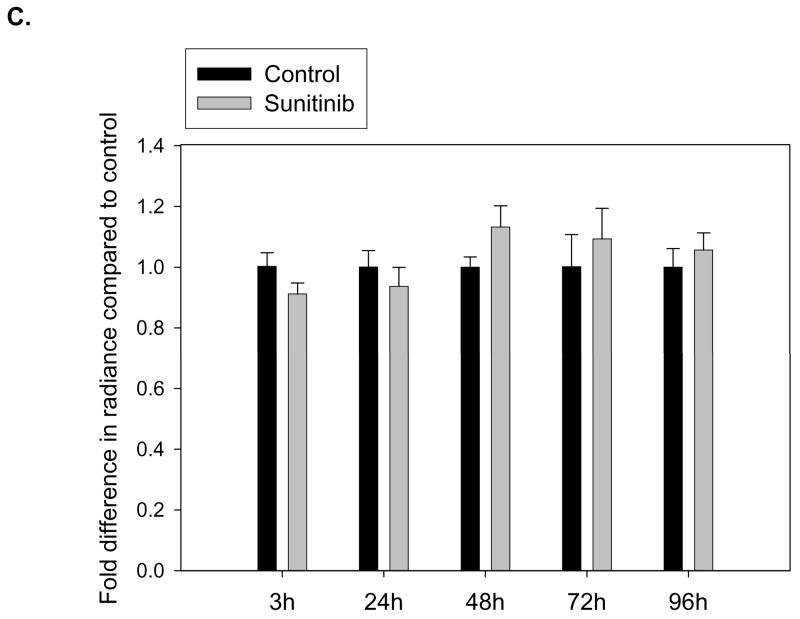
EGEVGLG peptide does not exhibit increased binding to tumors that do not respond to sunitinib therapy. A, HUVECs were co-cultured with MDA-MB-231, B16, or BxPC3 cells to simulate tumor vasculature. After treatment with 0.5 ug/mL sunitinib for 1 hour, incubation with fluorescently labeled EGEVGLG peptide for another hour, HUVECs were imaged for binding of peptide (*P < 0.05). B, Shown is tumor growth delay study of B16 melanoma and BxPC3 pancreatic tumor cells. The tumors were implanted in the hind limbs of nude mice (n=3 per group) and given once daily treatments of sunitinib at 40 mg/kg or a vehicle control for five consecutive days. C, D Peptide-dye complexes were injected 4 hours after the fifth day of treatment and no preferential binding of the peptide was observed between sunitininb-treated and untreated groups in C B16 or D BxC3 tumors.
In nude mice, these B16 or BxPC3 tumors were implanted and treated with either vehicle control or 40 mg/kg of sunitinib once daily for five consecutive days. The tumor growth rates in both tumor models showed no significant difference in tumor volumes between treated and untreated groups (Fig. 5B). The EGEVGLG peptide-dye complex was injected 4 hours after the third day of treatment. No significant difference was observed between the radiance of the sunitinib-treated and untreated groups (Fig. 5C–D). Neither of these two tumor models responded to treatment with sunitinib, and this correlated with no preferential binding of the EGEVGLG peptide to these treated tumors.
DISCUSSION
The goal of this study was to discover a means to rapidly and noninvasively assess cancer response to the TKI sunitinib. Sequence analysis of phage recovered from in vivo biopanning of MDA-MB-231 cells showed the following distribution: EGEVGLG, SSAVL, MRRSVGS, FGVR, VLI, SAGSVAL, and GFWEGGL. The first four peptides were also isolated from in vivo biopanning of MCF-7 cells (Table 1). Further investigation on the most active peptide, EGEVGLG, using immunohistochemical and in vitro co-culture analyses show that while the peptide appears to bind to the vascular endothelium, a necessary interaction between vascular endothelium and tumor must occur for EGEVGLG to bind to its putative receptor (Fig. 1). Imaging studies utilizing the EGEVGLG peptide-dye complex demonstrate that the peptide preferentially binds to sunitinib-treated ER/PR positive (MCF-7) and ER/PR negative (MDA-MB-435) breast tumors (Fig. 3 and 4). These two different cancer tumor models showed differential binding between the sunitinib-treated groups and untreated control groups in a dose-dependent fashion. In five days or less following peptide injection into treated, tumor-bearing mice, a significant difference in peptide binding as indicated by increased tumor radiance was observable between the treated and the untreated groups. Also, we found that the peptide is capable of differentiating between responding and non-responding tumor types. The EGEVGLG peptide binds to a treated, responding cancer like MDA-MB-435 but not to the treated, nonresponding cancer cell lines B16 and BxPC3 (Fig. 5).
