SUMMARY
Mirtrons are alternative precursors for micro-RNA biogenesis that were recently described in invertebrates. These short hairpin introns use splicing to bypass Drosha cleavage, which is otherwise essential for the generation of canonical animal microRNAs. Using computational and experimental strategies, we now establish that mammals have mirtrons as well. We identified 3 mirtrons that are well conserved and expressed in diverse mammals, 16 primate-specific mirtrons, and 46 candidates supported by limited cloning evidence in primates. As with some fly and worm mirtrons, the existence of well-conserved mammalian mirtrons indicates their relatively ancient incorporation into endogenous regulatory pathways. However, as worms, flies, and mammals each have different sets of mirtrons, we hypothesize that different animals may have independently evolved the capacity for this hybrid small RNA pathway. This notion is supported by our observation of several clade-specific features of mammalian and invertebrate mirtrons.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22 nucleotide (nt) RNAs that typically repress the activity of complementary messenger RNAs (Lai, 2003). Canonical animal miRNAs derive from longer primary transcripts bearing hairpin structures, which are processed in a stepwise fashion by the RNase III enzymes Drosha and Dicer. In the nucleus, Drosha cleaves near the hairpin base to release the pre-miRNA hairpin (Lee et al., 2003). Following its export to the cytoplasm, Dicer cleaves on the loop side of the hairpin to generate an miRNA:miRNA* duplex, one strand of which is preferentially incorporated into a silencing complex (Du and Zamore, 2005).
An alternative nuclear pathway for miRNA biogenesis was recently described in invertebrates (Okamura et al., 2007; Ruby et al., 2007a). Short introns with hairpin potential, termed mirtrons, can be spliced and debranched into pre-miRNA hairpin mimics that appear to bypass Drosha cleavage. Debranched mirtrons access the canonical miRNA pathway during nuclear export, and are then cleaved by Dicer and incorporated into silencing complexes (Okamura et al., 2007; Ruby et al., 2007a).
Mirtrons were found only in nematodes and flies thus far. It was suggested that the evolutionary emergence of invertebrate mirtrons was aided by the sheer number of short introns whose length is typical of pre-miRNA hairpins (Ruby et al., 2007a). The relative proportion of such introns in different species is flies > worms > mammals (Lim and Burge, 2001; Yandell et al., 2006). However, because mammals have many more introns than do worms and flies, the difference in absolute numbers of short introns among these species is less substantial.
In this study, we addressed the possibility that mirtrons might exist in mammals. Using computational methods, we identified a small set of mammalian short hairpin introns as possible well-conserved mirtron candidates. Cloned ∼22 nt RNA products from the ends of three of these candidates were present in multiple small RNA libraries from human, macaque, chimpanzee, rat, and/or mouse, validating the existence of conserved mammalian mirtrons. Emboldened by these findings, we analyzed whether more “newly evolved” mirtrons could be detected, as these comprise the majority of identified fly and worm mirtrons. Indeed, by analyzing large-scale primate small RNA data sets, we could confidently classify 16 additional primate-specific mirtrons from human and macaque brain; nearly 50 additional candidates were supported by more tentative evidence (one to two clones). These findings indicate that mirtrons constitute a substantial and highly dynamic class of regulatory RNA in both invertebrates and vertebrates. Curiously, we identified several basic distinctions between mirtrons from these different clades, suggesting that this alternative strategy to generate microRNAs may have arisen more than once during animal evolution.
RESULTS AND DISCUSSION
Computational Survey for Well-Conserved Mammalian Mirtrons
At least some invertebrate mirtrons have been well conserved during fly or worm evolution. These exhibit characteristic features that reflect their status as microRNA-class genes (Lai et al., 2003), namely that they are short, straight, hairpin introns that exhibit preferential conservation of the 5′ and 3′ terminal segments relative to the central intronic region (Okamura et al., 2007; Ruby et al., 2007a). In other words, the miRNA/miRNA* sequences of mirtron hairpins are much more conserved than their terminal loops. A forward analysis of all Drosophila introns that exhibit these properties across eight or more sequenced Drosophilids revealed only those mirtrons that were cloned previously (W.-J.C. and E.C.L., unpublished data), suggesting that there is a fairly limited repertoire of well-conserved mirtrons in flies (Okamura et al., 2007; Ruby et al., 2007a).
