Abstract
A recently developed high-throughput technique that allows multi-locus microsatellite analysis of individual miracidia of Schistosoma mansoni was used to assess the levels of genetic diversity and population structure in 12 infrapopulations of the parasite, each infrapopulation derived from an infected school child from the Mwea area, central Kenya. The mean number of alleles per locus was in the range 8.22 – 10.22, expected heterozygosity in Hardy-Weinberg equilibrium was 0.68 – 0.70, and pair wise FST values ranged from 0.16 −3.98% for the 12 infrapopulations. Although the genetic diversity within each infrapopulation of S. mansoni in this area was generally high, low levels of genetic structure were observed, suggestive of high levels of gene flow among infrapopulations. Private alleles were found in 8 of the 12 infrapopulation, the highest number of private alleles recorded per infrapopulation was 3. Our data suggest that the level of gene flow among infrapopulations of S. mansoni in Mwea is extremely high thus, providing opportunity for spread of rare alleles, including those that may confer character traits such as drug resistance and virulence.
Keywords: Schistosoma mansoni, genetic variation, population genetics, schistosomiasis, Kenya, microsatellite repeats
1. Introduction
Schistosoma mansoni causes human intestinal schistosomiasis in Africa, west Asia, parts of South America, and the Caribbean region (Savioli et al., 1997; Morgan et al., 2005). Human intestinal schistosomiasis is a debilitating disease characterized by a variety of symptoms including abdominal pain, diarrhoea and in severe cases hepatosplenomegaly and oesophageal varices (van der Werf et al., 2003). Worldwide, over 200 million people, mostly children, harbour schistosome parasites (Bundy, 1997; Chitsulo et al., 2000; Steinmann et al., 2006).
Schistosomes undergo sexual reproduction in the definitive hosts (humans, other primates, rodents) and this type of reproduction allows for reassortment and the perpetuation of parasite genotypic diversity. The eggs laid by the adult female worm pass in the host’s faeces and each will hatch in water to release a miracidium. These free-swimming larvae must find and penetrate an appropriate freshwater snail, in the case of S. mansoni, a snail of the genus Biomphalaria. Once it has penetrated the snail, the miracidium transforms into a mother sporocyst which produces multiple daughter sporocysts through asexual reproduction. These in turn produce cercariae which are released into water and are infective to human hosts, thus completing the life cycle.
Although S. mansoni is considered a uniform species morphologically, differences between parasite strains or populations have been observed in a number of biological characteristics such as infectivity (Thiong’o et al., 1997; Sire et al., 1999), virulence (Imbert-Establet and Combes, 1986), response to treatment (Dias et al., 1982; Gryseels et al., 2006) or fecundity (Imbert- Establet and Combes, 1986). By investigating mitochondrial sequence diversity, Morgan et al. (2005) provided a global overview of the genetic diversity inherent in S. mansoni, identifying five distinct lineages mostly separated by geographic distance, and that revealed considerable genetic variation particularly among East African specimens. Understanding the genetic structure of S. mansoni will lead to a better understanding of the variation in natural populations and the transmission dynamics of schistosomes between hosts and across geography. However, the intravascular location of adult schistosomes makes them routinely unavailable for study thus hindering population genetic studies of S. mansoni in the human definitive host. Most of the population genetic studies of S. mansoni undertaken so far have relied upon parasite materials obtained by indirect sampling procedures that involve passage of the parasite through laboratory animals (Minchella et al., 1995; Barral et al., 1996; Sire et al., 1999; Prugnolle et al., 2002; Morgan et al., 2003; Wang et al., 2006; Agola et al., 2006). Such procedures present ethical and practical problems, and may also result in sampling biases such as bottlenecking, and/or host-induced selection pressure (Gower et al., 2007).
Multi-locus microsatellite analysis of individual schistosome miracidia obtained from infected human hosts has recently been used successfully to characterize S. mansoni populations (Gower et al., 2007; Steinauer et al., 2008; Beltran et al., 2008), circumventing the problems associated with indirect sampling procedures and also providing a new overall approach to study the biology of S. mansoni in the human host.
S. mansoni is endemic in Kenya (Ministry of Health, Kenya, unpublished data), and recent studies of the parasites using adult worms derived from passages through laboratory animals have shown significant genetic diversity in the Kenyan S. mansoni populations, and that there is evidence of restricted gene flow among geographic locations (Morgan et al., 2003, 2005; Agola et al., 2006). Using the sampling procedure for schistosome miracidia recently described by Steinauer et al. (2008), genetic diversity and population structure of S. mansoni was examined using 9 microsatellite loci in infected school children living in the Mwea area, central Kenya, where intense rice farming is undertaken under irrigation, and S. mansoni prevalence is 47% (Kihara et al., 2007). Children are of particular relevance to sample in this respect because they play a major role in transmission of schistosomiasis, and in general, they are more at risk to morbidity and mortality than adults (Waiyaki, 1987; Gillespie, 2001).
