Abstract
HIV/SIV are thought to infect minimally activated CD4+ T cells after viral entry. Not much is known about why SIV selectively targets these cells. Here we show that CD4+ T cells that express high levels of the α4β7 heterodimer are preferentially infected very early during the course of SIV infection. At day 2–4 post infection, α4+β7hiCD4+ T cells had ∼ 5x more SIV-gag DNA than β7−CD4+ T cells. α4+β7hiCD4+ T cells displayed a predominantly central memory (CD45RA−CD28+CCR7+) and resting (CD25−CD69−HLA-DR−Ki-67−) phenotype. Though the expression of detectable CCR5 was variable on α4+β7hi and β7−CD4+ T cells, both CCR5+ and CCR5− subsets of α4+β7hi and β7−CD4+ T cells were found to express sufficient levels of CCR5 mRNA suggesting that both these subsets could be efficiently infected by SIV. In line with this, we found similar levels of SIV infection in β7−CD4+CCR5+ and β7−CD4+CCR5− T cells. α4β7hiCD4+ T cells were found to harbor most Th-17 cells that were significantly depleted during acute SIV infection. Taken together, our results show that resting memory α4+β7hiCD4+ T cells in blood are preferentially depleted during acute SIV infection, and the loss of these cells alters the balance between Th-17 and Th-1 responses thereby contributing to disease pathogenesis.
Keywords: HIV, SIV, simian, immunodeficiency, α4β7, Mucosa, CD4, Gut, Intestine, homing
Introduction
Acute human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infections are characterized by a massive loss of memory CD4+ T cells1–7, a process that is most pronounced in mucosal tissues as these tissues are enriched for memory CD4+ T cells. Like in non-natural hosts, severe depletion of CD4+ T cells have been reported in natural hosts such as sooty mangabeys and African green monkeys during acute SIV infection8,9. Surprisingly, the kinetics of infection in memory CD4+ T cells that reach a peak ∼ 10 days after SIV challenge, appear to be independent of the route of infection4,5.
The factors determining which target cells are infected initially, and those responsible for the delay between initial infection and the explosive levels of viral replication observed 10 days later, remain unclear. Studies5 have shown that prior to day 7 pi < 5–10% of CD4+ T cells in either the mucosal tissues or periphery are infected, whereas by day 10 pi most memory CD4+ T cells are infected and carry viral DNA. Li et al4 demonstrated that a few founder population of CD4+ T cells in the gut associated lymphoid tissue (GALT) are infected initially during SIV infection. Interestingly, these founder cells were minimally activated CD4+ T cells (CD25–CD69–Ki-67–) and had a predominantly resting phenotype. It is not clear why SIV targets these CD4+ T cells during the early phase of viral infection.
Recent in vitro studies10 have shown that CD4+ T cells that express high levels of the α4β7 (α4+β7hi) mucosal homing receptor can bind to HIV, and SIVsmm with high affinity making these cells more susceptible to infection. Arthos et al10 suggested that expression of high levels of α4β7 on CD4+ T cells likely offers a selective advantage to the virus, hence these cells may be the earliest cells targeted and infected by HIV. The loss of these mucosal homing T helper (Th) cells severely compromises the ability of the mucosal immune system. In line with this, recent studies have shown that mucosal Th–17 CD4+ T cells play a critical role in maintaining the integrity of the mucosal barrier11. Others have shown that alterations in the Th-17 and Th-1 responses contribute to disease progression12–14.
We hypothesized that if HIV selectively targeted the α4+β7hiCD4+ T cells during the early phase of infection, then these cells would be preferentially infected very early during the course of infection. We tested our hypothesis in the SIVmac251 infected rhesus macaque model by quantifying the cell-associated SIV DNA in α4+β7hiCD4+ T cells from peripheral blood during the early phase of SIV infection and compared them to SIV DNA levels in CD4+ T cells that either lacked (β7–) or expressed intermediate levels of α4β7 (α4+β7int). Previous studies15 have shown that HIV specific CD4+ T cells that were preferentially infected carried 2–5x more viral DNA than EBV or CMV specific CD4+ T cells. To determine if the differentiated and activated nature of α4+β7hiCD4+ T cells contributes to their susceptibility to SIV infection, we evaluated the expression of phenotypic (CD28, CD45RA and CCR7) and activation (CD25, CD69, HLA-DR, and Ki-67) markers on these cells. Additionally, to evaluate if preferential infection and depletion of α4+β7hiCD4+ T cells led to alterations in the balance between Th-17 and Th-1 type responses, we evaluated the expression of IL-17 and IFNγ in α4+β7hiCD4+ T cells and compared them to β7–CD4+ T cells.
Results
α4+β7hiCD4+ T cells are a minor population of peripheral CD4+ T cells
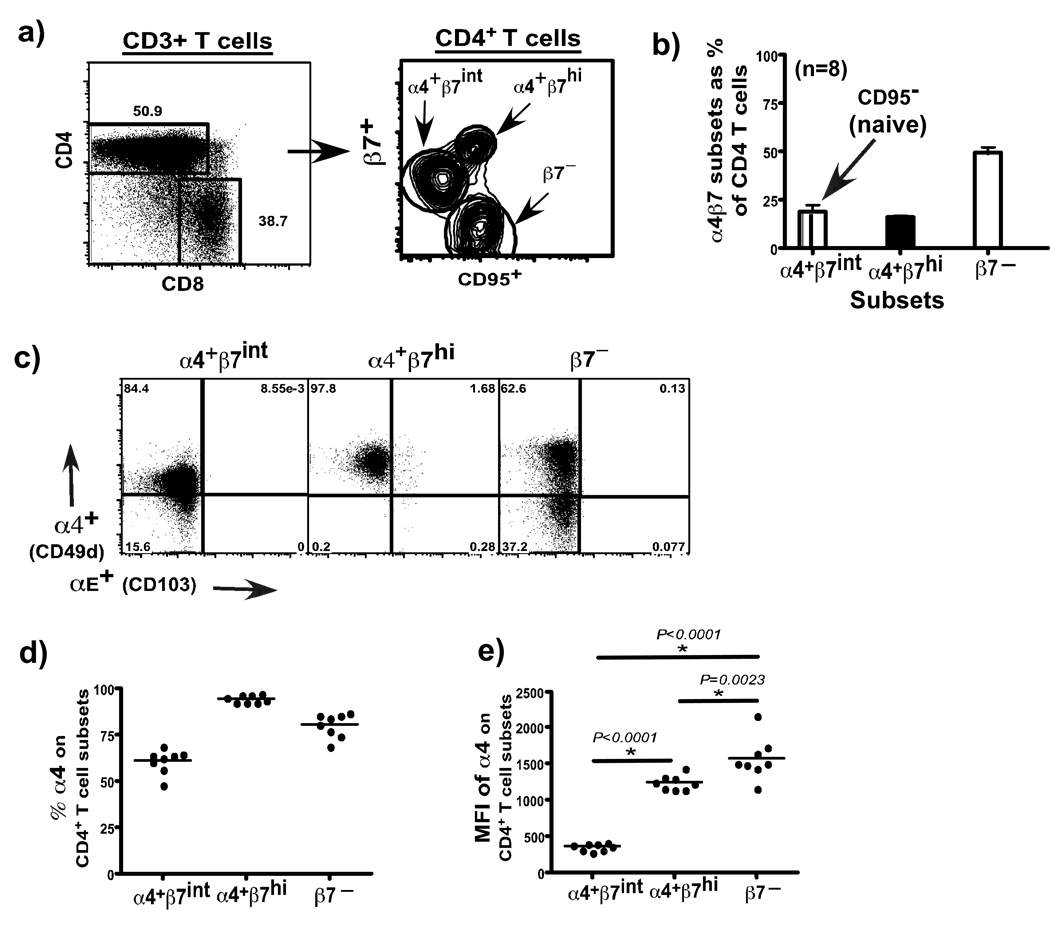
We used an antibody against the β7 receptor to identify different subsets of CD4+ T cells. Since β7 can form a heterodimer with α4 (CD49d) or αE (CD103)16, we first evaluated the expression of these receptors on β7 subsets in peripheral blood of healthy rhesus macaques. Our results showed that ∼50% of peripheral blood CD4+ T cells expressed the β7 receptor, whereas the rest of the 50% lacked β7 expression. Approximately 20% of CD4+ T cells that expressed the β7 receptor did so at high levels and had a β7hi phenotype, whereas ∼ 30% of them expressed intermediate levels of β7 receptor and had a β7int phenotype (Fig. 1a). More than 95% of β7hi, and ∼65% of β7int CD4+ T cells co-expressed the α4 receptor (Fig. 1b), and these are referred to as α4+β7hi and α4+β7int subsets in the text. Studies have shown that cells expressing high levels of the α4β7 efficiently traffic to mucosal tissues16.