We asked whether these simple features might yield candidate evidence for mammalian mirtrons. In brief, we extracted 25,935 RefSeq/Ensembl introns 50–200 nt in length from the UCSC Genome Browser (Kuhn et al., 2007) and identified conserved mammalian introns that exhibit a “saddle-shaped” conservation profile, then used RNAfold (Hofacker, 2003) and RNAshapes (Steffen et al., 2006) to identify those introns with straight hairpin structures in both primate and nonprimate orthologs (see Experimental Procedures). This yielded 13 candidates for well-conserved mammalian mirtrons (see Figures S1 and S2 in the Supplemental Data available with this article online), of which some appeared less compelling than others, due to hairpin conservation in relatively few species and/or relatively high free energy.
We then asked whether the cloned products of any of these mirtron hairpin candidates were present in collections of mammalian small RNAs (Berezikov et al., 2006a, 2006b). Indeed, multiple reads corresponding precisely to both the 5′ and 3′ ends of host introns (i.e., miRNA/miRNA*) were found in human, chimpanzee, rat, and/or mouse small RNA data sets for three loci (mir-877, mir-1224, and mir-1225, Figures 1A and 2 and Figures S1 and S4). As with invertebrate mirtrons, mammalian mirtrons generally lacked the pairing between their flanking exons needed for recognition by the Drosha/DGCR8 complex (Figure 1 and Figure S1); where pairing was found, it was typically not conserved and followed codon wobble rules.
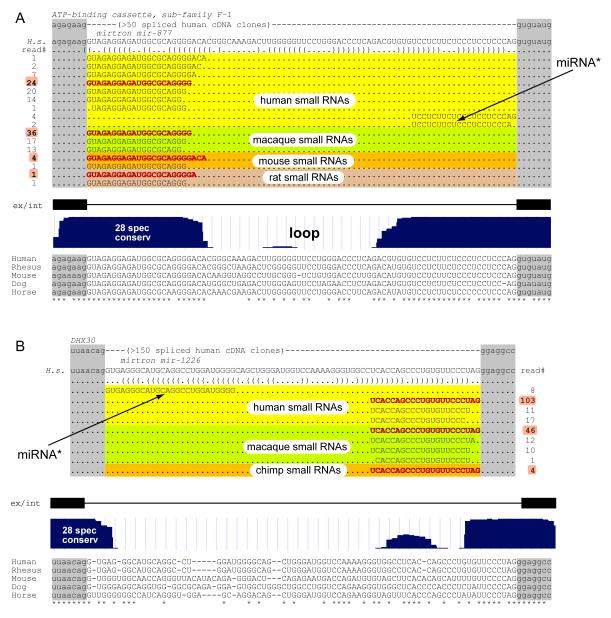
Figure 1. Examples of Mammalian Mirtrons.
(A) A well-conserved mammalian mirtron. (Top) The 13th intron of the ATP-binding cassette F-1 gene harbors the mirtron mir-877. This intron is bounded by consensus splice donor and acceptor sequences, and efficient processing of this intron was evidenced by the existence of over 50 spliced cDNA clones in EST databases. The hairpin structure of this mirtron is indicated with bracket notation. Human small RNAs corresponding precisely to the 5′ and 3′ ends of the intron were identified, as were 5′ small RNAs from macaque, mouse, and rat. Cloning frequencies define the left arm product of mir-877 as its “miRNA” and the right arm product as the “miRNA*.” (Bottom) Evolutionary characteristics of this mirtron. Sequence alignment and conservation track were obtained from http://genome.ucsc.edu. mir-877 is highly conserved among diverse eutherian species but exhibits accelerated divergence within the loop region.
(B) A primate-specific mirtron. (Top) The 21st intron of the putative helicase DHX30 gene harbors the mirtron mir-1226. Notation and layout are as described in (A). In this case, cloning frequencies define its right arm product as the miRNA and its left arm product as the miRNA*. (Bottom) This mirtron is identifiable only in primates; the conservation of its 3′-most terminal sequence in other mammals likely reflects the pressure to maintain splice recognition determinants.
Figure 2. Nineteen Confidently Annotated Mammalian Mirtron Loci.
These are divided into three categories: mirtrons that are conserved among diverse mammals and cloned from two or more species (three genes), mirtrons that are conserved among diverse primates and cloned from two or more species (two genes), and mirtrons that were cloned from independent libraries from a single species or three or more times from any single library (14 genes). The mirtron hairpin structures are designated with bracket notation, and exon-intron structure with “ >” and “+” notation. The cloned species are capitalized and highlighted green. Supplementary figures provide more detailed information on the cloned species (Figure S3), their tissue subtype of origin (Figure S4), and possible orthologs in other primates (Figure S5). In addition, information on 46 additional mirtron candidates is found in Figure S6.
