Summary
Targeting virulence factors has gained increasing attention as a potential approach to new antibiotics. Small molecule inhibitors of virulence have been shown to change the course of disease in whole organism infection models. Recently, key advances in the field include the identification of novel targets within cell signaling pathways, a new class of anti-virulence compounds that target bacterial defenses against host immunity, and a growing body of in vivo data to support the general approach of anti-virulence therapies. Additionally, there has been a distinct trend toward developing broader spectrum anti-virulence compounds, in particular agents with activity against diverse Gram-negative organisms. Herein we provide an update on the status of the field with a focus on recent advancements.
Introduction
Since the first use of penicillin in the 1940s, clinical drug resistance has quickly followed the introduction of any new antibiotic. Highly resistant bacteria, including methicillin-resistant Staphylococcus aureus [1], extended-spectrum beta-lactamase producing Gram-negative organisms [2], and extensively drug resistant tuberculosis [3] now pose an increasing threat to public health with limited treatment options. New antimicrobial agents are clearly needed; however, recent approaches to drug discovery have been unsuccessful [4]. New paradigms for therapeutics are warranted, including strategies that target bacterial virulence in the battle against resistant organisms.
Targeting in vitro essential genes, in vivo essential genes, or virulence factors
The goal of any antibiotic is clearance or prevention of infection within the context of the host. However, most traditional antibiotics were identified based on their in vitro antimicrobial activity under laboratory culture conditions. As a result, most antibiotics target processes essential for in vitro growth, with the implicit assumption that the same processes are also essential for in vivo infection. New work questions the validity of this assumption, as exemplified in studies of fatty acid biosynthesis (FAB) inhibitors. Recent interest in targeting FAB as a strategy for antibiotic discovery is based on both evidence for its essentiality under traditional laboratory growth conditions and knowledge that isoniazid, a potent antituberculosis drug, targets mycolic acid biosynthesis [5]. Thus, great excitement surrounded the identification of the natural product platensimycin and its derivatives as FabF/B inhibitors [6,7]. A recent study however, suggests that FAB may not be equally essential in vivo where organisms are able to scavenge fatty acids from their host microenvironment. Inhibitors of the biosynthetic enzymes FabI and FabB did not impair growth of Streptococcus agalactiae in the presence of unsaturated fatty acids, which are present in human serum. Additionally, strains lacking FabI or FabB were not attenuated in a mouse model of neonatal meningitis [8]. These results cast doubt on the relevance of fatty acid biosynthesis as an antimicrobial target and bring into sharp relief the potential disparity between requirements for in vitro and in vivo bacterial survival.
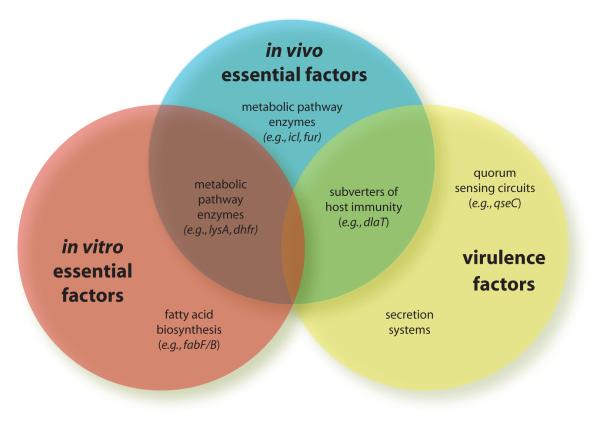
Bacterial functions that are required to cause disease in vivo can fall into two categories: those required for in vivo survival — which may or may not also be essential in vitro — and those required to cause tissue damage and disease, which are classically considered to be virulence factors (Fig. 1). In the first category, in vivo essential genes frequently fall along metabolic pathways that make or scavenge for required nutrients that are scarce within the host microenvironment. Those nutrients or their precursors may be readily available in culture media, obviating those pathways in vitro. For example, Mycobacterium tuberculosis deficient in both isocitrate lyase isozymes grows similarly to wild-type strains in standard culture media, but grows poorly in macrophages and is rapidly cleared in infected mice [9]. Other genes that are required in vivo include those that scavenge iron within the host, where levels may be low. As an example, Vibrio cholerae strains unable to produce the siderophore vibriobactin cannot colonize the intestine or cause diarrhea in a mouse infection model, yet grow normally in vitro [10]. Isocitrate lyase and the biosynthetic enzymes that produce vibriobactin would thus be considered essential in vivo but not in vitro, and would be potentially good targets for antibiotic development.