2. Materials and Methods
2.1. Study population
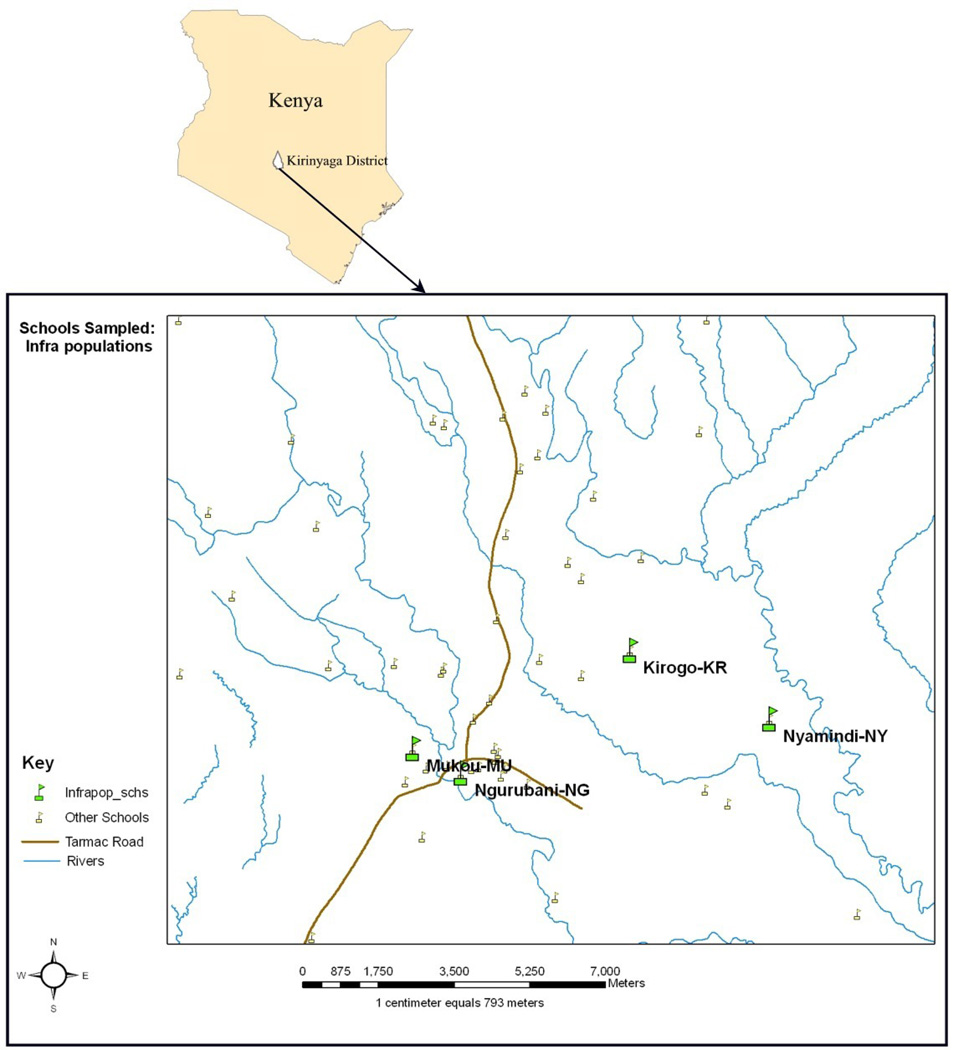
The study population comprised school children aged 6–15 years old living in the rice farming irrigation scheme of Mwea in central Kenya, located approximately 100km north-west of Nairobi (Fig 1). A model schistosomiasis control project based on use of praziquantel has been established in Mwea as part of the school health program in the area. Only children diagnosed to have S. mansoni infection based on a Kato-Katz procedure (Katz et al., 1972), and whose parents had given their consent for participation in the study were sampled. A total of twelve children from four schools were recruited into the study. School was chosen as a sampling unit based on the fact that children presumably get infected near school grounds and also because schools reflect the distinct geographical area from which children originate.
Figure 1.
Map showing the sampling site of S. mansoni in Mwea, Kirinyaga district Central Kenya
2.2. Fecal sample collection, isolation of S. mansoni eggs and miracidia collection
A fecal sample of approximately 150 gm was collected in a plastic container from each participating child. To isolate S. mansoni eggs, the fecal sample was placed in a screw capped tube and normal saline added to completely cover the sample. The tube was shaken thoroughly to prepare a uniform suspension. The suspension was then filtered through a series of nested sieves of sizes 710, 425, 212, 45 µm. The eggs were then washed off the last sieve using distilled water into a 1-litre conical flask and distilled water was added to fill up the flask. Parasite egg hatching was induced by exposure to bright light. To collect the hatched miracidia, the flask was covered with a black cloth for 30 min to allow the miracidia (which are phototropic) to swim to the top of the flask. Individual miracidia were then collected directly into each wells of a 96 well plate.
2.3. DNA extraction
DNA was extracted from schistosomes (miracidia) using a modified method described by Truett et al. (2000). In the modified procedure, 96 well PCR plates were used for DNA extraction instead of PCR tubes. Briefly, 5µl of a HotSHOT lysis reagent (25mM NaOH, 0.2mM disodium ethylenediaminetetraacetic acid (EDTA), pH 12) was added into each well of a 96 well PCR plate, sealed with pierceable aluminum tape, and chilled during specimen collection. An individual S. mansoni miracidium was transferred in a 3 µl volume of water using a 10 µl micropipette into a PCR plate well containing the lysis reagent by piercing the foil with the pipette tip. Pipetting through the foil eliminated the possibility of adding miracidia into the same well twice. Once the miracidia were in the wells, the plates were centrifuged for 20 seconds at 16000 × g, sealed with a silicone sealing mat, and heated in a PCR machine with a heated lid at 95°C for 30 minutes. The plates were periodically removed from the PCR machine and centrifuged to ensure that the liquid containing the miracidia remained at the bottom of the wells. After heating, the plates were cooled, centrifuged, and 5µl of HotSHOT neutralizing reagent (40mM Tris–HCl at pH 5) added into each well. The plates were then sealed and stored below −20°C until DNA amplification was performed. Using this procedure many samples could be processed very rapidly.
2.4. Microsatellite markers
A total of 9 previously published microsatellite loci were investigated: AF325695 (Curtis et al., 2001), AF202965, AF202966, AF202968, L46951, M85305, R95529 (Durand et al., 2000), AI395184 and BF936409 (Silva et al., 2006).
2.5. PCR amplification and genotyping
DNA amplification was carried out in a multiplex PCR format using two panels, P17 and P22, as described by Steinauer et al. (2008). Panel P17 used primers for AF325695, AF202965, AF202966, AF202968 and L46951 while P22 used primers for AI395184, BF936409, M85305 and R95529. The QIAGEN Multiplex PCR Kit (Qiagen) was used for PCR amplifications in 5.8 µl reactions according to the manufacturer’s directions. The thermal cycling profile included an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of 45 s at 94 °C, annealing temperature specific for each multiplex panel (52 °C for P 17, 48 °C for P 22), 30 s at 72 °C and a final step of 7 min at 72 °C using an Eppendorf Mastercycler Systems thermocycler. Primers were labeled with 6-FAM, HEX and NED dyes (Applied Biosystems). Multiplexed PCR products were diluted in N, N’-dimethyl formamide with Genescan® – 500 [ROX 500] as an internal size standard and genotyped using an ABI3100 automated sequencer (Applied Biosystems) and scored with GeneMapper® v. 4.0 (Applied Biosystems) software. All genotype calls were verified manually.