The mirtrons mir-877, mir-1224, and mir-1225 were clearly maintained as hairpins in mammals as diverse as rodents, dog, and horse, indicating their persistence over at least ∼80 million years of eutherian evolution (Figures S1 and S2). We note that small RNAs from the mir-877 locus were recently cloned independently by Tuschl and colleagues, who annotated it as a canonical miRNA gene (Landgraf et al., 2007). Its reclassification as a mirtron is akin to that of nematode mir-62, which was only recently recognized as a mirtron gene (Ruby et al., 2007a). We also note that two of the most abundantly cloned mirtron products were derived from mir-877 and mir-1224 (Figure S4), which were also two of the most perfectly conserved predicted mirtrons. This parallels the finding that the most highly expressed invertebrate mirtrons are also the most highly conserved ones (Okamura et al., 2007; Ruby et al., 2007a), as is also generally the case for canonical animal miRNAs (Berezikov et al., 2006b; Ruby et al., 2007b).
A Plethora of Primate-Specific Mirtrons
Although some are well conserved, most invertebrate mirtrons arose quite recently during Drosophilid and nematode radiation (Okamura et al., 2007; Ruby et al., 2007a); thus, the consideration of evolutionary conservation does not aid their computational identification. However, newly evolved miRNAs have emerged through high-throughput small RNA sequencing efforts. In D. melanogaster, adult heads expressed a high diversity of mirtrons and canonical miRNAs (Ruby et al., 2007b). This is consistent with the fact that brains harbor an exceptional diversity of neurons, a cell type that intrinsically has exceptional needs for translational regulation. We therefore mined a data set of 30 additional small RNA libraries from 15 matched anatomical regions of human and rhesus macaque brains (Figure S3), represented by 18,000–45,000 sequences each (E.B. and E.C., unpublished data).
In addition to revealing cloned evidence for mirtrons mir-877, mir-1224, and mir-1225 in macaque, analysis of these small RNA data sets yielded another 16 mirtrons expressed in primate brains with evidence justifying official nomenclature (Figure 2 and Figure S4). We considered minimum evidence to be the recovery of clones from independent libraries, or at least three clones from any individual library. In several cases, higher levels of evidence were attained, including their cloning from multiple species (i.e., mir-1226 and mir-1227 both from human and macaque), the isolation of many clones (i.e., mir-1229, 16 clones from 12 different libraries), and/or the isolation of both miRNA and miRNA* species (i.e., mir-1227 and mir-1228). These mirtrons appeared to be phylogenetically restricted to primates, with some presenting conserved hairpin structures in human/rhesus/chimp, and others that were restricted to a primate subset. We have summarized the sequences and secondary structures of the orthologous primate mirtronic introns in Figure S5.
Finally, we classified 46 additional hairpin introns from human (23 loci), macaque (16 loci), chimpanzee (3 loci), or mouse (4 loci) as mirtron candidates (Figure S6). The greater number of human and macaque candidates was due in part to the deeper sampling of human and macaque brains. A few of these candidates were cloned three or more times, but we considered their candidacy tentative because of an atypical intronic extension of 8–10 nt on one side of the hairpin (i.e., macaque_block210826 [3 reads/2 libs], and human_block172399 [3 reads/1 lib]). In Drosophila, at least one conserved mirtron-like locus (mir-1017) exhibits a long intronic extension on one side of the hairpin (Ruby et al., 2007a), suggesting that such “half-mirtron” loci might have one side defined by splicing and the other by exonucleolytic digestion. Of the remaining candidates, five (human_block107544, chimp_block23965, macaque_block550558, macaque_block137121, and mouse_block283) were sequenced twice while the rest were defined by single reads. Many of these candidate mirtrons exhibit compelling extended hairpin structures; thus, we anticipate that at least some of them (along with some of the uncloned, conserved, computational candidates) will eventually be validated by additional sequencing.
Most Short RNAs from Mammalian Intron Termini Derive from Mirtrons
The fact that at least three cloned mirtron loci have been highly conserved during mammalian evolution is evidence that vertebrate mirtrons can have regulatory functions that are subject to stringent constraint. Still, as mammalian mirtrons were not reported from previous sequencing efforts, we questioned whether some of these sequences might trivially represent intron degradation products, as opposed to bona fide regulatory RNAs. Certainly, this could apply especially to some members of our tentative “candidate” set. However, several lines of evidence argue against this being a major explanation.