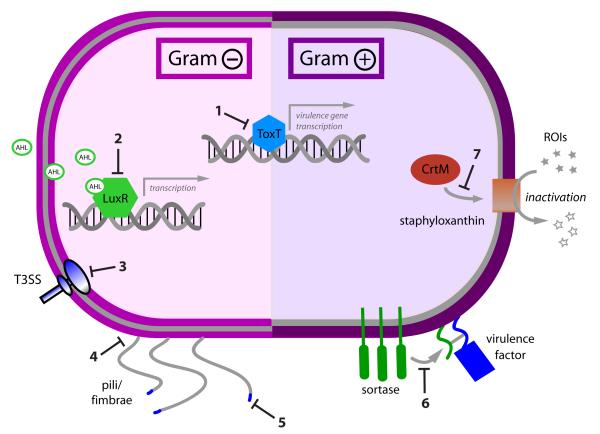
Figure 1. Virulence factors that represent potential targets for novel therapeutics in Gram-positive and Gram-negative bacteria.
1. toxin gene transcription, e.g. ToxT-dependent transcription of cholera toxin and toxin coregulated pilus in V. cholera 2. Quorum sensing, e.g. acyl-homoserine lactone (AHL) binding to transcriptional regulator LuxR in Gram-negative organisms 3. Secretion systems e.g. type III secretion in S. typhimurium 4. Adhesion e.g. pilus assembly in E. coli 5. Adhesion, e.g. carbohydrate binding motifs on pili in E. coli 6. Adhesion, e.g. sortase activity in S. aureus 7. Subverters of host immunity, e.g. CrtM production of staphyloxanthin in S. aureus
The second category of bacterial functions required to cause disease in vivo includes proteins that are classically referred to as virulence factors because they contribute directly to disease pathogenesis. While in vivo essential genes do not actively interact with host cells or functions, virulence factors actively damage host cells or interfere with host cell functions. For example, Salmonella effector proteins SopE and SopB, secreted into host cells through a type III secretion (T3S) machinery, reorganize the eukaryotic actin cytoskeleton, modulating bacterial uptake [11]. More subtly, some virulence factors may interfere with host immune functions. In M. tuberculosis, for example, dihydrolipoamide transferase (DlaT) neutralizes reactive nitrogen intermediates, key components of host immunity by reducing peroxynitrites [12]. Because of the active mechanism by which it subverts host function, we would consider it to be a virulence mechanism.
Distinguishing between in vivo essential functions and virulence mechanisms can sometimes be challenging. Since they both can effect in vivo bacterial survival, targeting either one is a viable therapeutic strategy. However, the remainder of this review will focus on targeting specific virulence factors as novel therapeutic strategies.
Pros and Cons of targeting virulence
Targeting virulence factors has several theoretical advantages over standard antibiotic treatment. First, a resistant clone’s survival advantage in the presence of traditional antibiotic drives selection for that clone. Theoretically, non-bactericidal drugs may not similarly select for resistance. If the targeted virulence factor is not essential for survival in vivo, mutations resulting in resistance should have no impact on relative bacterial fitness [13]. Second, because many virulence factors are organism-specific and virulence-targeting drugs are unlikely to be bactericidal, host commensal flora would be minimally impacted. Preserving commensals would reduce the risk of both secondary infections with organisms such as Clostridium difficile and colonization with drug-resistant organisms. Finally, the narrow spectrum of some anti-virulence therapies, while criticized as a potential drawback, can also be advantageous. Using new, limited spectrum antibiotics where clinically appropriate could restrict the use of broader spectrum antibiotics to instances of necessity, slowing the evolution of resistance to broad-spectrum agents.