2.6. Data analysis
The mean number of alleles per locus (MNA) detected in each infrapopulation and the expected heterozygosities were used as indicators of the genetic polymorphism within the populations under study. Differences both for the mean number of allele and the expected heterozygosity values were tested for samples of each population using Freiedman non parametric tests. The MNA and allele frequencies were calculated using the program MICROSATELLITE TOOLKIT (http:// animalgenomics.ucd.ie/sdepark/ms-toolkit/). Both the expected and observed heterozygosities were also calculated using MICROSATELLITE TOOLKIT and their statistical significance tested using the chi-square test and an α̍ of 0.05. GENEPOP 3.3 (Raymond and Rousset, 1997) was used to test for deviations from Hardy-Weinberg equilibrium (HW) using exact tests, testing the hypothesis that observed diploid genotypes are the product of random union of gametes. An exact test for linkage disequilibrium between pairs of loci was performed using the Markov chain method in GENEPOP web version 3.4 (Raymond and Rousset, 1997). A sequential Bonferroni correction was also performed to control type 1 error. MICRO-CHECKER software (Van Oosterhout et al., 2004) was used to identify loci that potentially are affected by presence of null alleles, long allele dropout, or scoring errors due to stuttering. Frequency of null alleles was estimated from the heterozygosity deficiency as described by Brookfield (1996) using the formula (HE - HO) / (1 + HE.). Mean estimates of FIS for each population and pairwise Fst between all population pairs were also calculated following the method of Weir and Cockerham (1984). Deviation of FIsand Fst values from zero was tested using a permutation test. All F statistics were carried out using FSTAT 2.9.3.2 (Goudet, 2000). The recoding method of Meirmans (2006) was used to standardize all measures of Fst and only standardized measures of Fst are reported. For PCA (Principle Component Analysis), genetic distance was calculated from the allele data and the genetic distance was plotted as PCA using GenAlEx (Peakall and Smouse, 2006). Phylogenetic trees were constructed using the neighbor-joining (NJ) method (Saitou and Nei, 1987) from matrices of Nei’s genetic, DA, distances (Nei et al., 1983). Bootstrap tests (Felsenstein, 1985) were performed with 1000 replicate resampling. These analyses were done using MEGA (Kumar et al., 2004). Correlation between genetic distance and geographic distance was implemented using GENEPOP 3.3 and the significance of the correlation coefficient was tested using the Mantel test (Mantel, 1967). In order to determine the usefulness of the different markers to distinguish between the twelve infrapopulations the statistical significance of the FST for each of the 9 loci and for each infrapopulation pair was calculated. An unbiased estimate of the P-value of a log-likelihood (G-based) exact test was performed using GENEPOP. Individual population assignment was also undertaken using GENECLASS 1.0.02 (Cornuet et al., 1999).
3. Results
3.1. Genetic variability
Schistosomes were defined at two hierarchical levels: 1) all miracidia from an individual patient were designated as an “infrapopulation”, 2) miracidia from all the patients within a school were treated as a “population” in the analysis. Twelve different infrapopulations were analysed, totaling 904 individual S. mansoni miracidia, each of which was examined at 9 loci. Total number of alleles per locus ranged from 2 for AF202966 to 25 for L46951. The largest mean number of alleles was found in the NYG infrapopulation (10.22), while the lowest mean number of alleles occurred in the NGC and MU infrapopulations (8.22). Private alleles were found in eight of the twelve infrapopulations, and all occurred at frequencies above 5%. In all infrapopulations, the multilocus probability tests showed departure from Hardy-Weinberg equilibrium (P < 0.001). Expected heterozygosity ranged from 68% in NGC to 70% in both the NYA and NYD infrapopulations (Table 1) and there were no significant differences between the expected and observed heterozygosities (x2 = 0.1849, DF = 1, P = 0.6671). Friedman non parametric test for both the mean number of alleles and the expected heterozygosity values were nonsignificant. Following bonferroni correction, all locus pair tests were found not to exhibit linkage disequilibrium.
Table 1.
