Abstract
An inherited variant on chromosome 8q24, rs6983267, is significantly associated with cancer pathogenesis. We present evidence that this region is a transcriptional enhancer, that the risk region physically interacts with the MYC proto-oncogene, and that the alleles of rs6983267 differentially bind transcription factor 7-like 2 (TCF7L2). These data provide strong support for a biological mechanism underlying this non-protein coding risk variant.
Several independent genetic association studies identify the single nucleotide polymorphism (SNP) rs6983267 as being strongly associated with colorectal cancer (CRC), with the G allele conferring increased risk 1–3. Resequencing and detailed determination of linkage disequilibrium in this region suggests that rs6983267 is likely to be the causal variant for disease 4. However, the variant is located in a gene desert and the nearest annotated gene, the MYC proto-oncogene, is ~335 kb telomeric from the risk region. Since the risk region is intergenic, the molecular mechanisms underlying disease causation are unknown. Therefore, we sought to understand the functional role of the 8q24 risk region and its relationship to MYC in CRC.
To evaluate the transcriptional landscape of 8q24, we designed a high resolution (5 bp median probe spacing) tiling microarray covering the chromosome 8 region of interest. The microarray was hybridized with labeled cDNA produced from total RNA isolated from five histologically normal human colon specimens. Cross hybridization artifacts, which could be erroneously interpreted as novel transcription, were rigorously addressed using a co-hybridization strategy with matched, labeled genomic DNA from the same five individuals (Online Methods). Our analysis strongly suggests that, other than MYC itself, the genomic interval containing the colorectal risk variant is largely devoid of transcription. (Supplementary Figure 1).
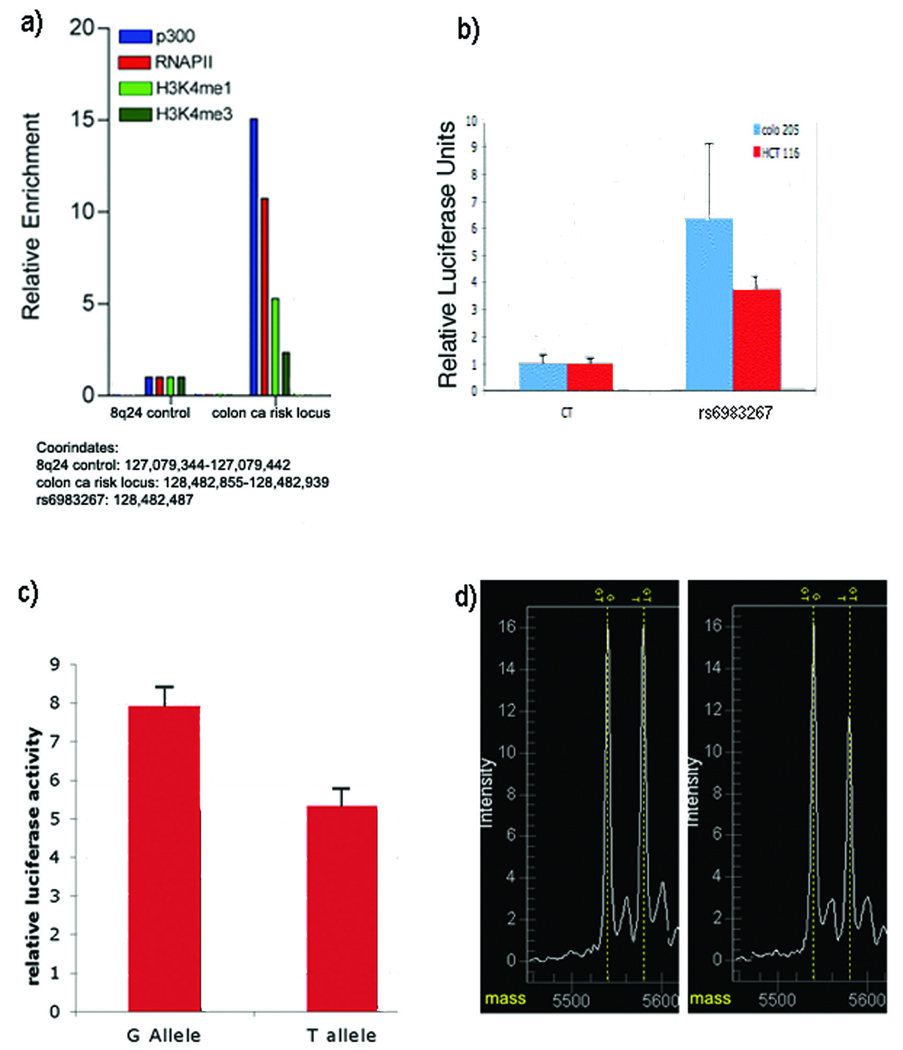
Given the absence of expression, we assessed whether this region acts as an enhancer. Site-specific ChIP assays were performed with antibodies recently demonstrated to mark enhancers, including p300, H3K4me1, and H3K4me3 in the CRC cell line Colo205 5,6. Colo205 is heterozygous for rs6983267. The emergent chromatin signature (Fig. 1a) supports enhancer function, showing differential enrichment of H3K4me1 relative to H3Kme35 as well as enrichment of p300 and RNApII compared to a control region 6. To evaluate enhancer activity, a 1,538 bp region flanking the SNP was cloned into a reporter vector (Online Methods). Luciferase assays demonstrate that the 8q24 CRC risk region functions as an enhancer (Fig. 1b). To evaluate whether enhancer activity was allele specific, the G risk allele was compared to the T allele in a luciferase reporter assay. Significantly increased reporter activity was observed with the G allele (P=0.024) (Fig. 1c). These data are consistent with the reporter data generated by Tuupanen et al 7. Thus, epigenetic and reporter assay data demonstrate that the CRC risk region functions as an enhancer, and that the CRC risk variant drives luciferase activity in an allele specific manner.