While there are multiple potential advantages to virulence inhibitors as therapeutics, questions about their utility remain. Whether they will work best as prophylactic agents, solo therapeutic agents, or therapeutic agents in conjunction with conventional antibiotics has yet to be determined. Currently, because such inhibitors are quite organism specific, their widespread application would be dependent on either the development of real-time diagnostic tests or the development of broader spectrum anti-virulence agents. Such efforts are ongoing and detailed below.
Examples of targeting virulence
An increasing number of virulence inhibitors have been described as potential therapeutics. In Gram-negative organisms, efforts have focused on inhibiting cell-to-cell communication networks, secretion systems, and pilus-mediated adhesion. In Gram-positive organisms, inhibition of bacterial adhesion has been of recent interest. In both Gram-positive and Gram-negative organisms, toxin production and mechanisms to evade host immunity are potential targets. (Fig. 2; Table 1) Recently emerging themes include a trend toward the discovery of more “broad-spectrum anti-virulence” agents and increasing proof of efficacy in whole organism infection models.
Figure 2. Overlap between in vivo essential factors, in vitro essential factors, and virulence factors.
Categorization of example bacterial targets.
Table 1.
Examples of small molecule inhibitors of virulence factor targets.
| Target pathway | Inhibitor | Studied organisms |
Mechanism of action |
In vivo/ex vivo effect |
References |
|---|---|---|---|---|---|
| Quorum sensing |
LED209 | EHEC F. tularensis S. typhimurium |
Inhibition of QseC phosphorylation and activation |
Improved mouse survival after infection w/ F. tularensis or S. typhimurium |
25 |
| Secretion systems |
2-imino-5- arylidene thiazolidinone |
S. typhimurium Y. enterocolitica F. novocida Pseudomonas spp. |
T3S: inhibition of needle complex formation |
Decreased macrophage lysis after infection with S. typhimurium; Decreased tobacco leaf tissue damage after infection with P. syringae |
27 |
| Adhesion | 2-substituted- pyridones |
E. coli | Inhibition of pilus assembly |
Decreased hemagglutination |
30-32 |
| Toxin production |
Virstatin | V. cholera | Inhibition of cholera toxin and toxin coregulated pilus expression |
Decreased colonization in infant mouse model of infection |
43 |
| Staphyloxanthin biosynthesis |
BPH-652 | S. aureus | Inhibition of CrtM production of staphyloxanthin |
Decreased S. aureus survival in mouse infection |
48 |
Cell-to-cell signaling
Bacterial cell-to-cell communication through quorum sensing circuits in response to changing population density regulates the expression of virulence-associated genes in both Gram-positive and Gram-negative pathogens. Multiple quorum sensing networks may exist within a given organism. For example, acyl-homoserine lactones (AHL), produced by homologues of synthase LuxI, bind to homologues of transcription activator LuxR in Gram-negative bacteria, leading to expression of virulence-associated genes. Inhibition of AHL binding to LuxR has been pursued as a potential antibiotic strategy, with molecules in several classes identified as inhibitors [13-16]. In particular, exploration of structure-activity relationships for furanones [17-19] and other synthetic AHL analogues [20-22] are the subject of ongoing investigation, as is the isolation of new natural products, such as the venom alkaloid solenopsin, with similar activity [23].
Two recent studies show that blocking quorum sensing can have a protective effect in animal infection models. In addition to having two LuxI/LuxR systems (LasI/R, RhlI/R), Pseudomonas aeruginosa has a LysR-type transcriptional regulator, MvfR, activated by quinoline ligands. Lesic et al. recently identified 3 anthranilic acid analogues that decreased production of quinolines and subsequent expression of MvfR-induced genes. Given several hours after inoculation, all three compounds improved mouse survival and decreased bacterial spread in a murine model of post-thermal-injury infection. As a preliminary indication that they may have a broader spectrum of activity, all three were additionally shown to reduce quinoline production in two Burkholderia species [24].