First, our libraries were constructed to select for 5′ phosphates and therefore against degradation products. Second, the size bias for 21–24 nt RNAs and multiple instances of cloned miRNA/miRNA* pairs were indicative of Dicer cleavage. Third, we observed that the number of mirtron clones recovered was not strictly proportional to the number of host ESTs found (Figure S7). Abundant mirtrons such as mir-877 and mir-1226 had many host ESTs, as might be expected if intronic small RNAs are coexpressed with their hosts (Baskerville and Bartel, 2005). In contrast, mir-1225, which has been highly conserved over mammalian evolution and was cloned cross-species, had relatively few clones compared to EST clones (i.e., underrepresented). Conversely, mir-1224, again a very highly conserved locus and cloned cross-species, had a similar number of reads as mir-877 but many fewer host ESTs (i.e., overrepresented). The lack of a strict correlation supports that mirtronic RNAs are not recovered simply as a degradation byproduct of the splicing of abundant mRNAs. Instead, it is consistent with the notion that the half-life of mirtronic small RNAs is influenced by their association with effector complexes, and thus may differ from the half-life of their host mRNAs.
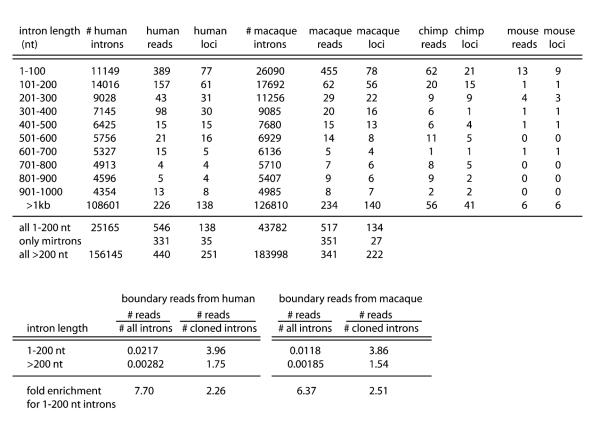
We probed this further by comparing the number of annotated human and macaque introns across 100 nt length increments with the number of human or macaque reads corresponding to the 5′ or 3′ termini of introns (“boundary reads”). We found that short introns (1–100 nt, and to a lesser extent 101–200 nt), were highly enriched for boundary reads (Figure 3). In particular, 138 short human introns 1–200 nt in length generated 55% of all boundary reads, while the remaining reads derived from 251 loci. This represented a 2.26-fold enrichment for cloned fragments to arise from short introns relative to introns of other sizes. However, because short introns comprise only 16% of all introns, this represented a 7.7-fold enrichment in reads per short intron versus all other introns. Analysis of macaque produced a similar picture: short introns generated 60.3% of all boundary reads, yielding a 2.51-fold enrichment when normalized as reads per cloned locus and a 6.37-fold enrichment when normalized for the number of short introns. We also observed that in both human and macaque, ∼60% of all boundary reads from short introns derive from our officially annotated or candidate mirtron loci. Therefore, cloned intron boundary RNAs are quite preferentially associated with short hairpin introns.
Figure 3. Short Hairpin Introns Are the Predominant Source of Cloned Intron-Terminal Small RNAs in Diverse Mammals.
Human and macaque introns were binned into 100 nt intervals. We then binned all small RNA reads derived from intron termini by intron length, excluding introns that also generated nonboundary reads (thus excluding cloned small RNAs arising from unannotated intronic noncoding RNA genes such as tRNAs or snoRNAs). It is evident that a majority of intron-terminal small RNAs in human, macaque, chimp, and mouse derive from 1–200 nt introns, and that most of these derive in turn from hairpin introns that we annotated as mirtrons or mirtron candidates.
Similar trends were evident in chimp and mouse, although the smaller number of mirtronic small RNAs in these species limited our ability to assess enrichment values confidently. Taken together, we can conclude that short introns are significantly biased to generate cloned small RNAs in different mammals, and the majority of these are derived from hairpin precursors. While we do not claim that all the cloned mirtrons have functional endogenous targets—indeed, many of the tentative candidates could be the result of fortuitous processing—the cloning, size distribution, evolutionary properties, and preferred derivation from short hairpins all support the idea that mirtrons are miRNA-pathway-derived regulatory RNAs in mammals.