Figure 1. Phenotypic analysis of CD3+CD4+ T cells in peripheral blood from healthy rhesus macaques (n = 8).
(a) Representative contour plots and (b) relative proportions of CD95+β7hi (α4+β7hi), CD95− β7int (α4+β7int), and CD95+β7− (β7−) subsets of CD4+ T cells. (c) Expression of α4 and αE integrins on α4+β7hi, α4+β7int, β7−CD4+ T cell subsets. (d) Relative proportions and (e) mean fluorescence intensity (MFI) of α4 expression on α4+β7hi, α4+β7int, β7−CD4+ T cell subsets. MFI was determined using Flowjo 8.6 software after gating on each subset.
On the other hand, CD4+ T cells that lacked β7 expression contained a mix of α4+ (∼75%) and α4– (∼25%) cells (Fig. 1c), and these subsets together are referred to as β7− CD4+ T cell subsets. α4 associates with β1 to form the α4β1 heterodimer, and binds to VCAM-1 (Vascular cell adhesion molecule-1) on activated endothelial cells which is involved in trafficking of T cells to peripheral lymphoid tissues17–20. Interestingly, β7−CD4 + T cell subsets expressed α4 integrin at a significantly higher density than α4β7hiCD4+ T cell subsets (Fig. 1e). Our results suggest that most of the β7−CD4+ T cell subsets likely home to peripheral lymphoid tissues.
All of the α4+β7hi and β7–CD4+ T cell subsets expressed CD95 indicating that these were memory CD4+ T cells. On the other hand, α4+β7intCD4+ T cells lacked CD95 expression indicative of a naïve phenotype. Previous studies have shown that rhesus macaque T cells expressing CD95 were memory T cells, whereas T cells that lacked CD95 were naïve T cells21. As our analysis was confined to peripheral blood, it is possible that the pattern of α4β7 expression may vary from that seen in GALT.
α4+β7hiCD4+ T cells express a central memory phenotype
Next we evaluated the phenotype of α4+β7hiCD4+ T cell subsets based on the co-expression of α4 and β7 integrins. Memory CD4+ T cells that express the α4β7 heterodimer appear on the diagonal as the antibodies to α4 and β7 bind to the same heterodimerized molecule, and co-labels with the Act-1 clone that recognizes an heterodimerized epitope of α4 and β722 (Suppl. Fig. 1a).
Three major subsets of memory CD4+ T cells could be delineated based on the expression of these two integrin molecules namely, α4+β7hi subset and two subsets of β7– CD4+ T cells namely, α4+β7– and α4–β7– subsets (Fig. 2a–b). Similar proportions of the 3 subsets are found within human memory CD4 T cells (personal communication; Mario Roederer, Suppl. Fig. 1e).
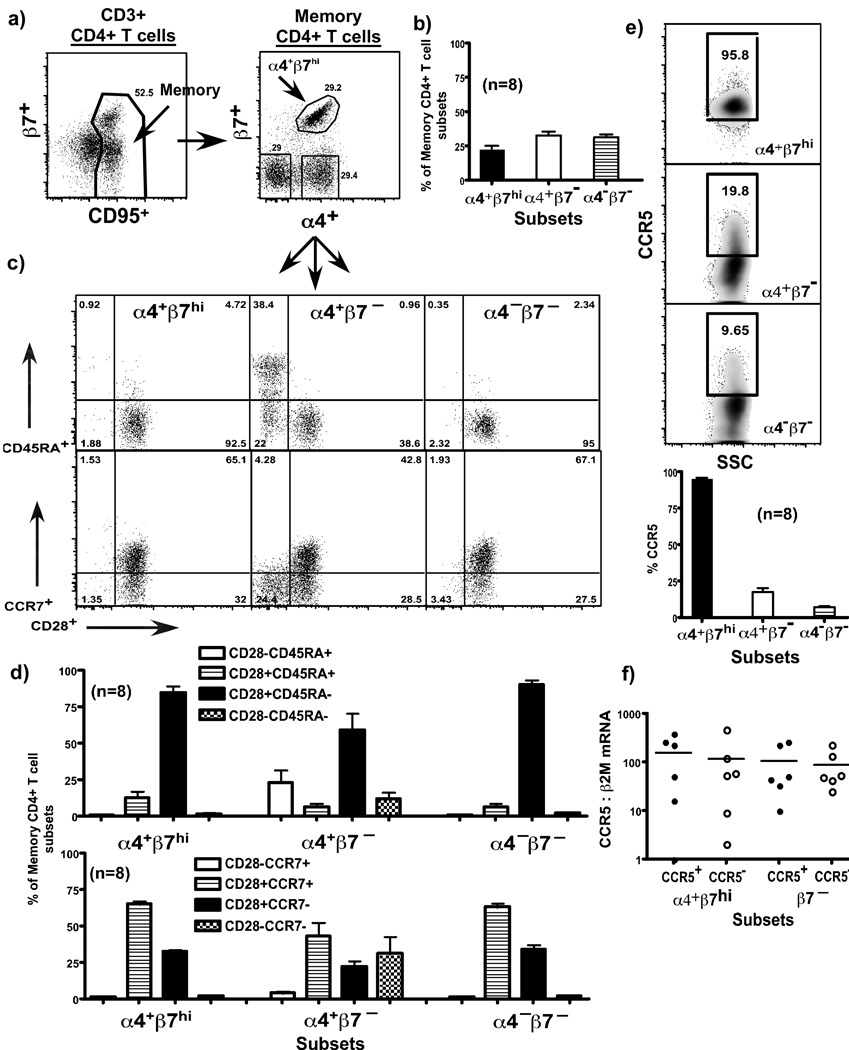
Figure 2. α4+β7hiiCD4+ T cell subsets display a predominantly central memory phenotype (n = 8).
Costaining for α4 and β7 identifies memory CD4 T cells that express the α4β7 heterodimer along a diagonal. (a) Representative dot plots and (b) relative proportions of α4+β7hi, α4+β7− and α4−β7− subsets of CD4+ T cells. (c) Representative dot plots and (d) relative proportions of α4+β7hi, α4+β7− and α4−β7− subsets of CD4+ T cells expressing CD45RA, CD28 and CCR7. (e) Representative dot plots and relative proportions of α4+β7hi, α4+β7− and α4−β7− subsets of CD4+ T cells expressing CCR5. Most α4+β7hi subsets express detectable levels of surface CCR5 (f) Relative expression of CCR5 mRNA in CCR5+ and CCR5− subsets of α4+β7hi and β7− CD4+ T cells (n = 6).
Majority of the α4+β7hiCD4+ T cell subsets expressed a central memory phenotype (C28+CCR7+CD45RA–), whereas most of the effector memory CD4+ T cells (CD28–CCR7–CD45RA+) are found within the α4+β7– subsets (Fig. 2 c–d).