In different Gram-negative organisms, Rasko et al. used a high-throughput screen to identify N-phenyl-4-{[(phenylamino)thioxomethyl]amino}-benzenesulfonamide, LED209, as an inhibitor of autophosphorylation and subsequent activation of QseC, a membrane kinase important for response to both host adrenergic molecules and another quorum sensing molecule (AI-3). LED209 blocked expression of QseC-regulated virulence-associated genes in enterohemorrhagic Escherichia coli (EHEC), Salmonella typhimurium, and Francisella tularensis. Additionally, the compound partially protected mice from death following infection with S. typhimurium or F. tularensis [25]. LED209’s activity demonstrates the possibility of quorum sensing-targeting therapies being broad in spectrum across Gram-negative pathogens.
Secretion systems
Bacterial secretion systems that deliver toxins to host cells have similarly been identified as potential targets for antimicrobial development [13,15,16,26]}. In a whole cell screen of small molecules, Felise et al. identified a thiazolidinone compound that inhibited type III secretion (T3S) system needle complex assembly in S. typhimurium. The thiazolidinone protected bone marrow-derived macrophages from lysis after infection with S. typhimurium and reduced the virulence of Pseudomonas syringae in a plant infection model. Importantly, the inhibitor showed a broad spectrum of activity in Gram-negative bacteria, blocking not only T3S, but also type II secretion (T2S) in Pseudomonas and type IV (T4P) secretion in Francisella [27]. In another study, a compound initially identified as an inhibitor of Yersinia T3S also inhibited T3S in Chlamydia [28]. Success in using these inhibitors across species of bacteria and classes of secretion systems adds to the growing body of evidence that broad-spectrum anti-virulence therapies are an achievable goal.
An alternative strategy employed for disrupting T3S has been inhibition of effector protein function. Arnoldo et al. identified an inhibitor of Exoenzyme S (ExoS), a T3S bacterial effector in P. aeruginosa with ADP ribosyltransferase activity, which protected CHO cells from lysis. However, a notable limitation to strategies targeting specific bacterial effectors such as ExoS arises from the lack of universal expression of ExoS in P. aeruginosa strains; thus therapeutics aimed at a single effector may have too narrow a spectrum to play any meaningful clinical role.
Adhesion
Gram-negative organisms
In Gram-negative bacteria, pili or fimbrae mediate bacterial adhesion to host cells. For some diseases such as recurrent E. coli cystitis, blocking adhesion to epithelial cells could theoretically prevent clinical infection. Two approaches to blocking adhesion have been pursued (for review, see [29]). The first approach, typified by molecules called pilicides, interferes with assembly of the bacterial adhesion apparatus. 2-substituted-pyridones act on the usher-chaperone pathway to reduce the number of pili formed [30-32]. While the utility of pilicides has been demonstrated in in vitro assays of hemagglutination [31], pilicides have not yet been shown to be efficacious in whole organism infection models.
An alternative means of blocking adhesion is saturation of exposed carbohydrate-binding sites on bacterial pili, which mediate receptor binding to host cells. Mannose derivatives were shown to block E. coli adhesion to a bladder cell line, to interfere with biofilm formation in vitro, and to decrease bacterial recovery and intracellular invasion in a murine model of cystitis [33]. Compounds in a second class, glycodendrimers, were shown to block E. coli adhesion in ELISA assays [34]. While conceptually intriguing, the theory of blocking bacterial carbohydrate binding motifs to interfere with infection has yet to be translated into a clinically applicable form.
Gram-positive organisms
Sortases, present in many Gram-positive bacteria, localize to the extracellular membrane and anchor proteins, including virulence factors, to the cell wall through transpeptidation. Adhesion to host cells also relies upon intact sortase activity [35,36]. A recent in silico virtual screen for small molecules that bind to the SrtA active site identified several molecules that have activity in in vitro assays [37]. Thus, a number of inhibitors of sortases have been described, including derivatives of 5-phenyl proline [38] and the natural product aaptamine [39]. However, sortase has not yet been proven to be a useful target in any in vivo studies.