By correlating this difference in radiance with tumor growth delay in these experiments, the EGEVGLG peptide can predict response to sunitinib therapy early in the course of therapy in tumor bearing mice. An increase in peptide binding and tumor radiance detected within 72 hours of beginning sunitinib treatment correlates with a significant delay in tumor growth observed several days later (Fig. 3 and 4). This peptide’s specificity could potentially reduce the time necessary to assess cancer response to therapy to sunitinib and allow a physician to personalize a patient’s treatment regimen more rapidly. Furthermore, it could also eliminate the need for invasive procedures such as biopsies and less accurate methods such as tumor volume measurements to determine response.
Previous studies have shown that various receptors and adhesion molecules are induced in tumor microvasculature in response to therapy, making them potential targets for imaging tumor response and targeted drug delivery (11, 15–25). Thus, there is a strong incentive to identify the putative receptor to which the EGEVGLG peptide binds. Since the peptide binds to the HUVECs in the co-culture experiments as well as in the immunohistochemistry samples (Fig. 1), it suggests the target is a membrane receptor with an extracellular domain. Experiments designed to identify the putative cellular receptor for EGEVGLG and the other screened recombinant peptides are currently underway. By determining the properties of these lipid or protein receptors, peptides and/or antibodies that bind to them with an even higher affinity can be synthesized for use as more accurate biomarkers for tumor response to sunitinib.
In addition, the apparent specificity of EGEVGLG to tumors that are responsive to sunitinib could open up opportunities to explore novel forms of drug delivery (26, 27). Our finding that the EGEVGLG peptide seems to preferentially bind only to tumors responding to sunitinib suggests that conjugating it to a nanoparticle drug delivery system could be an excellent form of adjuvant therapy. This could allow for the delivery of chemotherapeutic agents to metastatic tumors by utilizing these recombinant peptides to target tumor vasculature responding to systemic therapy. The peptide could concentrate the chemotherapeutic drug in responding (cancerous) areas, potentially reducing systemic toxicity and allowing for an increased drug load to be administered to the subject. In the future, efforts to convert this technology from mice to humans will require implementation of many new studies to determine how the peptide interacts with many different systems, particularly the immune system.
Since the peptide can bind to responding cancer cells that have MDA-MB-435 cells stably transfected with GFP, we have developed a model to detect treatment response in metastatic tumors. In our experiments, the observed correlation between GFP radiance and tumor volume changes in the transfected tumor line (Fig. 2 and 3) suggests the potential of these experimental models as a viable means to assess tumor volume, and ultimately tumor response, in a metastatic setting. In vivo use of GFP-expressing cancer cells for imaging offers a robust method of visualizing tumors in situ (26, 27). This technique could potentially allow for the evaluation of the screened recombinant peptides’ ability to assess a tumor’s response to TKI therapy in metastatic tumor models.
In conclusion, we identified recombinant peptides that discern responding from non-responding tumors after treatment with sunitinib very early in the course of therapy (less than 5 days). This is platform technology that shows the principle that recombinant peptide biomarkers are effective at rapidly assessing cancer susceptibility to molecular targeted therapy.
Acknowledgments
This work was supported by Grant BC061828 (to R.D.) from the United States Department of Defense; Grant R01-CA112385 (to D.E.H.) from the United States National Institutes of Health; and Vanderbilt ICMIC grant P50CA128323 (to D.E.H.). We thank G. Mundy (Vanderbilt University) for the gift of the MDA-MB-435-GFP cell line; E. Ruoslahti (Burnham Institute) for the gift of T7 phage-based random peptide library; J. Huamani and A. Fu for technical support; and Z. Han for multiple suggestions and helpful discussions. R. Diaz is a recipient of the Leonard B. Holman Research Pathway fellowship from the American Board of Radiology.
Footnotes
Statement of translational relevance: We identified recombinant peptides that discern responding from non-responding tumors after treatment with sunitinib very early in the course of therapy (less than 5 days). The use of recombinant peptides to assess the pharmacodynamic response of cancer holds promise in minimizing the duration of ineffective treatment regimens in patients, potentially providing a more rapid and less invasive assessment of cancer response to systemic therapy. This is platform technology that shows the principle that recombinant peptide biomarkers are effective at rapidly assessing cancer susceptibility to molecular targeted therapy.