Interestingly, most (>95%) of the α4+β7hiCD4+ T cell subsets expressed detectable levels of CCR5 on their surface (Fig. 2e), whereas only a few (<20%) of the β7− CD4+ T cell subsets expressed detectable levels of CCR5. To determine if these subsets expressed variable levels of CCR5 mRNA, we sorted CCR5+ and CCR5− subsets of α4+β7hi and β7− CD4+ T cells from uninfected animals (Suppl. Fig. 1b) and quantified the levels of CCR5 mRNA in these subsets using a relative qPCR assay (Fig. 2f). Our results show that though a majority of the β7− CD4+ T cell subsets expressed little CCR5 on their surface they had significant levels of CCR5 mRNA that did not differ from that of the other memory CD4 T cell subsets.
α4+β7hiCD4+ T cells are preferentially infected during acute SIV infection
To determine if the differential expression of α4, and β7 integrin’s on CD4+ T cell subsets made them variably susceptible to infection and subsequent loss, we evaluated the dynamics of α4+β7hiCD4+ T cell subsets in peripheral blood and compared them to α4+β7int and β7–CD4+ T cell subsets.
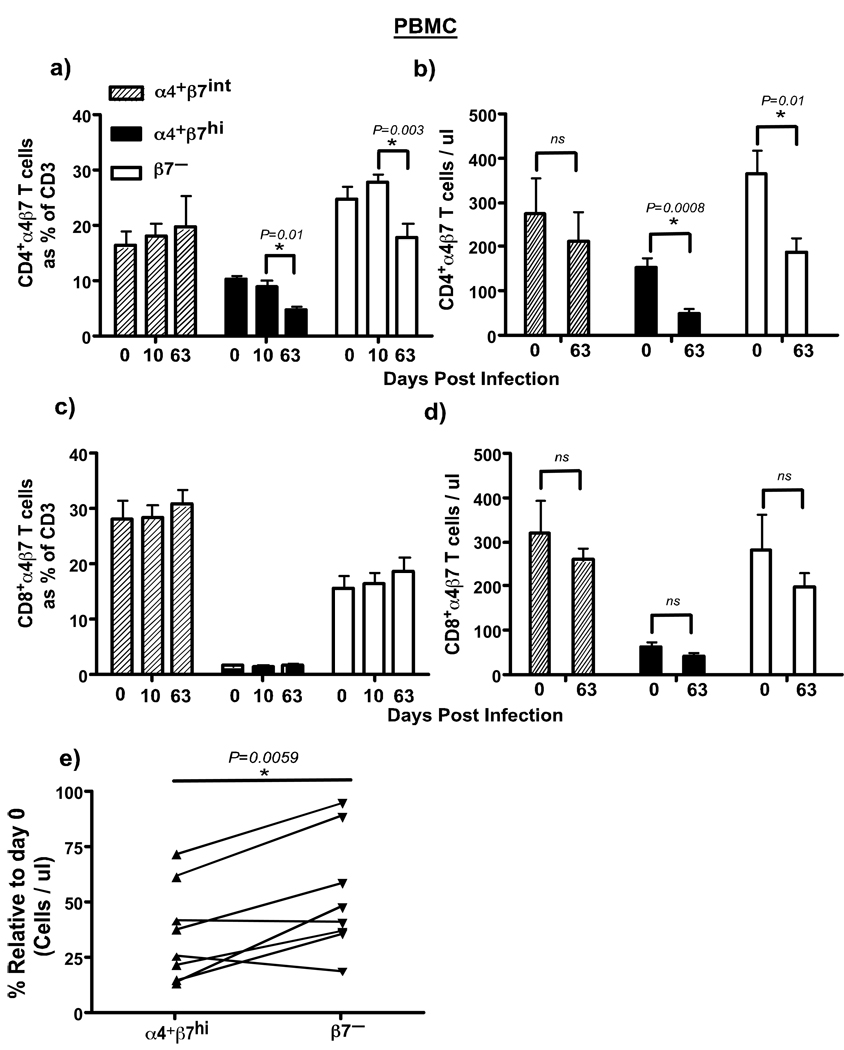
We observed a significant loss of both α4+β7hi and β7–CD4+ T cell subsets in peripheral blood at day 63 pi as compared to day 10 pi (Fig. 3a & b), whereas there was no major difference in either the frequency or absolute numbers of α4+β7intCD4+ T cell subsets or CD8 T cell subsets (Fig. 3a–d). The loss of both α4+β7hi and β7–CD4+ T cell subsets was not unexpected as previous studies5 have reported a significant loss of memory CD4+ T cells in peripheral blood during acute SIV infection; both α4+β7hi and β7–CD4+ T cell subsets have a memory phenotype. However, we observed a significantly higher loss of α4+β7hiCD4+ T cell subsets by day 63 pi as compared to the β7–CD4+ T cell subsets (Fig. 3e) suggesting that α4+β7hiCD4+ T cell subsets were preferentially depleted during the course of acute SIV infection.
Figure 3. Dynamics of α4 β 7 subsets of CD4+ T cells in peripheral blood during acute SIV infection.
Significant loss of α4β7hi and β7− CD4+ T cells is observed during acute SIV infection. Both frequency and absolute numbers are shown for (a–b) CD4+ and (c–d) CD8+ T cells (n = 8). (e) Higher numbers of α4+β7hiCD4+ T cells were destroyed by day 63 pi as compared to β7− CD4+ T cells. The % change in absolute numbers of α4+β7hi and β7−CD4+ T cells was calculated using each animals day 63 values relative to its day 0 values using day 0 values as 100%.
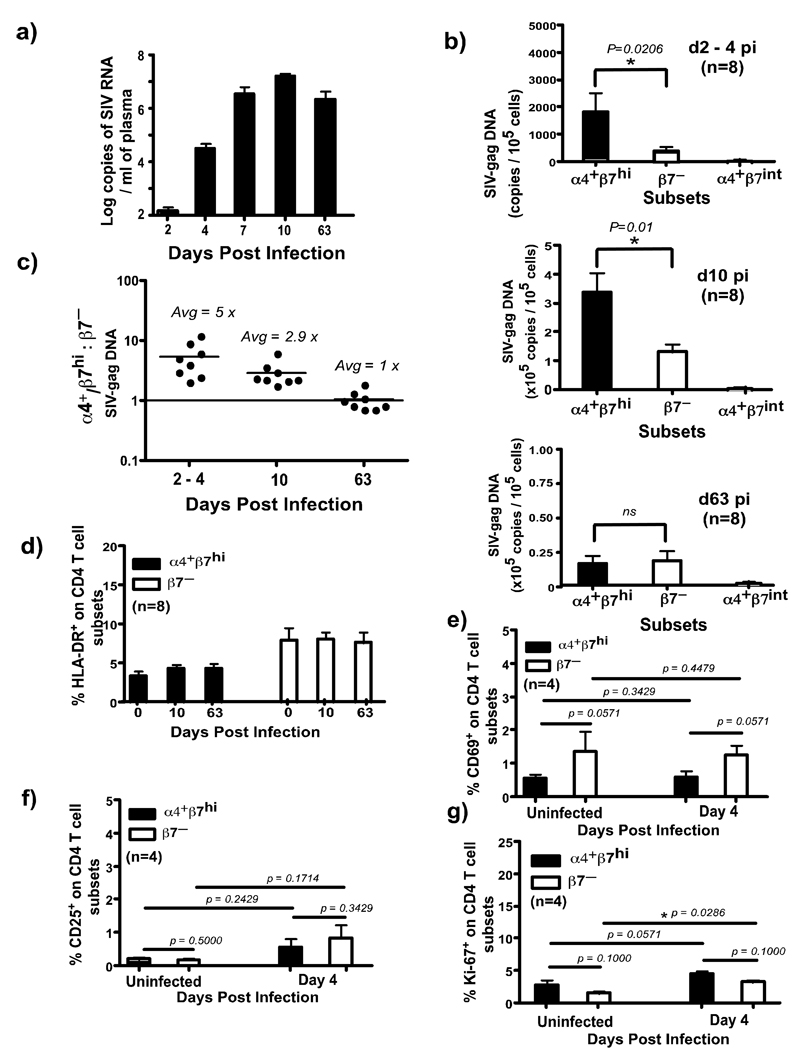
To determine if the preferential loss of α4+β7hiCD4+ T cell subsets was due to a higher level of viral infection in these subsets, we measured the level of SIV-gag DNA in α4+β7hiCD4+ T cell subsets and compared them to α4+β7int, and β7–CD4+ T cell subsets. α4+β7hi, α4+β7int, and β7–CD4+ T cell subsets, discriminated based on the expression of β7 and CD9521 (Supplementary Fig. 1c), were sorted from peripheral blood and used in a quantitative PCR assay to measure SIV-gag DNA. We have previously shown that this assay was highly sensitive for measuring viral infection in CD4+ T cells5. Measuring the level of SIV DNA in target cells allows us to quantify both productively and non-productively infected cells, hence is a more sensitive assay for quantifying viral infection in cells than RNA based assays that measures only productively infected cells. Previous studies have shown that most of the memory CD4+ T cells carried viral DNA during acute SIV infection, whereas only about 7–20% of them were productively infected4,5.