Biofilm formation
Many pathogenic bacteria form biofilms, which are extensive, structured communities on surfaces where they remain protected within an extracellular matrix. Biofilms present challenging problems in prosthetic device infections and may contribute to other infections, including recurrent pseudomonal pneumonia in cystic fibrosis patients and endocarditis. Biofilms are particularly problematic because the bacteria in biofilms display phenotypic drug tolerance [40], rendering them extremely difficult to clear. A recent high-throughput screen for inhibitors of P. aeruginosa biofilm formation identified 30 compounds in 6 structural classes with potent activity [41]. A library of triazole derivatives was also screened for activity against biofilm formation in P. aeruginosa, S. auerus, and Acinetobacter baumanii; several compounds were identified which inhibited biofilms in one or more of these bacteria [42]. Because biofilm formation relies on both quorum sensing circuits and adhesion, inhibitors of either of those processes should also impact biofilm formation.
Toxin production
Toxins often mediate the bulk of pathology seen in several types of bacterial infections. Because of their direct role in causing disease, toxin transcription, expression, and function would make excellent targets for novel antimicrobials. Inhibition of toxin transcription has proven efficacious. In a murine model of cholera infection, virstatin, a small molecule inhibitor of cholera toxin (CT) and toxin coregulated pilus (TCP) expression, blocked intestinal colonization with V. cholerae [43]. In another example, a synthetic methionyl-tRNA inhibitor, REP3123, improved survival in a hamster model of C. difficile infection [44]. C. difficile, a growing cause of morbidity and mortality with the recent spread of highly pathogenic strains [45], is a spore forming organism that produces toxins A and B, resulting in a prototypical colitis. REP3123 decreased levels of toxin A and B in vitro and blocked sporulation.
Because of concerns over its potential use as a weapon of bioterrorism and its toxin-based pathogenic mechanism, Bacillus anthracis has also been the focus of studies aimed at toxin inhibition. Lethal factor (LF), one of two toxins critical for anthrax pathogenesis, is a metalloproteinase that contributes directly to host cell death. A hydroxamate derivative inhibited LF protease activity in vitro, protected macrophages from cell death, and substantially decreased mortality in mouse and rabbit infection models [46]. This compound has been the subject of ongoing studies aimed at producing a marketable anti-anthrax drug [47]. While inhibitors of bacterial toxin production or function will likely have narrow spectrum activity, they hold considerable promise in select cases.
Inhibitors of bacterial defenses against host immunity
Although they contribute to disease less directly, factors that allow bacteria to overcome host immune defenses fit our definition of virulence factors and would make good therapeutic targets. Staphyloxanthin, produced by synthase CrtM, is a pigment that protects S. aureus from killing by reactive oxygen species by quenching free radicals. An isogenic strain deficient in crtM has significantly decreased survival when exposed to H2O2 or whole blood [48]. While treatment with a CrtM inhibitor, phosphonosulfonate BPH-652, had no impact on mouse nasal colonization, it successfully inhibited S. aureus growth in a murine model of intraperitoneal infection. A similar strategy of inhibiting mechanisms to evade host defenses was pursued in M. tuberculosis, with the discovery of D157070, (3-hydroxypropyl3-((Z)-5-((5-(2-chlorophenyl)furan2-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)benzoate, an inhibitor of DlaT-mediated neutralization of reactive nitrogen intermediates in non-replicating bacteria [49]. BPH-652 and D157070 thus represent a new class of anti-virulence compounds that interfere with bacterial mechanisms to subvert host immunity.