Differences between Mammalian and Invertebrate Mirtrons
Our studies reveal that primates have more mirtrons than do worms or flies; thus, mirtrons are a substantial source of regulatory RNAs in mammals. However, mammalian mirtrons exhibit several differences from invertebrate mirtrons, which collectively have implications for the genesis of mirtrons.
3′ versus 5′ miRNA
All invertebrate mirtrons with more than two cloned products generate 3′ dominant miRNAs (Ruby et al., 2007a). In contrast, several of the most highly expressed mammalian mirtrons clearly produce 5′ dominant species, with some 3′ miRNA* species representing only a few percent of clones from a given hairpin (i.e., mir-877, Figure 1A and Figure S4). We note that the corresponding 3′ mirtron species of 5′ dominant loci are often extremely pyrimidine rich. For example, miR-877* contains 19 consecutive pyrimidines before its terminal AG splice acceptor. This is consistent with location at 3′ intron ends, which are typically pyrimidine rich, but at odds with the sequence complexity typical of miRNAs. Therefore, at least some 5′ mirtron products are likely functional.
Importantly, we observed that the asymmetry of mammalian mirtron strand selection generally follows the thermodynamic rules proposed for canonical miRNA duplexes (Khvorova et al., 2003; Schwarz et al., 2003), which provides further support that they transit the miRNA biogenesis pathway. These analyses are summarized in Figure S8. A curious exception is mir-1226, which preferentially generates a 5′ miRNA, although its 3′ arm was expected to predominate. It may be that other factors can reverse miRNA strand selection.
5′ nt Identity
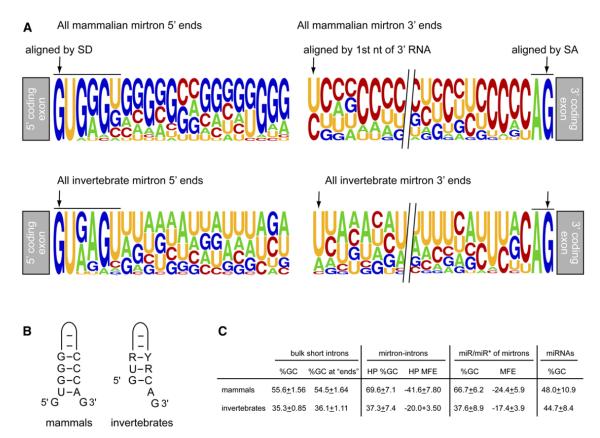
The 3′ products of mammalian mirtrons exhibit equal tendency to begin with either pyrimidine, which contrasts with the strong 5′ uridine bias of invertebrate mirtrons (Figure 4A). Approximately equal numbers of mammalian 3′ mirtron products start with U versus C, regardless of whether the 5′ or 3′ product was dominant (Figure 4 and Figure S4). Curiously, none of the 3′ mirtron species (cloned from 17 different loci) begin with an A or G, indicating a strong bias against 3′ mirtron products to begin with a purine, even in cases where the 3′ arm is not the dominant species (Figure 2 and Figure S4). However, animal mirtrons are united in that no cloned 3′ mirtron product from flies, worms, or mammals thus far begins with a G. Animal miRNAs are generally, but not exclusively (Figure S9), biased against 5′ G residues. The fact that 5′ mirtron products begin with a G makes their selection as miRNAs in mammals noteworthy.
Figure 4. Sequence and Structural Features of Mammalian and Invertebrate Mirtrons.
(A) Sequence logos of 5′ and 3′ mirtron products. Data represent 19 primate/mammalian mirtrons (this study) and 18 invertebrate (14 fly and 4 worm) mirtrons (Ruby et al., 2007a). (Top row) Mammalian mirtrons generate G-rich 5′ mirtron products and C-rich 3′ mirtron products. Alignment of the 3′ mirtron products by their first nucleotides shows an equal frequency of U and C residues. (Bottom row) Invertebrate mirtrons do not show such overall G:C bias, and their 3′ products are strongly biased toward 5′ U residues.
(B) Typical hairpin-end structures of mammalian and invertebrate mirtrons. These preferred end structures are also evident from the sequence logos presented in (A).