Summary of genetic variation at nine microsatellite loci in a) twelve infrapopulations of S. mansoni sampled in Mwea. (HE) is the expected heterozygosity,(HO) the observed heterozygosity, (A) is the mean number of alleles, (N) is the sample size and (P) is the number of private alleles FIS is the Weir and Cockerham (1984) estimate for inbreeding; b) similar values for "populations" arranged by school (See text)
| Co-ordinates |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| School sampled | Latitude/Longitude | Code | N | Loci typed | HE | HO | A | P | FIS |
| a) Infra-populations | |||||||||
| Nyamindi Pri. Sch. | 00° 40' S, 037° 24' E | NYA | 80 | 9 | 0.7002 ± 0.0866 | 0.6908 ± 0.0189 | 8.89* | 2 | 0.010 |
| NYB | 84 | 9 | 0.6896 ± 0.0864 | 0.6293 ± 0.0187 | 8.93* | 2 | 0.014 | ||
| NYC | 59 | 9 | 0.6920 ± 0.0889 | 0.6287 ± 0.0217 | 9.07* | 1 | 0.088 | ||
| NYD | 87 | 9 | 0.7034 ± 0.0876 | 0.6896 ± 0.0169 | 9.45* | 0 | 0.037 | ||
| NYE | 73 | 9 | 0.6850 ± 0.0910 | 0.5878 ± 0.0213 | 8.12* | 3 | 0.092 | ||
| NYF | 76 | 9 | 0.6859 ± 0.0809 | 0.6173 ± 0.0194 | 8.55* | 0 | −0.005 | ||
| NYG | 84 | 9 | 0.6826 ± 0.0922 | 0.6308 ± 0.0185 | 9.37* | 1 | 0.020 | ||
| Ngurubani Pri. Sch. | 00° 41' S, 037°21' E | NGA | 89 | 9 | 0.6951 ± 0.0856 | 0.6880 ± 0.0169 | 8.81* | 0 | 0.082 |
| NGB | 69 | 9 | 0.6828 ± 0.0916 | 0.6580 ± 0.0199 | 8.91* | 1 | 0.150 | ||
| NGC | 81 | 9 | 0.6761 ± 0.0869 | 0.6797 ± 0.0178 | 7.91* | 2 | 0.101 | ||
| Mukou Pri. Sch. | 00° 40' S, 037° 20' E | MU | 55 | 9 | 0.6981 ± 0.0859 | 0.6413 ± 0.0235 | 8.19* | 1 | 0.076 |
| Kirogo Pri. Sch. | 00° 39' S, 037° 23' E | KR | 67 | 9 | 0.6909 ± 0.0805 | 0.6389 ± 0.0219 | 8.46* | 0 | 0.076 |
| b) School populations | |||||||||
| Nyamindi Pri. Sch. | NY | 543 | 9 | 0.6965 ± 0.0874 | 0.6467 ± 0.0073 | 8.91* | 9 | 0.010 | |
| Ngurubani Pri. Sch. | NG | 238 | 9 | 0.6981 ± 0.0872 | 0.6759 ± 0.0105 | 9.48* | 3 | 0.082 | |
| Mukou Pri. Sch. | MU | 55 | 9 | 0.6981 ± 0.0859 | 0.6413 ± 0.0235 | 8.46* | 1 | 0.076 | |
| Kirogo Pri. Sch. | KR | 67 | 9 | 0.6909 ± 0.0805 | 0.6389 ± 0.0219 | 8.22* | 0 | 0.076 | |
NY= Nyamindi Pri. Sch.(NYA-NYG are infrapopulations); NG= Ngurubani Pri. Sch (NGA - NGC are infrapopulations);
MU= Mukou Pri. Sch.; KR= Kirogo Pri. Sch.
Mean number of alleles corrected for sample size
A total of four populations (schools) were analysed. To remove any bias, the mean number of alleles per locus was corrected for sample size by random sampling of 55 individual miracidia with 99 replicate replacements. The largest mean number of alleles was found in the NY (Nyamindi school) population (13.67), while the lowest mean number of alleles occurred in the MU (Mukou school) population (8.22). Private alleles were found in all populations except the KR (Kirogo school) population, and most of them occurred in the NY population (Table 1). In all populations, the multilocus probability tests showed departure from Hardy-Weinberg equilibrium (P < 0.001). Expected heterozygosity ranged from 69% in KR to 70% in the NY, NG (Ngurubani school) and MU populations (Table 1) and there were no significant differences between the expected and observed heterozygosities (x2 = 0.085, DF = 1, P = 0.7706).
3.2. Genetic distances and phylogeography
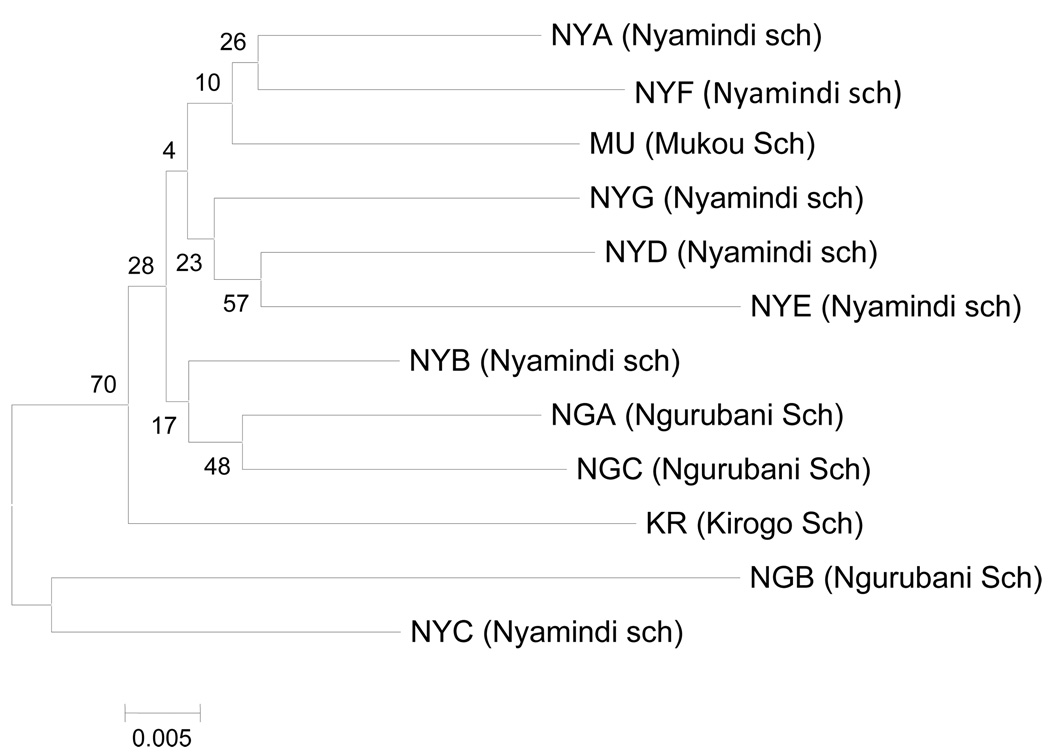
For infrapopulations, genetic distances (Da) were highest (0.1178) between NYD and NGB. The lowest Da was observed between NGC and the MU (0.051). A neighbor-joining tree based on Da genetic distances for the twelve infrapopulations of S. mansoni (Fig 2) showed that the NGB and NYC infrapopulations are separated from the other infrapopulations with a bootstrap support of 70%. For populations, genetic distances (DA) were highest (0.0994) between the MU population (Mukou primary school) and KR (Kirogo primary school). The lowest DA (0.0278) was observed between NG and NY populations.
Figure 2.
Unrooted neighbor-joining phylogenetic tree of 12 S. mansoni infrapopulations based on allele frequencies of nine microsatellite loci. The numbers shown are bootstrap values.