We found ∼5x more SIV-gag DNA in α4+β7hiCD4+ T cell subsets at day 2–4 pi as compared to β7–CD4+ T cell subsets suggesting that α4+β7hiCD4+ T cell subsets were preferentially infected very early during the course of infection (Fig. 4 b–c). By day 10 pi, there was a significant increase in the number of SIV-gag DNA copies in both subsets as compared to day 2–4 pi. However, like at day 2–4 pi, α4+β7hiCD4+ T cell subsets harbored ∼3x more SIV-gag DNA than β7–CD4+ T cell subsets (Fig. 4b–c).
Figure 4. α4+β7hiCD4+ T cells are preferentially infected during acute SIV infection.
(a) Plasma viral loads. Limit of detection is <30 copies /ml of plasma. (b) Levels of SIV-gag DNA were determined using qPCR assay for SIV-gag in sorted subsets of α4+β7hi, α4+β7int and β7− CD4+ T cells. α4+β7hiCD4+ T cells are preferentially infected at day 2–4 (n = 8) and day 10 pi (n = 8). (c) Ratio of SIV-gag DNA levels in α4+β7hi: β7−CD4+ T cells at day 2–4 (n = 8), 10 (n = 8) and 63 (n = 8) pi. The expression of (d) HLA-DR (n = 8) (e) CD69 (n = 4) (f) CD25 (n = 4) and (g) Ki-67 (n = 4) on α4+β7hi and β7−CD4+ T cells during the early phase infection.
By day 63 pi, however, the number of SIV-gag copies decreased significantly in both α4+β7hi and β7–CD4+ T cell subsets (Fig. 4b) indicating that most of the cells that were infected at day 10 pi, were destroyed by day 63 pi. Interestingly, unlike day 2–4 and 10 pi, both α4+β7hi and β7–CD4+ T cell subsets at day 63 pi had similar levels of SIV-gag DNA (Fig. 4c).
Few α4+β7hiCD4+ T cell subsets expressed HLA-DR (Fig. 4d) that did not change after infection. Previous reports23 have shown that there was no significant change in the expression of HLA-DR on memory CD4+ T cells during the early phase of acute SIV infection. Like HLA-DR, few α4+β7hiCD4+ T cell subsets expressed either CD25 (< 0.5%) or CD69 (< 0.5%) or Ki-67 (∼ 3%) prior to infection that did not change significantly at day 4 pi (Fig. 4d–g). Previous studies have shown that there was little or no change in the expression of HLA-DR or Ki-67 on memory CD4+ T cells at day 4, 7 and 10 pi as compared to preinfection values23, whereas others have shown that very few peripheral blood CD4+ T cells express CD25 or CD69 or Ki-67 during the acute phase of SIV infection24.
Unlike α4+β7hiCD4+ T cell subsets, higher proportions of β7–CD4+ T cell subsets expressed HLA-DR prior to infection, yet these cells were not preferentially infected. Likewise, β7–CD4+ T cell subsets were found to express significantly higher levels of Ki-67 at day 4 pi as compared to uninfected animals. Taken together, these findings indicate that preferential infection during the early phase of SIV infection is confined to α4+β7hiCD4+ T cell subsets that display a predominantly resting phenotype. This is in line with previous reports by Li et al4 who demonstrated that SIV primarily infects resting memory CD4+ T cells in the GALT that express a CD25–CD69–Ki-67– phenotype. It is important to point out that our results only represent snap shots of the changes occurring in vivo. As such it is difficult to rule out if α4+β7hiCD4+ T cell subsets were transiently activated at the time of infection.
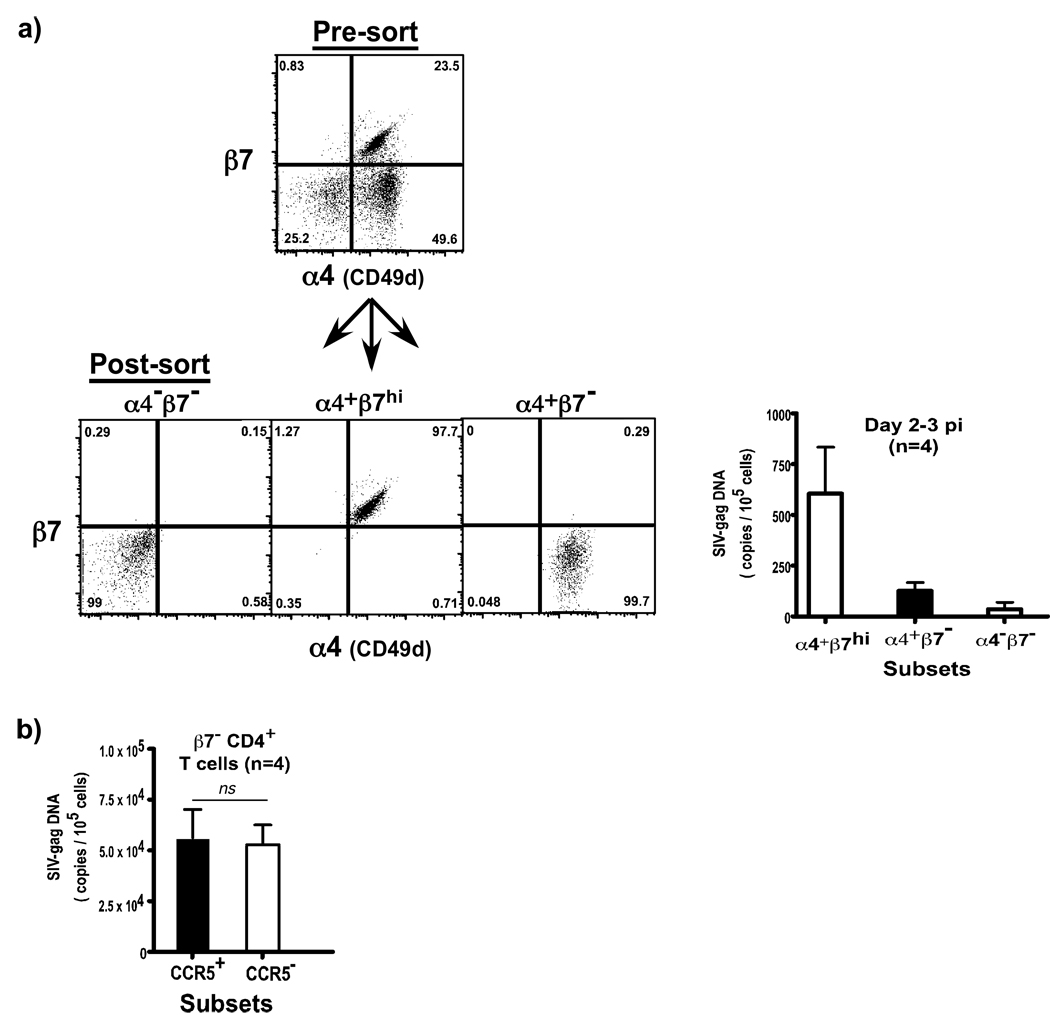
To confirm if SIV preferentially targeted CD4+ T cell subsets that expressed the α4β7 heterodimer during the early phase of SIV infection, we evaluated the levels of SIV-gag DNA in α4+β7hi, α4+β7−, and α4− β7− subsets of memory CD4+ T cells (Fig. 5a) using peripheral blood samples (day 2–3 pi) that were obtained from an unrelated study.