(C) Comparison of the nucleotide composition of mammalian and invertebrate mirtrons with bulk short introns in humans and flies. We analyzed the GC content of 13,453 human introns and 29,120 D. melanogaster introns, each 50–120 nt in length. We also analyzed their 5′-most and 3′-most 24 nt (intron “ends”) as a proxy for miRNA/miRNA* regions. GC content and minimum free energy (mfe, kcal/mol) of straight hairpin structures for the cloned mammalian and fly mirtrons were also assessed; where only one mirtron product was obtained, the miRNA* region was inferred by assuming a 2 nt 3′ overhang. Values are shown ±SD. For comparison, we show the GC content of all human and worm/fly (invertebrate) canonical miRNAs listed in miR-base Release 10.
Hairpin End Structure
None of the most highly cloned mammalian mirtrons exhibit a stem structure with a precise AG 3′ overhang to the hairpin, as is typical for highly expressed Drosophila and nematode mirtrons. In fact, of the 19 confidently annotated mammalian mirtrons, only three had precise AG overhangs adjacent to a terminal duplex. Instead, the most frequent configuration was for single nucleotide overhangs at both ends (seven loci, Figure S4) in which the U of the GU splice donor pairs with the A of the AG splice acceptor (Figure 4B). The distinct, preferred end configurations of mammalian and invertebrate mirtrons were evident from their sequence logos (Figure 4A). The unusual configuration of (3 nt-5′) + (2 nt-3′) hairpin overhangs also seemed to be compatible with efficient processing of mammalian mirtrons (i.e., mirtron mir-1226, Figure 1B). Nevertheless, the end of the miR-1226/miR-1226* duplex on the terminal loop side exhibits a 2 nt 3′ overhang, as expected for Dicer cleavage of this otherwise atypical hairpin.
These observations appear to extend the potential range of endogenous Dicer substrates, previously comprised mostly of Drosha products (pre-miRNA hairpins), Drosha mimics (mirtrons), or other Dicer products—all of which exhibit signature 2 nt 3′ overhangs. Still, our presumption that mammalian mirtrons require the canonical pre-miRNA export machinery, as shown for Drosophila mirtrons (Okamura et al., 2007), led us to investigate the structural constraint on pre-miRNA hairpin ends. We analyzed all miRbase miRNAs with annotated miRNA* species and calculated their hairpin end structures. With the caveat that the ends of some miRNA* species might be incorrectly annotated, this study showed that a number of deduced pre-miRNA hairpins are not predicted to have perfect 2 nt 3′ overhangs (Figure S10). Therefore, Exportin-5 may accept a broader range of small RNA hairpins than is often considered. Indeed, gel-shift analyses support the ability of Exportin-5 to bind to certain hairpins with noncanonical ends (Zeng and Cullen, 2004). Alternatively, other factors might participate in the export of both canonical pre-miRNAs and mirtrons.
GC Content
Mammalian mirtrons exhibited much higher GC content, and thus much lower free energy, than either invertebrate mirtrons or bulk human short introns (Figure 4C). Comparison of the 18 invertebrate mirtrons with the 29,120 D. melanogaster introns that are 50–120 nt in length showed that they had similar GC characteristics as bulk D. melanogaster short introns. In contrast, comparison of the 19 cloned primate mirtrons with all 13,453 human introns 50–120 nt in length showed that mammalian mirtrons are significantly enriched for high GC content compared to bulk human short introns (Figure 4C). These findings remained true when the miRNA/miRNA* portions of mirtrons were compared with matched lengths of 5′ and 3′ termini of short introns. In addition, the GC content of mammalian mirtrons was also much higher than that of canonical human miRNAs or invertebrate miRNAs (Figure 4C). It is conceivable that these characteristics might compensate in some way for the fact that mammalian mirtrons are frequently suboptimal mimics of Drosha products, in terms of hairpin end structure.
On the Evolutionary Emergence of Mirtrons and the Effect of Mirtrons on Evolution
The many differences between plant and animal miRNAs have been taken to indicate convergent evolution of miRNA pathways among divergent eukaryotes that share an ancestral RNA interference pathway. Similarly, the many distinctions between mammalian and invertebrate mirtrons might reflect independent acquisition of mirtron pathways in different animal clades. Consistent with this, while several mirtrons are highly conserved among Drosophilids (Okamura et al., 2007; Ruby et al., 2007a), nematodes (Ruby et al., 2007a), and mammals (this work), these animals do not collectively share any mirtrons that are clearly related by ancestry. This does not exclude a model in which mirtrons facilitated the evolution of a canonical animal miRNA pathway, prior to the evolution of a Drosha-type activity (Ruby et al., 2007a). However, in this scenario, it is necessary to posit that none of these ancient mirtrons evolved substantial functions and were all lost through evolution, or that all of them accumulated so many sequence changes that their ancestry is no longer apparent from sequence alignment. These scenarios are not easily reconciled with the fact that highly conserved mirtrons have subsequently emerged in three different animal lineages, nor with the fact that many canonical miRNAs have been retained completely unchanged from the bilaterian ancestor of invertebrates and vertebrates (Prochnik et al., 2007).