3.3. Population structure
For infrapopulations, all but one (NYF) had positive FIS values (Local fixation index which corresponds to the excess of homozygosity of individuals in a sub-population) that ranged from 0.01 in NYA to 0.15 in NGB (Table 1). Global testing showed that all but one infrapopulation (NYF) had positive FIS values that deviated significantly from Hardy-Weinberg equilibrium. The presence of null alleles was tested and the frequencies ranged from 0.05 to 0.056 which is insignificant (i.e < 0.10) (Brookfield, 1996). Pairwise FST values (measure of allele frequencies among subpopulations) are presented in Table 2 and the highest values were recorded between S. mansoni infrapopulations NYF and NYD (FST = 0.1241) while the lowest were between NYB and NYG (FST = 0.0023). Although FST values were low, pairwise FST between infrapopulations were significantly different (P < 0.001). We also tested the usefulness of different markers in infrapopulation differentiation. All markers were efficient at distinguishing between all the infrapopulations, and the level of differentiation, at all infrapopulation pairs, across the nine loci was significant (P < 0.001). To understand the clustering pattern of S. mansoni infrapopulations, we also carried out genetic distance based Principal Component Analysis (PCA) using GenAlEx. The PCA lacked clear geographical patterns suggesting the absence of strong substructure within the S. mansoni population within Mwea. Assignment of individual to populations using GENECLASS software also indicated the same pattern of random assignment of individuals to population further indicating a lack of genetic structure.
Table 2.
Population pairwise FST value (below diagonal) and the associated probability of genetic heterogeneity (above diagonal) among the Mwea schistosomes
| Populations | NYA | NYB | NYC | NYD | NYE | NYF | NYG | NGA | NGB | NGC | MU | KR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NYA | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| NYB | 0.0111 | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| NYC | 0.0120 | 0.0093 | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| NYD | 0.0377 | 0.0316 | 0.0340 | *** | *** | *** | *** | *** | *** | *** | *** | |
| NYE | 0.0137 | 0.0075 | 0.0095 | 0.0313 | *** | *** | *** | *** | *** | *** | *** | |
| NYF | 0.0107 | 0.0092 | 0.0098 | 0.0398 | 0.0194 | *** | *** | *** | *** | *** | *** | |
| NYG | 0.0213 | 0.0067 | 0.0093 | 0.0373 | 0.0147 | 0.0143 | *** | *** | *** | *** | *** | |
| NGA | 0.0136 | 0.0065 | 0.0062 | 0.0349 | 0.0106 | 0.0122 | 0.0111 | *** | *** | *** | *** | |
| NGB | 0.0131 | 0.0055 | 0.0056 | 0.0336 | 0.0077 | 0.0082 | 0.0065 | 0.0103 | *** | *** | *** | |
| NGC | 0.0170 | 0.0074 | 0.0113 | 0.0392 | 0.0093 | 0.0164 | 0.0170 | 0.0044 | 0.0049 | *** | *** | |
| MU | 0.0142 | 0.0038 | 0.0037 | 0.0343 | 0.0119 | 0.0113 | 0.0075 | 0.0053 | 0.0016 | 0.0075 | *** | |
| KR | 0.0182 | 0.0161 | 0.0154 | 0.0329 | 0.0189 | 0.0206 | 0.0139 | 0.0165 | 0.0102 | 0.0220 | 0.0157 |
FST values between populations (defined by schools) were generally low. The highest pairwise FST values were recorded between S. mansoni from Mukou school (MU) and Ngurubani school (NG) (FST = 0.0156) while the lowest were between Nyamindi school (NY) and Kirogo school (KR)(FST = 0.0061). All population pairs were significantly differentiated (P < 0.001) but the levels of differentiation were generally low (Table 2).
3.4. Isolation by distance
We applied the Mantel test to assess the correlation between genetic differentiation (FST/ 1- FST) and the logarithm of linear geographical distances (Ln distance) for all S. mansoni collection localities (schools). The analysis revealed a non-significant trend between both variables (r = 0.698; P = 0.383).
4. Discussion
S. mansoni infrapopulations (each infrapopulation derived from a single host) from Mwea in central Kenya have a high level of genetic diversity as seen by the large number of alleles detected in each infrapopulation, in agreement with observations of S. mansoni populations from different parts of Kenya (Morgan et al., 2005; Agola et al., 2006). Even though the present study was carried out on a small geographic scale, high levels of genetic diversity of the parasite were still observable. The level of genetic diversity in schistosome populations is likely to be influenced by a variety of factors. On the one hand, repeated exposure of children in a selected number of transmission sites can result in the recirculation of related populations of schistosomes. If infection of humans and snails occurs at isolated transmission sites, then this would lead to development of distinct schistosome lineages as mutation and drift affects isolated populations within hosts and across geography. Also, acquired immunity and /or incomplete host susceptibility may also serve to limit the variety of schistosome seen in an infrapopulation (Brouwer et al., 2001). The rapid turnover of the infected snails induces a temporal succession and renewal of the different larval genotypes within a site and spatial aggregation of infected snails concentrate cercariae with different genotypes within a limited standing water area favouring multigenotype infection of definitive hosts (Theron et al., 2004). On the other hand, opportunities for genetic exchange resulting from snail and especially human movements may lead to coexistence of a large number of genotypes in an area (Brouwer et al., 2001).