Figure 5. CD4+ T cells that express the α4β7 hetrodimer are preferentially infected in vivo.
Memory CD4+ T cells were sorted based on the co-expression of α4 and β7 integrins by flow cytometry, and used in the qPCR assay for SIV-gag DNA. (a) Sorting strategy and purity of sorted subsets are shown along with the level of SIV infection in α4+β7hi, α4+β7− and α4−β7− subsets at day 2–3 pi (n = 4). α4+β7hi subsets harbor significantly higher levels of SIV DNA at day 2–3 pi (n = 4; p = 0.0244) as compared to α4+β7− and α4−β7− subsets. Statistical analysis was performed using Kruskal-Wallis One Way Analysis of Variance (ANOVA) (b) There is no significant difference in the level of SIV-gag DNA between β7−CD4+CCR5+ and β7−CD4+CCR5− T cell subsets of memory CD4+ T cells from peripheral blood at day 7 &14 pi (day 7: n = 2, day 14: n = 2). β7− subsets were gated based on the lack of β7 expression and CD95, followed by sorting of CCR5+ and CCR5- subsets.
Our results showed that during the first few days after infection, the virus is largely confined to memory CD4+ T cells that express the α4β7 heterodimer; α4+β7hiCD4+ T cell subsets had significantly higher levels of SIV-gag DNA than either α4+β7– or α4–β7–CD4+ T cell subsets (Fig. 5a).
Taken together these results support the in vitro findings reported by Arthos et al10, and shows that SIVmac251, selectively target CD4+ T cells that express the α4β7 heterodimer.
Both β7–CD4+CCR5+ and β7–CD4+CCR5– T cell subsets are equally infected during acute SIV infection
To determine if detectable surface expression of CCR5 makes CD4+ T cells subsets more susceptible to SIV infection, we evaluated the levels of SIV infection in β7–CD4+CCR5+ T cell subsets and compared them to β7–CD4+CCR5– T cell subsets. Krzysiek at al25 demonstrated that CCR5+CD4+ T helper cells with non-lymphoid homing potential are preferentially depleted during HIV infection despite early treatment. Likewise, a number of studies have shown that CCR5+CD4+ T helper cells were preferentially depleted during HIV and SIV infection, whereas others have shown that both CCR5+ and CCR5–CD4+T cell subsets were infected during acute SIV infection1,5,26,27.
Since most (>95%) of the α4+β7hiCD4+ T cell subsets express detectable levels of CCR5 on their surface, we did not have sufficient samples to sort enough α4+β7hiCD4+CCR5– T cell subsets for the qPCR analysis. Hence we restricted our analysis to β7–CD4+ T cell subsets. We sorted β7–CD4+CCR5+ and β7–CD4+CCR5– T cell subsets from peripheral blood of SIV infected animals (day 7 &14 pi) obtained from an unrelated study, and quantified the levels of SIV-gag DNA by qPCR.
Our results (Fig. 5b) show that both β7–CD4+CCR5+ and β7–CD4+CCR5– T cell subsets had similar levels of SIV-gag DNA which did not differ significantly suggesting that preferential infection was not directly dependent on the surface level of CCR5 expression. As shown in Fig. 2f, CCR5– T cell subsets express sufficient levels of CCR5 message that likely enough to make these cells susceptible to SIV infection. These results support previous studies5 showing that there was no difference in the level of SIV infection in memory CD4+CCR5+ and memory CD4+CCR5– T cell subsets at day 3, 7, 10 and 14 pi.
α4+β7hiCD4+ T cell subsets harbor most Th-17 cells and are lost during acute SIV infection
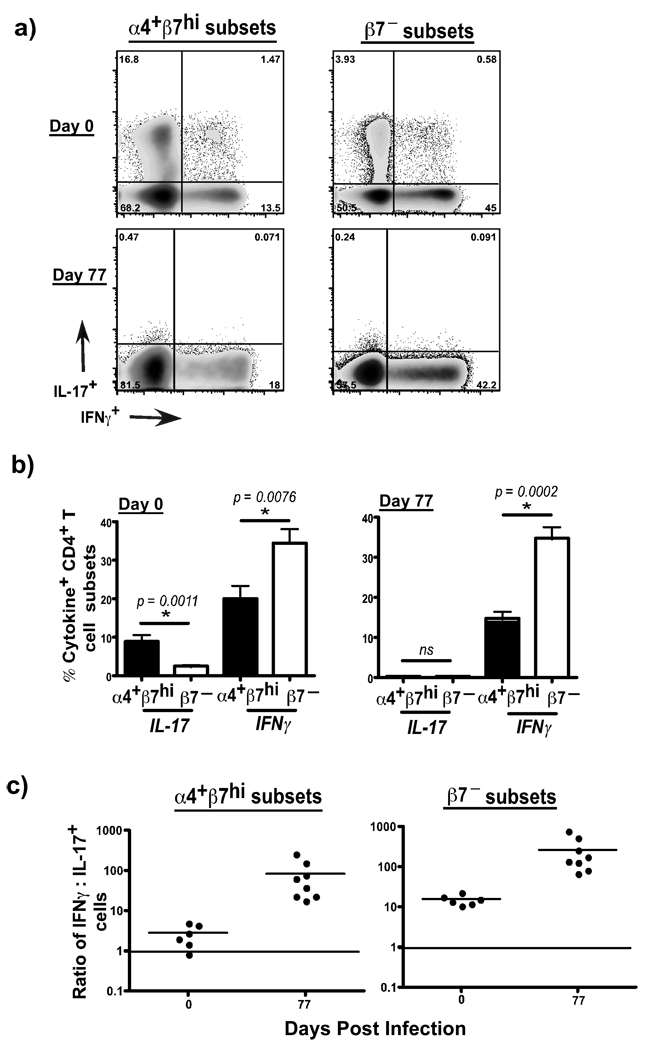
Previous studies have shown that mucosal CD4+ T cells are major producers of IL-17, and HIV infection was associated with alterations in the balance between Th-17 and Th-1 type responses11–14. To determine if the preferential loss of α4+β7hiCD4+ T cell subsets was accompanied by changes in the profile of Th-17 vs Th-1 type responses, we evaluated the production of IL-17 and IFNγ by α4+β7hiCD4+ T cell subsets and compared them to β7–CD4+ T cell subsets.
Our results showed that α4+β7hiCD4+ T cell subsets harbored significantly higher frequencies of Th-17 type cells whereas β7– CD4+ T cell subsets harbored higher proportions of IFNγ producing Th-1 type cells (Fig. 6a–b). Most of the Th-17 cells were found to be CD28+ memory CD4+ T cells (Suppl. Fig. 1d). The ratio of IFNγ: IL-17 producing cells was ∼ 2 within the α4β7hiCD4+ T cell subsets (Fig. 6c), whereas it was >10 within the β7− CD4+ T cell subsets indicating that α4β7hiCD4+ T cell subsets were the primary source of Th-17 responses. SIV infection was found to significantly skew the ratio to a predominantly Th-1 phenotype, as there was a significant depletion of Th-17 cells during early SIV infection.