Our findings also do not clearly support a model in which mirtrons arise in genomes strictly proportionally to the fraction of short introns whose size is comparable to pre-miRNA hairpins (Ruby et al., 2007a). The extant evidence demonstrates that primate brains express a greater number of mirtrons than do flies and worms put together, despite the fact that these invertebrates have more short introns (Lim and Burge, 2001; Yandell et al., 2006). In addition, because mammalian mirtrons have very high GC content relative to bulk mammalian short introns, they evidently do not comprise a random sampling of mammalian short introns (Figure 4C). Indeed, the differences in sequence composition and structure between mammalian mirtron and pre-miRNA hairpins (Figure 4C) further suggest that they are not simply pre-miRNA mimics, as appears to be the case for their invertebrate counterparts.
Overall, the observation of cloned products from many newly evolved mirtrons in diverse animal species suggests that the mirtron might represent an evolutionarily opportunistic and facile strategy for the birth of regulatory RNAs in animal species with a preexisting canonical miRNA pathway. This is conceptually similar to the notion that animals and plants may have evolved miRNA genes independently, building their respective pathways via an ancestral RNA interference pathway. The fact that a majority of D. melanogaster mirtrons arose quite recently during Drosophilid evolution, combined with the observation that miRNAs have relatively minimal requirements for target identification, suggested that mirtrons could have a palpable effect on insect speciation. Our parallel observation that primates, and specifically primate brains, express a strong diversity of processed mirtrons similarly suggests that they might also contribute to primate evolution and/or primate-specific behavior.
EXPERIMENTAL PROCEDURES
Computational Screen for Conserved Mammalian Mirtrons
From the UCSC Genome Browser (Kuhn et al., 2007), we extracted 21,883 RefSeq human introns 50–200 nt in length, and supplemented these with a nonredundant set of 4052 Ensembl-exclusive human introns 50–200 nt in length (many of which might be misannotated coding exons). We then identified introns for which at least 17 nt in the 5′-most 25 nt and 3′-most 25 nt exhibited phastCons score of >0.7 across 17 mammalian species. This yielded 220 and 223 conserved introns from Refseq and Ensembl-only intron data set, respectively. Of these, 89 RefSeq and 34 Ensembl introns exhibited a saddle shape conservation profile, in which a minimum of five continuous nucleotides exhibited phastCons score < 0.1 within the central region of the intron. Operationally, we required that the diverged region either overlapped the midpoint of the intron, or its closest boundary was no more than 5 nt away from the midpoint. In addition to selecting for candidates with microRNA-like evolutionary properties, saddle selection proved useful for removing misannotated coding regions from consideration.
The mammalian orthologs of these selected introns were then folded using RNAfold (Hofacker, 2003) and RNAshapes (Steffen et al., 2006). We used these algorithms because at least one Drosophila mirtron (mir-1015) is not predicted to adopt a straight hairpin in any alternative mfold structure, but is using either RNAshapes or RNAfold. The ability of RNAshapes to report a diversity of suboptimal minimum free energy structures proved useful to cull single arm, straight hairpin folds. We defined a potential mirtron candidate to be a straight arm hairpin in which at least 16 out of the 5′ terminal 30 nt and 17 out of the 3′ terminal nt were base paired to each other (these numbers were not the same because of the nonsymmetrical nature of many hairpins). Candidates with an overhang of >8 nt at either end were also excluded. Finally, we defined a conserved mammalian mirtron candidate as a locus for which orthologs of at least some primate and nonprimate introns satisfied the minimum hairpin criteria. Note that we did not set a lower limit on the minimum free energy of conserved hairpin candidates. This computational pipeline yielded 13 conserved mammalian mirtron candidates (Figure S1). Loci for which a greater number of orthologous candidates passed minimum criteria were deemed more compelling; therefore, we rank ordered the candidates by the number of species orthologs identified.