Schistosome populations were significantly structured at two hierarchical scales, across geography and among human hosts. To assess the levels of differentiation between the infrapopulations analysed in this study, FST was calculated for all possible infrapopulations pairs. FST was considered appropriate because genetic drift is assumed to be the main factor in genetic differentiation among closely related infrapopulations or for short term evolution (Reynolds et al., 1983; Weir, 1990). Generally, FST calculated between all infrapopulation pairs was low but significant (P < 0.001). One possible explanation for the differences among patients is the presence of family substructure. We did not see negative FIS values as expected if there was strong family structure in the samples. Hence there were very low levels of genetic differentiation within S. mansoni infrapopulations in the Mwea area, suggesting high levels of gene flow between all the populations analysed. However, some infrapopulations from school children from different schools had slightly higher FST value as compared to the other pair wise FST values indicating slightly restricted gene flow (0.0392; between the NYD and NGC populations). A major contributor to gene flow is most likely to be the free movement of the school children in the study area. These children potentially sample a number of transmission sites, thereby becoming ‘genetic mixing bowls’ for the parasite (Curtis and Minchella, 2000). The parasites also live long enough to integrate infections across multiple seasons. The low levels of genetic differentiation can also be attributed to the nature of human population sampled in this study. Cross-sectional surveys of communities that will include other age groups left out in the present study are more likely to reveal population structure than, for example, a population of schoolchildren (Curtis et al., 2002). Efforts were also made to determine whether there was a correlation between genetic differentiation and geographical distance. The calculated interpopulation similarity indices showed that the populations do not follow a pattern of isolation by distance. Similar studies in Brazil (Thiele et al., 2008) also observed in their study of S. mansoni population a lack of significant isolation by distance.
The PCA analysis with GenAlEx failed to find significant structuring within the data, in contrary to the other analyses. This apparent discordance suggests that although there is significant structure, it is not necessarily strong. This is further evidenced by the relatively low genetic distances between all the infrapopulations (0.049- 0.1178) compared to those detected in our previous studies (0.203- 0.609) of S. mansoni populations analysed across the country (Agola et al., 2006), with the highest being recorded between NGB and NYE; distances were also low between populations such as KR and the MU, suggesting that these populations probably share a common gene pool. We hypothesize that although there is significant genetic structure of schistosomes among geographic regions and hosts, extensive human host movement in the area, leads to gene flow among infrapopulations. Hence the human host in this area serves both as a means of dispersal and reservoir of genotypes.
The high levels of genetic diversity reported here are comparable with those that have been detected in other mammalian hosts such as in the rat population in Guadeloupe and also those reported in an earlier study of the genetic diversity of S. mansoni within Kenya (Barral et al., 1996; Agola et al., 2006). Despite the homogeneity of the genotypes observed in this area, with most alleles being shared among the infrapopulations, private alleles (alleles occurring exclusively in one population) were observed in most of the infrapopulations analysed except four infrapopulations (NYD, NYF, NGA and KR infrapopulations). Interestingly, these private alleles occurred at frequencies higher than 5%. We hypothesize that the presence of private alleles in these populations could be as a result of exposure to unique schistosome genotypes found exclusively at one infection foci. An alternative hypothesis could be that since we are samplings offspring and not the adult population, private alleles could arise as a result of few highly fecund worm pairs with unique alleles that are producing more offspring thus creating the high frequencies of private alleles. In all the infrapopulations analysed in the present study, the observed heterozygosity was much lower than expected, if the populations were in Hardy-Weinberg equilibrium. These results are comparable to those of Rodrigues et al. (2002) and Stohler et al. (2004) who detected heterozygote deficiency in field populations of S. mansoni. We hypothesize here that the Wahlund effect (sampling different sites that are combined to represent a single population) is the most probable cause of heterozygote deficiency in the populations analysed in this study. This again supports the idea that there is a lot of movement and sampling of the parasite by school children from several infection foci within the larger Mwea area. Alternatively, the possible cause of heterozygote deficiency could have been due to the presence of null alleles and/or scoring errors. The frequencies of null alleles in all the populations analysed, however, were found to be low (0.5–5.6%) and therefore unlikely to be the cause of the observed heterozygote deficiency in the S. mansoni infrapopulations in Mwea. Long allele drop out is a problem in scoring microsatellites particularly when the quantity of DNA is low, which is an inherent problem with genotyping miracidia but this type of error appeared to be minimal in this study because the analysis with MICROCHECKER did not reveal any problematic loci.
In terms of the potential for the evolution of drug resistance, high genetic diversity suggests that there may be a greater potential for such populations to harbor alleles that may confer drug resistance, and also that the effective population size is large, which would increase the probability of novel mutations that confer resistance. Large population sizes tend to prevent the fixation of such alleles unless they have a strong selective advantage, or population sizes become dramatically reduced, as predicted with large scale drug treatment. Because there is gene flow occurring among subpopulations (infrapopulations) in Mwea, there is an opportunity for the spread of rare alleles, some of which may confer traits such as drug resistance or virulence. It is also important to note that in Mwea there is an ongoing schistosomiasis control program involving mass treatment of school children with praziquantel. Finally, although we do not know if drug resistance alleles occur in this population, one of the important factors that determine how rapidly such alleles would increase in frequency is the size of the treated proportion of the population relative to the untreated proportion, termed “refugium” ( Sissay et al., 2006; Leathwick et al., 2008; Waghorn et al., 2008). Our findings suggest this refugium is relatively large because of the high rates of gene flow and lack of population substructure throughout Mwea and only a subset of school children are targeted for treatment.
Acknowledgements
We thank the children and teachers of Nyamindi, Kirogo, Ngurubani and Mukou primary schools in Mwea for their participation in the study and cooperation, the staff of the Division of Vector Borne Disease (DVBD), Mwea in particular, Mr. Njoroge for facilitating access to their lab facility during field sample collection, and Dr. Charles Mwandawiro, Jimmy Kihara and the staff of the Japan International Co-operation Agency (JICA)-supported Eastern and Southern Africa Centre of International Parasite Control (ECACIPAC) for allowing us access to their field sites and the children participating in their schistosomiasis control program. We are equally grateful to Mr. George Rosenberg and the staff of the Molecular Biology Facility in the Department of Biology, University of New Mexico (UNM) for help with genotyping. The technical assistance of Joseph Kinuthia, Geoffrey Maina and Stephen Kamau of the Centre for Biotechnology Research and Development of KEMRI is highly appreciated. This research was supported through NIH grant R01-A144913. Approvals for this study were obtained through KEMRI’s scientific and ethical review committees. This paper is being published with the approval of the Director, KEMRI.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
L. E. Agola, Centre for Biotechnology Research and Development, Kenya Medical Research Institute (KEMRI), P.O. Box 54840, Nairobi, Kenya.