Conclusions and future directions
While the concept of taking aim at virulence factors in a new generation of antibacterial agents is now established in theory, an increasing number of examples of successful applications of such a strategy in in vivo animal infection models supports the concept in reality. In addition, more potential targets and approaches are being described, as evidenced by the recently noted targeting of factors that subvert host immunity. While this type of approach awaits wider acceptance as a functional solution to the growing problem of resistance, objections such as the challenge of tracking clinical resistance to anti-virulence compounds becomes less concerning as we enter an era of sophisticated molecular diagnostics. In addition, concerns regarding the limited spectrum of activity of compounds targeting specific virulence mechanisms are being addressed by recent progress toward broader spectrum agents. As the need for new antibiotics becomes increasingly acute, there may be growing motivation to move from laboratory-based exploration of this principle toward viable drug candidates.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller LG, Kaplan SL. Staphylococcus aureus: A Community Pathogen. Infectious Disease Clinics of North America. 2009;23:35. doi: 10.1016/j.idc.2008.10.002. -+ [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Bliziotis IA, Kasiakou SK, Samonis G, Athanassopoulou P, Michalopoulos A. Outcome of infections due to pandrug-resistant (PDR) Gram-negative bacteria. Bmc Infectious Diseases. 2005;5:7. doi: 10.1186/1471-2334-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LoBue P. Extensively drug-resistant tuberculosis. Current Opinion in Infectious Diseases. 2009;22:167–173. doi: 10.1097/qco.0b013e3283229fab. [DOI] [PubMed] [Google Scholar]
- 4.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Reviews Drug Discovery. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 5.Heath RJ, White SW, Rock CO. Lipid biosynthesis as a target for antibacterial agents. Progress in Lipid Research. 2001;40:467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458:83–U85. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 9.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nature Medicine. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson DP, Payne SM. VIBRIO-CHOLERAE IRON TRANSPORT-SYSTEMS - ROLES OF HEME AND SIDEROPHORE IRON TRANSPORT IN VIRULENCE AND IDENTIFICATION OF A GENE ASSOCIATED WITH MULTIPLE IRON TRANSPORT-SYSTEMS. Infection and Immunity. 1994;62:5120–5125. doi: 10.1128/iai.62.11.5120-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan JE, Collmer A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 12.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 13.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nature Chemical Biology. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 14.Geske GD, O’Neill JC, Blackwell HE. Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria. Chemical Society Reviews. 2008;37:1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nature Reviews Microbiology. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hett EC, Hung DT. Chemical Biology Approaches to Host-Pathogen Interactions. Wiley Encyclopedia of Chemical Biology. 2008 [Google Scholar]
- 17.Kim C, Kim J, Park HY, Park HJ, Lee JH, Kim CK, Yoon J. Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa. Applied Microbiology and Biotechnology. 2008;80:37–47. doi: 10.1007/s00253-008-1474-6. [DOI] [PubMed] [Google Scholar]
- 18.Janssens JCA, Steenackers H, Robijns S, Gellens E, Levin J, Zhao H, Hermans K, De Coster D, Verhoeven TL, Marchal K, et al. Brominated Furanones Inhibit Biofilm Formation by Salmonella enterica Serovar Typhimurium. Applied and Environmental Microbiology. 2008;74:6639–6648. doi: 10.1128/AEM.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estephane J, Dauvergne J, Soulere L, Reverchon S, Queneau Y, Doutheau A. N-acyl-3-amino-5H-furanone derivatives as new inhibitors of LuxR-dependent quorum sensing: Synthesis, biological evaluation and binding mode study. Bioorganic & Medicinal Chemistry Letters. 2008;18:4321–4324. doi: 10.1016/j.bmcl.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 20.Geske GD, Mattmann ME, Blackwell HE. Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators. Bioorganic & Medicinal Chemistry Letters. 2008;18:5978–5981. doi: 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang WZ, Morohoshi T, Ikeda T, Chen L. Inhibition of Lux quorum-sensing system by synthetic N-acyl-L-homoserine lactone analogous. Acta Biochimica Et Biophysica Sinica. 2008;40:1023–1028. doi: 10.1111/j.1745-7270.2008.00490.x. [DOI] [PubMed] [Google Scholar]
- 22.Morohoshi T, Shiono T, Takidouchi K, Kato M, Kato N, Kato J, Ikeda T. Inhibition of quorum sensing in Serratia marcescens AS-1 by synthetic analogs of N-acylhomoserine lactone. Applied and Environmental Microbiology. 2007;73:6339–6344. doi: 10.1128/AEM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Kaufmann GF, Bowen JP, Arbiser JL, Janda KD, Solenopsin A. a venom alkaloid from the fire ant Solenopsis invicta, inhibits quorum-sensing signaling in Pseudomonas aeruginosa. Journal of Infectious Diseases. 2008;198:1198–1201. doi: 10.1086/591916. [DOI] [PubMed] [Google Scholar]
- 24**.Lesic B, Lepine F, Deziel E, Zhang JW, Zhang QH, Padfield K, Castonguay MH, Milot S, Stachel S, Tzika AA, et al. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. Plos Pathogens. 2007;3:1229–1239. doi: 10.1371/journal.ppat.0030126.Identification of anthranilic acid analogues that give in vivo protection against Pseudomonas aeruginosa infection by blocking quorum sensing. Analogues may also have broader spectrum activity, as shown by activity against Burkholdaria species.