In some cases, including mir-877, mir-1224, and mir-1225, we observed clear conservation of sequence and structure among most mammals. Terminal small RNAs from these three loci were each cloned multiple times in multiple species, and thus qualified as bona fide mirtrons. Most, but not all, of the remaining candidates passed minimum criteria in the three primate species surveyed (in addition to some number of nonprimate species). Detailed information on the sequences, secondary structures, and evolutionary profiles of the computational candidates are reported in Figure S1.
Although all of these candidates met minimum criteria, some were clearly less compelling. Because our strategy considered the pattern of nucleotide divergence and conservation of structural features, but not minimum free energy, some candidates had free energies that were atypically high by standards of the cloned mammalian mirtrons (i.e., NM_025160_1, NM_173474, NM_015232_11, and NM_002912_7). In other cases, the species that shared an apparently conserved, orthologous, hairpin intron were not necessary the most closely related species. For example, NM_002912_7 and NM_152345_9 had possible nonprimate candidate orthologs but did not pass minimum criteria in human. While some of these candidates may not be bona fide, we expect several of them to eventually be validated by additional sequencing.
Small RNA Library Construction and Sequencing
Rhesus macaque tissues from 15 different brain regions (Figure S3) were provided by Biomedical Primate Research Center (Rijswijk, The Netherlands). Human tissue from corresponding brain regions was obtained from the Netherlands Brain Bank (single female donor). Small RNA libraries were made by Vertis Biotechnology AG (Freising-Weihenstephan, Germany) as described (Berezikov et al., 2006b) and sequenced using the Genome Sequencer 20 system (454 Life Sciences, Branford, USA). Chimpanzee, mouse, and human small RNA libraries besides the 15 brain regions, as well as chimpanzee and mouse brain libraries, were described previously (Berezikov et al., 2006a, 2006b) and reanalyzed in this study.
Small RNA Data Analysis
Initial processing of sequencing data was performed as previously described (Berezikov et al., 2006a) with some modifications. After trimming of adaptor sequences, reads were mapped to genomes (NCBI 36, NCBI m36, MMUL 1.0, and PanTro 2.1 assemblies for human, mouse, macaque, and chimpanzee, respectively) using megablast software (ftp://ftp.ncbi.nlm.nih.gov/blast/). Reads that did not match perfectly to genomes were analyzed for the presence of extra A bases in 3′ ends of the reads, since pyrosequencing through poly(A) tails on the 454 system often results in calling of additional A bases in adjacent wells. In most cases, removal of these nonmatching As resulted in perfect matching of reads to genomes. In cases where this adjustment did not result in perfect match but at least 20 first bases of the read matched perfectly, nonmatching 3′ parts were trimmed and longest matches were considered as actual genome matches. The most frequently trimmed sequence was a single T base, which is consistent with previous observations on nontemplated modification of miRNAs (Landgraf et al., 2007). Genomic context of the mapped reads was annotated using Ensemble API and databases (http://www.ensembl.org, v.45), and reads that mapped within five bases from exon:intron boundaries of introns shorter than 500 bp were selected for further manual inspection. RNA folding predictions were performed using RNAfold (Hofacker, 2003) and RNAshapes (Steffen et al., 2006) software.
Supplementary Material
ACKNOWLEDGMENTS
We thank I. Kondova (BPRC, Rijswijk) for providing macaque material, and the Netherlands Brain Bank (NBB Amsterdam, head Dr. R. Ravid) for providing human samples. E.B. was supported by Horizon and VIDI grants (NWO). E.C.L. was supported by the Leukemia and Lymphoma Society, the Burroughs Wellcome Foundation, the V Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research, and the National Institutes of Health (GM083300).
Footnotes
Supplemental Data Supplemental Data include ten figures and can be found with this article online at http://www.molecule.org/cgi/content/full/28/2/328/DC1/.
REFERENCES
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006a;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006b;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. microRNAs: runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev. Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston W, Kellis M, Bartel DP, Lai EC. Biogenesis, expression and target predictions for an expanded set of microRNA genes in Drosophila. Genome Res. 2007b doi: 10.1101/gr.6597907. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- Yandell M, Mungall CJ, Smith C, Prochnik S, Kaminker J, Hartzell G, Lewis S, Rubin GM. Large-scale trends in the evolution of gene structures within 11 animal genomes. PLoS Comput. Biol. 2006;2:e15. doi: 10.1371/journal.pcbi.0020015. 10.1371/journal.pcbi.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.