M. L. Steinauer, Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA
D. N. Mburu, University of Manitoba/ Nairobi project, P.O. Box 19676, Nairobi, Kenya
B.N. Mungai, Centre for Biotechnology Research and Development, Kenya Medical Research Institute (KEMRI), P.O. Box 54840, Nairobi, Kenya
I.N. Mwangi, Centre for Biotechnology Research and Development, Kenya Medical Research Institute (KEMRI), P.O. Box 54840, Nairobi, Kenya
G.N. Magoma, Department of Biochemistry, Jomo Kenyatta University of Agriculture and Technology (JKUAT), P.O. Box 43844, Nairobi, Kenya
E. S. Loker, Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA
G. M. Mkoji, Centre for Biotechnology Research and Development, Kenya Medical Research Institute (KEMRI), P.O. Box 54840, Nairobi, Kenya
Reference
- Agola LE, Mburu DN, DeJong RJ, Mungai BN, Muluvi GM, Njagi EN, Loker ES, Mkoji GM. Microsatellite typing reveals strong genetic structure of Schistosoma mansoni from localities in Kenya. Infect Genet Evol. 2006;6:484–490. doi: 10.1016/j.meegid.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Barral V, Morand S, Pointier JP, Theron A. Distribution of schistosome genetic diversity within naturally infected Rattus rattus detected by RAPD markers. Parasitology. 1996;113:511–517. doi: 10.1017/s003118200006755x. [DOI] [PubMed] [Google Scholar]
- Beltran S, Galinier R, Allienne JF, Boissier J. Cheap, rapid and efficient DNA extraction method to perform multilocus microsatellite genotyping on all Schistosoma mansoni stages. Memorias Do Instituto Oswaldo Cruz. 2008;103:501–503. doi: 10.1590/s0074-02762008000500017. [DOI] [PubMed] [Google Scholar]
- Brookfield JF. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996;5:453–455. doi: 10.1111/j.1365-294x.1996.tb00336.x. [DOI] [PubMed] [Google Scholar]
- Brouwer KC, Ndhlovu P, Munatsi A, Shiff CJ. Genetic diversity of a population of Schistosoma haematobium derived from schoolchildren in east central Zimbabwe. J. Parasitol. 2001;87:762–769. doi: 10.1645/0022-3395(2001)087[0762:GDOAPO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bundy DAP. This wormy world – then and now. Parasitol. Today. 1997;13:407–408. [Google Scholar]
- Chitsulo L, Engels D, Montresor A, Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41–51. doi: 10.1016/s0001-706x(00)00122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournet JM, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis J, Minchella DJ. Schistosome population genetic structure: when clumping worms is not just splitting hairs. Parasitol. Today. 2000;16:68–71. doi: 10.1016/s0169-4758(99)01553-7. [DOI] [PubMed] [Google Scholar]
- Curtis JS, Kristen PL, Minchella DJ. Microsatellite loci in human blood fluke Schistosoma mansoni and their utility for other Schistosome species. Mol. Ecol. Notes. 2001;1:143–145. [Google Scholar]
- Curtis J, Sorensen RE, Minchella DJ. Schistosome genetic diversity: the implications of population structure as detected with microsatellite markers. Parasitology. 2002;125:51–59. doi: 10.1017/s0031182002002020. [DOI] [PubMed] [Google Scholar]
- Dias LCS, Jesus PR, Deberaldini ER. Use of praziquantel in patients with Schistosoma mansoni previously treated with oxaminiquine and/ or hycanthone: resistance of Schistosoma mansoni to schistosomicidal agents. Trans. R. Soc. Trop. Med. Hyg. 1982;76:652–659. doi: 10.1016/0035-9203(82)90235-8. [DOI] [PubMed] [Google Scholar]
- Durand P, Sire C, Theron A. Isolation of microsatellite markers in the digenetic trematode Schistosoma mansoni from Guadeloupe island. Mol. Ecol. 2000;9:997–998. doi: 10.1046/j.1365-294x.2000.00939-4.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gillespie S. Intestinal nematodes. In: Gillespie SH, Pearson RD, editors. Principles and Practice of Clinical Parasitology. Chichester, England: John Wiley and Sons Ltd; 2001. pp. 561–583. [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (Version 2.9.3) 2000 http://www.unil.ch/software/fstat.html. [Google Scholar]
- Gower CM, Shrivastava J, Lamberton PH, Rollinson D, Webster BL, Emery A, Kabatereine NB, Webster JP. Development and application of an ethically and epidemiologically advantageous assay for the multi-locus microsatellite analysis of Schistosoma mansoni. Parasitology. 2007;134:523–536. doi: 10.1017/S0031182006001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- Imbert-Establet D, Combes C. Schistosoma mansoni: comparison of a Caribbean and African strain and experimental crossing based on compatibility with intermediate hosts and Rattus rattus. Exp. Parasitol. 1986;61:210–218. doi: 10.1016/0014-4894(86)90154-2. [DOI] [PubMed] [Google Scholar]
- Katz N, Chaves A, Pellegrino J. A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni. Rev. Inst. Med. Trop. Sao Paulo. 1972;14:397–400. [PubMed] [Google Scholar]
- Kihara JH, Muhoho N, Njomo D, Mwobobia IK, Josyline K, Mitsui Y, Awazawa T, Amano T, Mwandawiro C. Drug efficacy of praziquantel and albendazole in school children in Mwea Division, Central Province, Kenya. Acta Trop. 2007;102:165–171. doi: 10.1016/j.actatropica.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Leathwick DM, Miller CM, Atkinson DS, Haack NA, Waghorn TS, Oliver AM. Managing anthelmintic resistance: Untreated adult ewes as a source of unselected parasites, and their role in reducing parasite populations. New Zealand Veterinary Journal. 2008;56:184–195. doi: 10.1080/00480169.2008.36832. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- Minchella DJ, Sollenberger KM, Pereira de Souza C. Distribution of schistosome genetic diversity within molluscan intermediate hosts. Parasitology. 1995;111:217–220. doi: 10.1017/s0031182000064970. [DOI] [PubMed] [Google Scholar]