Figure 6. α4+β7hiCD4+ T cells harbor significantly higher frequencies of Th-17 cells that are lost after SIV infection.
α4+β7hiCD4+ T cell subsets have a significantly higher capacity to produce IL-17 as compared to β7−CD4+ T cell subsets. Expression of IL-17 and IFNγ was determined by flow cytometry using PBMC from uninfected (n = 6) and SIV infected animals (n = 8) following stimulation with PMA/Ionomycin for 4 hours. (a) Representative dotplots showing IL-17 and IFNγ production α4+β7hi and β7−CD4+ T cell subsets at day 0 and day 77 pi. (b) IL-17 and IFNγ production α4+β7hi and β7−CD4+ T cell subsets at day 0 (n = 6) and day 77 pi (n = 8). (c) Ratio of IFNγ: IL-17 producing α4+β7hi and β7−CD4+ T cell subsets at day 0 and day 77 pi.
Discussion
Early SIV infection have been shown to target founder populations of CD4+ T cells4. These cells are minimally activated, and play a central role in early viral replication and dissemination. Not much is known about why SIV specifically targets these founder populations of CD4+ T cells. Given the highly activated microenvironment in mucosal tissues, and the enrichment of target cells in these tissues one would expect massive infection and replication to occur after viral entry. However, there appears to be a substantial delay of about 7–8 days before infection explodes out of the early founder population of cells to infect and destroy most of the memory CD4+ T cell compartment4,5. Arthos et al10 using in vitro studies demonstrated that HIV, and SIVsmm selectively targeted CD4+ T cells that expressed the α4β7 heterodimer suggesting that these cells could be earliest targets for viral infection in vivo. It is not known if HIV preferentially targets memory CD4+ T cells that express the α4β7 receptor in vivo.
Using the SIVmac251 infection model, we show that memory CD4+ T cells that expressed high levels of the α4β7 heterodimer are preferentially infected during the early phase of viral infection; α4+β7hiCD4+ T cells harbored ∼5x more SIV-gag DNA as compared to β7–CD4+ T cells. Surprisingly, α4+β7hiCD4+ T cells were found to be predominantly resting T cells (CD25–CD69–HLA-DR–Ki67–) with a central memory (CD28+CD45RA–CCR7+) phenotype that did not significantly change during the early phase of viral infection. Li et al4 reported that during the early phase of infection, SIV infection in the GALT is confined to resting CD4+ T cells that were CD25–CD69–Ki67–.
Both CCR5+ and CCR5– subsets of α4+β7hiCD4+ and (37–CD4+CCR5+T cells were found to have significant levels of CCR5 mRNA suggesting that all the memory CD4+ T cells subsets have sufficient CCR5 for infection by SIV. In line with this, both β7– CD4+CCR5+ and β7–CD4+CCR5– T cells had similar levels of SIV infection. It is likely that once SIV binds to the cells, the high levels of CCR5 expression enables SIV to more efficiently infect α4+β7hiCD4+ T cells, especially during the early phase of infection, when viral loads are very low, as seen at day 2–4 pi.
By day 7–10 pi, there was a significant increase in the extent of infection in both α4+β7hi and β7–CD4+ T cells, most likely as a consequence of the massive viral replication occurring during this period. However, α4+β7hiCD4+ T cell subsets were found to harbor ∼3x more SIV DNA than β7–CD4+ T cell subsets.
Interestingly, α4+β7hiCD4+ T cells were found to contain ∼3.5 to 4 copies of SIV / cell at day 10 pi suggesting that there were likely more than 1 virion / cell. The assay used to quantify SIV infection in cells is stringently standardized using a non-expressing SIV infected cell line that carries only a single copy for SIV DNA5. As such we do not think that these high copy numbers are assay related. It is more likely that the expression of the α4β7 receptor along with CD4 and high levels of CCR5 makes α4+β7hiCD4+ T cells more susceptible to super infection with SIV. Previous studies have reported multiply infected CD4+ T cells in HIV infected subjects. Jung et al28 using fluorescent in situ hybridization assays showed that CD4+ T cells in the spleen of HIV infected subjects carried 4–6 proviruses per infected cell. Future studies will attempt to address this question in more detail.
By day 63 pi, the extent of infection had equalized between the two subsets suggesting that there was a significantly higher loss of infected α4+β7hiCD4+ T cells following peak viral infection. In line with this conclusion, a significantly greater number of α4+β7hiCD4+ T cell subsets were lost between day 0 and 63 pi as compared to the β7–CD4+ T cell subsets.
A number of factors likely contribute to the amplification of infection in β7–CD4+ T cell subsets. Acute SIV infection has been shown to be associated with acute immune activation23,24 leading to the production of numerous pro-inflammatory mediators such as IL-1523. Likewise, acute HIV infection is associated with an increase in numerous pro-inflammatory cytokines such as TNFα, IFNγ, IL-6, IL8, MCP-1, IFNα etc29. These factors likely play a role in making CD4+ T cells more susceptible to infection. Eberly et al23 showed that there was a significant increase in the production of IL-15 between day 7–14 pi that was found to correlate with infection in memory CD4+ T cells during the early stages of infection. Others have shown that efficiency of infection was greatly influenced by the density of CD4, but not CCR5 expression on the surface of target cells30–33.
It is also possible that during periods of high viremia, as seen during the ramp-up phase of acute viral infection, the threshold for efficient infection is lower than during the pre-ramp phase, when viral loads are very low. Thus, even though β7–CD4+ T cell subsets inherently expressed low levels of CCR5, it was likely enough for SIV to efficiently infect these cells between day 7–14 pi when plasma viral loads are as high as 7–8 logs / ml.
Previous studies4,5 have shown that <5% of memory CD4 T cells were infected prior to day 7 pi, whereas by day 10–14 pi there was a significant increase in infection of these cells. Though these memory CD4+ T cells lacked detectable CCR5 on their surface, they were found to have 20x more CCR5 mRNA as compared to naïve CD4+ T cells that was sufficient for them to be infected by SIV. In line with these studies, we found significant levels of CCR5 mRNA in all the subsets of α4+β7hi and β7–CD4+ T cells. Others have shown that, minimal expression of CCR5 was sufficient for infection of CD4+ T cells30,32,33. Eberly et al23 demonstrated that acute SIV infection was associated with little or no upregulation of CCR5 expression on memory CD4+ T cells during the first 10 days of infection, yet there was a significant increase in the level of SIV infection in these cells between day 7 and 10 pi. Taken together, these studies indicate that when viral loads are high, minimal levels of CCR5 is likely sufficient to support efficient infection of memory CD4+ T cell subsets.
Interestingly, α4+β7hiCD4+ T cell subsets harbored a significantly higher potential to produce IL-17 suggesting that most of the T helper-17 (Th-17) cells are found within these mucosal homing T cell subsets. The preferential loss of these subsets during acute SIV infection likely cripples the ability of the immune system to maintain homeostasis in mucosal tissues. On the other hand, most of the IFNγ producing T helper (Th-1) cells were found to be β7–CD4+ T cell subsets, and fewer of these cells were lost suggesting to a skewing of the T helper response to a Th-1 phenotype. Recent studies11 have shown that Th-17 responses play a central role in protecting the mucosal barrier function, and the loss of Th-17 cells was associated with systemic dissemination of Salmonella in SIV infected rhesus macaques. Cecchinato et al13 showed that altered balance between Th-17 and Th-1 cells in mucosal sites predicted AIDS progression in SIV infected rhesus macaques. On the other hand, Favre et al14 showed that pathogenic SIV infection was associated with altered balance between Th-17 and T regulatory populations. Likewise, Brenchley et al12 demonstrated that Th-17 cells were differentially depleted in pathogenic vs non-pathogenic lentiviral infections. Macal et al34 showed that restoration of mucosal CD4 T cells was associated with enhanced Th-17 responses in HIV infected patients. Taken together, these studies suggest that preferential infection and depletion of α4β7hiCD4+ T cell subsets significantly skews T helper responses towards a Th-1 phenotype thereby contributing to disease progression.