- 25**.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei SG, Roth M, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354.High-throughput screen discovery of a histidine kinase inhibitor that blocks quorum sensing in multiple Gram-negative bacteria. In vivo efficacy shown in mouse models of infection with Francisella tularensis and Salmonella typhimurium.
- 26.Keyser P, Elofsson M, Rosell S, Wolf-Watz H. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. Journal of Internal Medicine. 2008;264:17–29. doi: 10.1111/j.1365-2796.2008.01941.x. [DOI] [PubMed] [Google Scholar]
- 27**.Felise HB, Nguyen HV, Pfuetzner RA, Barry KC, Jackson SR, Blanc MP, Bronstein PA, Kline T, Miller SI. An Inhibitor of Gram-Negative Bacterial Virulence Protein Secretion. Cell Host & Microbe. 2008;4:325–336. doi: 10.1016/j.chom.2008.08.001.Identification of a broad-spectrum inhibitor of Gram-negative bacterial secretion with evidence of in vivo effect against plant pathogen Pseudomonas syringae.
- 28.Wolf K, Betts HJ, Chellas-Gery B, Hower S, Linton CN, Fields KA. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Molecular Microbiology. 2006;61:1543–1555. doi: 10.1111/j.1365-2958.2006.05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobrindt U, Hacker J. Targeting virulence traits: potential strategies to combat extraintestinal pathogenic E-coli infections. Current Opinion in Microbiology. 2008;11:409–413. doi: 10.1016/j.mib.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Aberg V, Das P, Chorell E, Hedenstrom M, Pinkner JS, Hultgren SJ, Almqvist F. Carboxylic acid isosteres improve the activity of ring-fused 2-pyridones that inhibit pilus biogenesis in E-coli. Bioorganic & Medicinal Chemistry Letters. 2008;18:3536–3540. doi: 10.1016/j.bmcl.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aberg V, Fallman E, Axner O, Uhlin BE, Hultgren SJ, Almqvist F. Pilicides regulate pili expression in E-coli without affecting the functional properties of the pilus rod. Molecular Biosystems. 2007;3:214–218. doi: 10.1039/b613441f. [DOI] [PubMed] [Google Scholar]
- 32.Pemberton N, Pinkner JS, Edvinsson S, Hultgren SJ, Almqvist F. Synthesis and evaluation of dihydroimidazolo and dihydrooxazolo ring-fused 2-pyridones - targeting pilus biogenesis in uropathogenic bacteria. Tetrahedron. 2008;64:9368–9376. [Google Scholar]
- 33.Wellens A, Garofalo C, Nguyen H, Van Gerven N, Slattegard R, Hernalsteens JP, Wyns L, Oscarson S, De Greve H, Hultgren S, et al. Intervening with urinary tract infections using anti-adhesivesbased on the crystal structure of the FimH-soligomannose-3 complex. PLoS One. 2008;3:e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Touaibia M, Roy R. Glycodendrimers as anti-adhesion drugs against type 1 fimbriated E-coli uropathogenic infections. Mini-Reviews in Medicinal Chemistry. 2007;7:1270–1283. doi: 10.2174/138955707782795610. [DOI] [PubMed] [Google Scholar]
- 35.Suree N, Jung ME, Clubb RT. Recent advances towards new anti-infective agents that inhibit cell surface protein anchoring in Staphylococcus aureus and other Gram-Positive pathogens. Mini-Reviews in Medicinal Chemistry. 2007;7:991–1000. doi: 10.2174/138955707782110097. [DOI] [PubMed] [Google Scholar]