- Morgan JA, DeJong RJ, Lwambo NJ, Mungai BN, Mkoji GM, Loker ES. First report of a natural hybrid between Schistosoma mansoni and S. rodhaini. J Parasitol. 2003;89:416–418. doi: 10.1645/0022-3395(2003)089[0416:FROANH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Morgan JA, Dejong RJ, Adeoye GO, Ansa ED, Barbosa CS, Bremond P, Cesari IM, Charbonnel N, Correa LR, Coulibaly G, D'Andrea PS, De Souza CP, Doenhoff MJ, File S, Idris MA, Incani RN, Jarne P, Karanja DM, Kazibwe F, Kpikpi J, Lwambo NJ, Mabaye A, Magalhaes LA, Makundi A, Mone H, Mouahid G, Muchemi GM, Mungai BN, Sene M, Southgate V, Tchuente LA, Theron A, Yousif F, Zanotti-Magalhaes EM, Mkoji GM, Loker ES. Origin and diversification of the human parasite Schistosoma mansoni. Mol. Ecol. 2005;14:3889–3902. doi: 10.1111/j.1365-294X.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data II. Gene frequency data. J. Mol. Evol. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Ota T. Pennsylvania: Pennsylvania State University; 1993. Dispan: Genetic Distance and Phylogenetic analysis. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetics software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, De Meeus T, Durand P, Sire C, Theron A. Sex-specific genetic structure in Schistosoma mansoni: evolutionary and epidemiological implications. Mol. Ecol. 2002;11:1231–1238. doi: 10.1046/j.1365-294x.2002.01518.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. J. Hered. Vol. 86. 1997. GENEPOP (Version 1.2): a population genetics software for exact tests and ecumenicism; pp. 248–249. GENEPOP (version 3.3) http://wbiomed.edu.au/genepop/. Updated from Raymond M and Rousset F., 1995. [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the Coancestry Coefficient: Basis for a Short-Term Genetic Distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NB, Coura Filho P, de Souza CP, Jannoti Passos LK, Dias-Neto E, Romanha AJ. Populational structure of Schistosoma mansoni assessed by DNA microsatellites. Int. J. Parasitol. 2002;32:843–851. doi: 10.1016/s0020-7519(02)00031-0. [DOI] [PubMed] [Google Scholar]
- Savioli L, Renganathan E, Montresor A, Davis A, Behbehani K. Control of schistosomiasis--a global picture. Parasitol. Today. 1997;13:444–448. doi: 10.1016/s0169-4758(97)01141-1. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Silva LK, Liu S, Blanton RE. Microsatellite analysis of pooled Schistosoma mansoni DNA: an approach for studies of parasite populations. Parasitology. 2006;132:331–338. doi: 10.1017/S0031182005009066. [DOI] [PubMed] [Google Scholar]
- Sire C, Durand P, Pointier JP, Theron A. Genetic diversity and recruitment pattern of Schistosoma mansoni in a Biomphalaria glabrata snail population: a field study using random-amplified polymorphic DNA markers. J. Parasitol. 1999;85:436–441. [PubMed] [Google Scholar]
- Sissay MM, Asefa A, Uggla A, Waller PJ. Anthelmintic resistance of nematode parasites of small ruminants in eastern Ethiopia: Exploitation of refugia to restore anthelmintic efficacy. Veterinary Parasitology. 2006;135:337–346. doi: 10.1016/j.vetpar.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Steinauer LM, Agola LE, Mwangi IN, Mkoji GM, Loker ES. Molecular epidemiology of Schistosoma mansoni: A robust, high-throughput method to assess multiple microsatellite markers from individual miracidia. Infect. Genet. Evol. 2008;8:68–73. doi: 10.1016/j.meegid.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect. Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- Stohler RA, Curtis J, Minchella DJ. A comparison of microsatellite polymorphism and heterozygosity among field and laboratory populations of Schistosoma mansoni. Int. J. Parasitol. 2004;34:595–601. doi: 10.1016/j.ijpara.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Theron A, Sire C, Rognon A, Prugnolle F, Durand P. Molecular ecology of Schistosoma mansoni transmission inferred from the genetic composition of larval and adult infrapopulations within intermediate and definitive hosts. Parasitology. 2004;129(5):571–585. doi: 10.1017/s0031182004005943. [DOI] [PubMed] [Google Scholar]
- Thiele EA, Sorensen RE, Gazzinelli A, Minchella DJ. Genetic diversity and population structuring of Schistosoma mansoni in a Brazilian village. Int. J. Parasitol. 2008;38:389–399. doi: 10.1016/j.ijpara.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiongo FW, Madsen H, Ouma JH, Andreassen J, Christensen NO. Host-parasite relationships in infections with two Kenyan isolates of Schistosoma mansoni in NMRI mice. J. Parasitol. 1997;83:330–332. [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, de Vlas SJ, Brooker S, Looman CW, Nagelkerke NJ, Habbema JD, Engels D. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86:125–139. doi: 10.1016/s0001-706x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Waghorn TS, Leathwick DM, Miller CM, Atkinson DS. Brave or gullible: Testing the concept that leaving susceptible parasites in refugia will slow the development of anthelmintic resistance. New Zealand Veterinary Journal. 2008;56:158–163. doi: 10.1080/00480169.2008.36828. [DOI] [PubMed] [Google Scholar]
- Waiyaki PG. FAO Food and Agricultural Organization of the United Nations, Rome, Italy: FAO; 1987. The history of irrigation development in Kenya and the associated spread of schistosomiasis. In: Effects of agricultural development on vector-borne diseases, eds. PEEM (WHO/FAO/UNEP Panel of Experts on Environmental Management for Vector Control). AGL/MISC/12/87. [Google Scholar]
- Wang TP, Shrivastava J, Johansen MV, Zhang SQ, Wang FF, Webster JP. Does multiple hosts mean multiple parasites? Population genetic structure of Schistosoma japonicum between definitive host species. Int. J. Parasitol. 2006;36:1317–1325. doi: 10.1016/j.ijpara.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Weir BS. Sunderland, Massachusetts, USA: Sinaeuer Associates; 1990. Genetic Data Analysis. [Google Scholar]