In conclusion, our results provide in vivo evidence that, during the early phase of SIV infection, SIVmac251 selectively targets α4+β7hiCD4+ T cell subsets. These cells are predominantly central memory T cells with a resting phenotype and resemble the founder populations of CD4+ T cells described by Li et al4. The inherently high level of CCR5 expression likely allows SIV to more efficiently infect the α4+β7hiCD4+ T cell subsets very early during the course of viral infection. Development of therapeutic strategies targeted at preventing the early infection of memory CD4+ T cell subsets that express high levels of α4β7 heterodimer may significantly aid in the early control of viral infection and contribute to the maintenance of homeostasis in mucosal tissues.
Materials and Methods
Animals & infection
Mamu*A01neg rhesus macaques (Macaca mulatta) of Indian origin (n = 8) housed at Advanced Bioscience Laboratories Inc., MD were used in this study. Animals were housed in accordance with American Association for Accreditation of Laboratory Animal Care guidelines and were sero-negative for SIV, simian retrovirus and simian T-cell leukemia virus type-1. All the animals were infected with 100 animal-infectious doses of uncloned pathogenic SIVmac251 intravenously; Peripheral blood was collected at different time points (day 0, 2, 4, 10 and 63) after challenge, and Complete blood counts (CBC) data was obtained at day 0 and 63 pi. No CBC data was collected at other time points, hence absolute counts for various subsets could only be determined at day 0 and 63 pi. Additionally, peripheral blood samples were also obtained from 14 SIVmac251 infected rhesus macaques at day 2–3 (n = 4) and 7–14 pi (n = 10), 77 (n = 8) and from uninfected animals (n = 6 – 8).
Tissue samples
PBMC were isolated by density gradient centrifugation as previously described5. Plasma viral loads were determined by real-time PCR (ABI Prism 7700 sequence detection system, Applied Biosystems) using reverse-transcribed viral RNA as templates using methods previously described35 (limit of detection < 30 copies / ml of plasma).
Antibodies and flow cytometry
All antibodies, except for CD103, used in this study were obtained from BD Biosciences (San Diego, CA), and titrated using rhesus macaque PBMC. For phenotypic analysis and sorting of CD4+ T cell subsets cells were labeled simultaneously with the following combinations of antibodies: CD3-Cy7APC, CD8-Alexa-700, CD4-APC, CD95-FITC, CCR5-PE, HLA-DR-TexasRed-PE and Integrin β7 (FIB504 clone)-Cy5-PE. FIB504 clone has been shown to cross react with rhesus macaques10,21. β7 integrin forms a heterodimer with α4 (CD49d) or αE (CD103) integrins, and plays a central role in homing of T cells to mucosal tissues36–39. To delineate α4+β7+CD4+ T cell subsets from αEβ7+ CD4+ T cell subsets, cells were labeled with CD3-Cy7APC, CD8-Alexa-700, CD4-APC, CD95-FITC, CD49d–PE, CD103-FITC (clone 2G5 from Beckman Coulter) and Integrin β7-Cy5-PE. Clone 2G5 has been shown to cross react with rhesus macaque mucosal homing T cells, and expressed by intraepithelial lymphocytes40,41. To evaluate the phenotype of α4β7 subsets of CD4+ T cells, cells were labeled with a panel of CD3-Cy7APC, CD8-Qdot-605, CD4-APC, CD95-FITC, CD49d–PE and β7-Cy5-PE, CD45RA-TR-PE, CD28-Cy7-PE and CCR7-Alexa-680. The expression of activation markers was determined by using a panel of CD3-Cy7APC, CD8-Alexa-700, CD4-TR-PE, CD95-FITC, CD49d–PE, β7-Cy5-PE, CD69 or CD25-APC.
To determine which α4β7 subsets were preferentially infected, α4β7 subsets of CD4+ T cells (discriminated based on the expression of β7 and CD9521) were sorted, and subjected to qPCR assay for measuring SIV-gag DNA as described previously5,15. To determine if CD4+ T cells that expressed the α4β7 heterodimer were preferentially infected, PBMC were labeled with: CD3-Cy7APC, CD8-Alexa-700, CD4-APC, CD95-FITC, CD49d–PE and β7-Cy5-PE. Additionally, KI-67 expression was evaluated using a panel of CD3-Cy7APC, CD8-Alexa-700, CD4-APC, CD95-PE and β7-Cy5-PE. After surface labeling, cells were fixed / permeabilized using the Cytofix/perm kit (BD Biosciences, San Diego, CA) and labeled with Ki-67-FITC.
To evaluate the ability of α4β7 subsets of CD4+ T cells to produce IL-17 and IFNγ, PBMC were stimulated with Phorbol myristate acetate (PMA) at 10 ng / ml and ionomycin at 500 ng / ml in the presence of Brefeldin-A (1 uM) for 4 hours. Cells were harvested and labeled with CD3-Cy7APC, CD8-Alexa-700, CD4-Qdot-605, CD95-APC, and β7-Cy5-PE. After surface labeling, cells were fixed / permeabilized using the Cytofix/perm kit and labeled with IL-17-PE (e-Biosciences) and IFNγ-FITC (BD Biosciences).
Labeled cells were fixed in 0.5% paraformaldehyde, and analyzed using a modified Becton Dickinson Aria sorter.
qPCR assay
T-cell-associated viral DNA was measured by a quantitative PCR assay for SIV gag using a Perkin-Elmer ABI 7700 instrument as previously described5,15 using SIV gag primers and probe as described by Lifson et al42. Cell numbers were quantified simultaneously using rhesus macaque Albumin specific primers and probe using previously described protocols5.The assay was calibrated using a cell line that carried a single copy of proviral SIV DNA as shown previously5. Sorted samples were lysed in Proteinase-K and used for PCR analysis. We have previously shown that this assay was highly sensitive, and can be used to successfully measure SIV-DNA in single cells5.
CCR5 mRNA levels were determined using primer/probes as described previously5, and normalized to β2M levels in sorted subsets. Data are shown relative to α4+β7intCD4+ (naïve) T cells.
Data analysis
Flow cytometric data was analyzed using FlowJo version 8.6 (Tree Star, Inc., Ashland, OR). Statistical analysis was performed with GraphPad Prism Version 4.0 software (GraphPad Prism Software, Inc. San Diego, CA) using non-parametric tests. As the data did not exhibit normal distribution due to small sample size, comparisons between groups were performed using one-tailed Mann-Whitney U test, and multiple comparisons between groups was performed using the Kruskal-Wallis One Way Analysis of Variance (ANOVA). P < 0.05 was considered significant.
Supplementary Material
(a) FACs plots showing that nearly all memory CD4+ T cells that express ACT-1 costains with α4 and β7, and most of the α7hi subsets costain with ACT-1. ACT-1 clone (The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: α4–β7 monoclonal antibody (cat# 11718) from Dr. A. A. Ansari) recognizes an epitope on the α4β7 heterodimer22 (b) Sorting strategy used for sorting CCR5+ and CCR5− subsets of a4+β7hi and β7−CD4+ T cells. FACs plots showing post-sort purity is shown. (c) Gating strategy used for sorting the a4+β7hi, a4+β7int, and α7− subsets of CD4+ T cells for qPCR analysis. Post-sort purity FACs plots are shown. (d) IL-17 producing CD4+ T cells display a predominantly CD28+ phenotype. Representative dotplots showing the expression of IL-17 by a4+β7hi and β7−CD4+ T cells. Cells were stimulated with PMA/ionomycin for 4 hours and stained with a panel of markers that included CD28 and IL-17.