- 36.Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacological Reviews. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 37.Chenna BC, Shinkre BA, King JR, Lucius AL, Narayana SVL, Velu SE. Identification of novel inhibitors of bacterial surface enzyme Staphylococcus aureus Sortase A. Bioorganic & Medicinal Chemistry Letters. 2008;18:380–385. doi: 10.1016/j.bmcl.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 38.Kudryavtsev KV, Bentley ML, McCafferty DG. Probing of the cis-5-phenyl proline scaffold as a platform for the synthesis of mechanism-based inhibitors of the Staphylococcus aureus sortase SrtA isoform. Bioorganic & Medicinal Chemistry. 2009;17:2886–2893. doi: 10.1016/j.bmc.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang KH, Chung SC, Shin J, Lee SH, Kim TI, Lee HS, Oh KB. Aaptamines as sortase A inhibitors from the tropical sponge Aaptos aaptos. Bioorganic & Medicinal Chemistry Letters. 2007;17:5366–5369. doi: 10.1016/j.bmcl.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Lewis K. Persister cells, dormancy and infectious disease. Nature Reviews Microbiology. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 41.Junker LM, Clardy J. High-throughput screens for small-molecule inhibitors of Pseudomonas aeruginosa biofilm development. Antimicrobial Agents and Chemotherapy. 2007;51:3582–3590. doi: 10.1128/AAC.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huigens RW, Rogers SA, Steinhauer AT, Melander C. Inhibition of Acinetobacter baumannii, Staphylococcus aureus and Pseudomonas aeruginosa biofilm formation with a class of TAGE-triazole conjugates. Organic & Biomolecular Chemistry. 2009;7:794–802. doi: 10.1039/b817926c. [DOI] [PubMed] [Google Scholar]
- 43**.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310:670–674. doi: 10.1126/science.1116739.Describes a small molecule inhibitor of cholera toxin transcription with efficacy in a mouse model of infection.
- 44.Ochsner UA, Bell SJ, O’Leary AL, Hoang T, Stone KC, Young CL, Critchley IA, Janjic N. Inhibitory effect of REP3123 on toxin and spore formation in Clostridium difficile, and in vivo efficacy in a hamster gastrointestinal infection model. Journal of Antimicrobial Chemotherapy. 2009;63:964–971. doi: 10.1093/jac/dkp042. [DOI] [PubMed] [Google Scholar]
- 45.Loo VG. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality (vol 353, pg 2442, 2005) New England Journal of Medicine. 2006;354:2200–2200. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 46.Shoop WL, Xiong Y, Wiltsie J, Woods A, Guo J, Pivnichny JV, Felcetto T, Michael BF, Bansal A, Cummings RT, et al. Anthrax lethal factor inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7958–7963. doi: 10.1073/pnas.0502159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong YS, Wiltsie J, Woods A, Guo J, Pivnichny JV, Tang W, Bansal A, Cummings RT, Cunningham BR, Friedlander AM, et al. The discovery of a potent and selective lethal factor inhibitor for adjunct therapy of anthrax infection. Bioorganic & Medicinal Chemistry Letters. 2006;16:964–968. doi: 10.1016/j.bmcl.2005.10.088. [DOI] [PubMed] [Google Scholar]
- 48**.Liu CI, Liu GY, Song YC, Yin FL, Hensler ME, Jeng WY, Nizet V, Wang AHJ, Oldfield E. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–1394. doi: 10.1126/science.1153018.Discovery of an antivirulence compound that targets Staphylococcus aureus protections from the host immune system. Shown to have a protective effect in vivo.
- 49**.Bryk R, Gold B, Venugopal A, Singh J, Samy R, Pupek K, Cao H, Popescu C, Gurney M, Hotha S, et al. Selective killing of nonreplicating mycobacteria. Cell Host & Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003.Includes identification of a molecule that blocks mycobacterial protections from the host immune system. Efficacy shown in a mouse model of infection.