References
- 1.Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 2.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 3.Cullinane C, Dorow DS, Kansara M, et al. An in vivo tumor model exploiting metabolic response as a biomarker for targeted drug development. Cancer research. 2005;65:9633–6. doi: 10.1158/0008-5472.CAN-05-2285. [DOI] [PubMed] [Google Scholar]
- 4.Kumar R, Chauhan A, Zhuang H, Chandra P, Schnall M, Alavi A. Clinicopathologic factors associated with false negative FDG-PET in primary breast cancer. Breast Cancer Res Treat. 2006;98:267–74. doi: 10.1007/s10549-006-9159-2. [DOI] [PubMed] [Google Scholar]
- 5.Bakheet SM, Powe J, Kandil A, Ezzat A, Rostom A, Amartey J. F-18 FDG uptake in breast infection and inflammation. Clin Nucl Med. 2000;25:100–3. doi: 10.1097/00003072-200002000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Avril N, Dose J, Janicke F, et al. Metabolic characterization of breast tumors with positron emission tomography using F-18 fluorodeoxyglucose. J Clin Oncol. 1996;14:1848–57. doi: 10.1200/JCO.1996.14.6.1848. [DOI] [PubMed] [Google Scholar]
- 7.Han Z, Fu A, Wang H, et al. Noninvasive assessment of cancer response to therapy. Nature medicine. 2008;14:343–9. doi: 10.1038/nm1691. [DOI] [PubMed] [Google Scholar]
- 8.Arap W, Kolonin MG, Trepel M, et al. Steps toward mapping the human vasculature by phage display. Nature medicine. 2002;8:121–7. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–6. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 10.Ruoslahti E. Vascular zip codes in angiogenesis and metastasis. Biochem Soc Trans. 2004;32:397–402. doi: 10.1042/BST0320397. [DOI] [PubMed] [Google Scholar]
- 11.Hallahan D, Geng L, Qu S, et al. Integrin-mediated targeting of drug delivery to irradiated tumor blood vessels. Cancer Cell. 2003;3:63–74. doi: 10.1016/s1535-6108(02)00238-6. [DOI] [PubMed] [Google Scholar]
- 12.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–90. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 13.Mendel DB, Laird AD, Xin X, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9:327–37. [PubMed] [Google Scholar]
- 14.Sun L, Liang C, Shirazian S, et al. Discovery of 5-[5-fluoro-2-oxo-1,2- dihydroindol-(3Z)-ylidenemethyl]-2,4- dimethyl-1H-pyrrole-3-carboxylic acid (2-diethylaminoethyl)amide, a novel tyrosine kinase inhibitor targeting vascular endothelial and platelet-derived growth factor receptor tyrosine kinase. J Med Chem. 2003;46:1116–9. doi: 10.1021/jm0204183. [DOI] [PubMed] [Google Scholar]
- 15.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–80. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 16.Baillie CT, Winslet MC, Bradley NJ. Tumour vasculature--a potential therapeutic target. British journal of cancer. 1995;72:257–67. doi: 10.1038/bjc.1995.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks PC, Montgomery AM, Rosenfeld M, et al. Integrin alpha v beta 3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell. 1994;79:1157–64. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 18.Burg MA, Pasqualini R, Arap W, Ruoslahti E, Stallcup WB. NG2 proteoglycan-binding peptides target tumor neovasculature. Cancer research. 1999;59:2869–74. [PubMed] [Google Scholar]
- 19.Ellerby HM, Arap W, Ellerby LM, et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nature medicine. 1999;5:1032–8. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 20.Fox SB, Harris AL. Markers of tumor angiogenesis: clinical applications in prognosis and anti-angiogenic therapy. Investigational new drugs. 1997;15:15–28. doi: 10.1023/a:1005714527315. [DOI] [PubMed] [Google Scholar]
- 21.Hallahan D, Clark ET, Kuchibhotla J, Gewertz BL, Collins T. E-selectin gene induction by ionizing radiation is independent of cytokine induction. Biochemical and biophysical research communications. 1995;217:784–95. doi: 10.1006/bbrc.1995.2841. [DOI] [PubMed] [Google Scholar]
- 22.Hallahan D, Kuchibhotla J, Wyble C. Cell adhesion molecules mediate radiation-induced leukocyte adhesion to the vascular endothelium. Cancer research. 1996;56:5150–5. [PubMed] [Google Scholar]
- 23.Hallahan DE, Staba-Hogan MJ, Virudachalam S, Kolchinsky A. X-ray-induced P-selectin localization to the lumen of tumor blood vessels. Cancer research. 1998;58:5216–20. [PubMed] [Google Scholar]
- 24.Hallahan DE, Virudachalam S. Accumulation of P-selectin in the lumen of irradiated blood vessels. Radiation research. 1999;152:6–13. [PubMed] [Google Scholar]
- 25.Molema G, de Leij LF, Meijer DK. Tumor vascular endothelium: barrier or target in tumor directed drug delivery and immunotherapy. Pharmaceutical research. 1997;14:2–10. doi: 10.1023/a:1012038930172. [DOI] [PubMed] [Google Scholar]
- 26.Oyajobi BO, Munoz S, Kakonen R, et al. Detection of myeloma in skeleton of mice by whole-body optical fluorescence imaging. Molecular cancer therapeutics. 2007;6:1701–8. doi: 10.1158/1535-7163.MCT-07-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyruchaud O, Winding B, Pecheur I, Serre CM, Delmas P, Clezardin P. Early detection of bone metastases in a murine model using fluorescent human breast cancer cells: application to the use of the bisphosphonate zoledronic acid in the treatment of osteolytic lesions. J Bone Miner Res. 2001;16:2027–34. doi: 10.1359/jbmr.2001.16.11.2027. [DOI] [PubMed] [Google Scholar]