Acknowledgments
We thank Nancy Miller at the SVEU of NIAID for help with the animals; Karen Wolcott and Kateryna Lund at the Biomedical Instrumentation Core facility at USUHS for help with flow cytometry; Dr. Deborah Weiss and Jim Treece at ABL, Inc, Rockville, MD for expert assistance with the animals.
This project was supported by Grant Number K22AI07812 from the National Institutes of Allergy and Infectious diseases (NIAID) & Grant number R21DE018339 from the National Institute of Dental and Craniofacial Research (NIDCR) to JJM, and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contracts N01-CO-12400 and HHSN266200400088C. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID or NIDCR or NCI or the National Institutes of Health.
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Brenchley JM, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. The Journal of experimental medicine. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guadalupe M, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. Journal of virology. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kewenig S, et al. Rapid mucosal CD4(+) T-cell depletion and enteropathy in simian immunodeficiency virus-infected rhesus macaques. Gastroenterology. 1999;116:1115–1123. doi: 10.1016/s0016-5085(99)70014-4. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 5.Mattapallil JJ, et al. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 6.Mehandru S, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. The Journal of experimental medicine. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veazey RS, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 8.Gordon SN, et al. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J Immunol. 2007;179:3026–3034. doi: 10.4049/jimmunol.179.5.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandrea IV, et al. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol. 2007;179:3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthos J, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 11.Raffatellu M, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008 doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenchley JM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecchinato V, et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal immunology. 2008;1:279–288. doi: 10.1038/mi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favre D, et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS pathogens. 2009;5:e1000295. doi: 10.1371/journal.ppat.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douek DC, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 16.Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 17.Chan BM, Elices MJ, Murphy E, Hemler ME. Adhesion to vascular cell adhesion molecule 1 and fibronectin. Comparison of alpha 4 beta 1 (VLA-4) and alpha 4 beta 7 on the human B cell line JY. The Journal of biological chemistry. 1992;267:8366–8370. [PubMed] [Google Scholar]
- 18.Hemler ME, Huang C, Schwarz LThe, VLA proteinfamily. Characterization of five distinct cell surface heterodimers each with a common 130,000 molecular weight beta subunit. The Journal of biological chemistry. 1987;262:3300–3309. [PubMed] [Google Scholar]
- 19.Rose DM, Han J, Ginsberg MH. Alpha4 integrins and the immune response. Immunological reviews. 2002;186:118–124. doi: 10.1034/j.1600-065x.2002.18611.x. [DOI] [PubMed] [Google Scholar]
- 20.Takada Y, Elices MJ, Crouse C, Hemler ME. The primary structure of the alpha 4 subunit of VLA-4: homology to other integrins and a possible cell-cell adhesion function. The EMBO journal. 1989;8:1361–1368. doi: 10.1002/j.1460-2075.1989.tb03516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitcher CJ, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 22.Lazarovits AI, et al. Lymphocyte activation antigens. I. A monoclonal antibody, anti-Act I, defines a new late lymphocyte activation antigen. J Immunol. 1984;133:1857–1862. [PubMed] [Google Scholar]
- 23.Eberly MD, et al. Increased IL-15 production is associated with higher susceptibility of memory CD4 T cells to simian immunodeficiency virus during acute infection. J Immunol. 2009;182:1439–1448. doi: 10.4049/jimmunol.182.3.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur A, Hale CL, Ramanujan S, Jain RK, Johnson RP. Differential dynamics of CD4(+) and CD8(+) T-lymphocyte proliferation and activation in acute simian immunodeficiency virus infection. Journal of virology. 2000;74:8413–8424. doi: 10.1128/jvi.74.18.8413-8424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzysiek R, et al. Preferential and persistent depletion of CCR5+ T-helper lymphocytes with nonlymphoid homing potential despite early treatment of primary HIV infection. Blood. 2001;98:3169–3171. doi: 10.1182/blood.v98.10.3169. [DOI] [PubMed] [Google Scholar]
- 26.Veazey RS, et al. Dynamics of CCR5 expression by CD4(+) T cells in lymphoid tissues during simian immunodeficiency virus infection. Journal of virology. 2000;74:11001–11007. doi: 10.1128/jvi.74.23.11001-11007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veazey RS, Marx PA, Lackner AA. Vaginal CD4+ T cells express high levels of CCR5 and are rapidly depleted in simian immunodeficiency virus infection. J Infect Dis. 2003;187:769–776. doi: 10.1086/368386. [DOI] [PubMed] [Google Scholar]
- 28.Jung A, et al. Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 29.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viraemia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. Journal of virology. 2009 doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak SL, Kuhmann SE, Platt EJ, Kabat D. Roles of CD4 and coreceptors in binding, endocytosis, and proteolysis of gp120 envelope glycoproteins derived from human immunodeficiency virus type 1. The Journal of biological chemistry. 1999;274:23499–23507. doi: 10.1074/jbc.274.33.23499. [DOI] [PubMed] [Google Scholar]
- 31.Kozak SL, et al. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. Journal of virology. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesenti E, et al. Role of CD4 and CCR5 levels in the susceptibility of primary macrophages to infection by CCR5-dependent HIV type 1 isolates. AIDS Res Hum Retroviruses. 1999;15:983–987. doi: 10.1089/088922299310494. [DOI] [PubMed] [Google Scholar]
- 33.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. Journal of virology. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macal M, et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal immunology. 2008;1:475–488. doi: 10.1038/mi.2008.35. [DOI] [PubMed] [Google Scholar]
- 35.Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 36.Andrew DP, Rott LS, Kilshaw PJ, Butcher EC. Distribution of alpha 4 beta 7 and alpha E beta 7 integrins on thymocytes, intestinal epithelial lymphocytes and peripheral lymphocytes. European journal of immunology. 1996;26:897–905. doi: 10.1002/eji.1830260427. [DOI] [PubMed] [Google Scholar]
- 37.Berlin C, et al. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 38.Briskin M, et al. Human mucosal addressin cell adhesion molecule-1 is preferentially expressed in intestinal tract and associated lymphoid tissue. The American journal of pathology. 1997;151:97–110. [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell DJ, Butcher EC. Rapid acquisition of tissue-specific homing phenotypes by CD4(+) T cells activated in cutaneous or mucosal lymphoid tissues. The Journal of experimental medicine. 2002;195:135–141. doi: 10.1084/jem.20011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattapallil JJ, Letvin NL, Roederer M. T-cell dynamics during acute SIV infection. AIDS (London, England) 2004;18:13–23. doi: 10.1097/00002030-200401020-00002. [DOI] [PubMed] [Google Scholar]
- 41.Veazey RS, et al. Characterization of gut-associated lymphoid tissue (GALT) of normal rhesus macaques. Clinical immunology and immunopathology. 1997;82:230–242. doi: 10.1006/clin.1996.4318. [DOI] [PubMed] [Google Scholar]
- 42.Lifson JD, et al. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. Journal of virology. 2001;75:10187–10199. doi: 10.1128/JVI.75.21.10187-10199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) FACs plots showing that nearly all memory CD4+ T cells that express ACT-1 costains with α4 and β7, and most of the α7hi subsets costain with ACT-1. ACT-1 clone (The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: α4–β7 monoclonal antibody (cat# 11718) from Dr. A. A. Ansari) recognizes an epitope on the α4β7 heterodimer22 (b) Sorting strategy used for sorting CCR5+ and CCR5− subsets of a4+β7hi and β7−CD4+ T cells. FACs plots showing post-sort purity is shown. (c) Gating strategy used for sorting the a4+β7hi, a4+β7int, and α7− subsets of CD4+ T cells for qPCR analysis. Post-sort purity FACs plots are shown. (d) IL-17 producing CD4+ T cells display a predominantly CD28+ phenotype. Representative dotplots showing the expression of IL-17 by a4+β7hi and β7−CD4+ T cells. Cells were stimulated with PMA/ionomycin for 4 hours and stained with a panel of markers that included CD28 and IL-17.