Figure 1.
Enhancer activity at the colon cancer risk locus and allele specific differences in enhancer activity and TCF7L2 binding. a) ChIP assay on Colo205, demonstrating a pattern consistent with enhancer activity. b) luciferase reporter assay demonstrating enhancer activity in two CRC lines. c) a representative luciferase assay showing increased enhancer activity of G over T alleles; performed on a total of 18 clones (9 G and 9 T over 3 days) (P=0.024). d) mass spectrometry plots from Sequenom analysis, showing preferential binding of TCF7L2 to risk allele (G) in immunoprecipitated DNA, as evidenced by differential peak heights (right panel) compared to control input DNA (left panel) (P=1.1×10−5).
The rs6983267 SNP (underlined base) is located within a (T-cell factor) TCF consensus binding sequence (A/T)(A/T)CAA(A/T)GG. Indeed, the main transcriptional effector of Wnt signaling and the partner for β-catenin co-activation in CRC is the sequence-specific DNA-binding protein TCF7L2 8–10. Genome wide ChIP-chip analysis in the LS174T cell line (a CRC cell line heterozygous for rs6983267) identified a strong and specific signal for TCF7L2 binding coincident with the risk region (Supplementary Figure 2).
We used a mass spectrometry-based platform to quantify the relative abundance of G and T alleles in TCF7L2 immunoprecipitates from heterozygote LS174T cells. This technology can accurately measure the ratio of allelic peak heights, a reliable proxy for relative allelic load (Online Methods). The G allele was significantly overrepresented in TCF7L2 immunoprecipated DNA, but not in the input (P=1.14×10−5), demonstrating differential and increased binding of TCF7L2 to the risk allele at rs6983267 (Fig. 1d).
MYC is one of the strongest candidate genes in the 8q24 region. MYC is a target gene of the Wnt signaling pathway, a key pathway in colorectal cancer pathogenesis 11,12. Strikingly, solitary deletion of MYC rescues the tumor phenotype that follows APC gene deletion in mice, establishing MYC as a critical mediator of early steps in Wnt pathway-mediated colorectal tumorigenesis 13. A prior study demonstrated no association between MYC protein expression in tumors and risk allele status 2. We find no association between rs6983267 and MYC mRNA expression levels in either normal colon or paired tumor samples in 110 individuals (Supplementary Figure 3).
Steady state levels of RNA or protein at a single point in time may not adequately capture MYC’s role in tumorigenesis. Since the magnitude of risk elevation is modest for common alleles and the clinical expression of disease is relatively late in life, expression differences driving risk can occur at any timepoint prior to the disease onset. Therefore, we sought complementary methods to examine MYC’s role in CRC etiology.
Although the MYC gene lies at a considerable linear DNA distance from rs6983267, we posited that the enhancer region physically interacts with the MYC locus and used the chromosome conformation capture (3C) technique to test this hypothesis. 3C assesses if a candidate fragment (e.g., the MYC promoter) interacts physically with a region of interest (e.g., the genomic region that contains rs6983267) 14. To rigorously quantify ligation fragments, we employed a novel competitive PCR approach using the Sequenom platform (Online Methods, Supplementary Figure 4, and Supplementary Figure 5).
A constant fragment containing rs6983267 was interrogated against a series of 10 target fragments (Fig. 2a) in two heterozygous CRC cell lines- Colo205 and LS174T- as well as in a control fibroblast cell line- LL24 (homozygous -GG- for the risk variant). As expected, increased interaction frequency was observed between the constant and the adjacent fragment (3 kb) in both the CRC cell lines as well as the fibroblast line; such interaction was almost completely absent by fragment 3, located 95 kb away (Fig. 2b). In CRC lines, strong interactions were observed between rs6983267 and fragments encompassing the promoter and the first half of MYC (fragments 4, 5, and 6) (Fig. 2b), but not in the fibroblast line (Supplementary Figure 6). Notably, the magnitude of the peak interaction between the rs6983267 and MYC regions in CRC lines is as abundant as that between rs6983267 and its adjacent fragment 1. Thus, the rs6983267 risk region physically interacts with MYC in CRC cell lines, and does so in a tissue-specific manner. Further experiments analyzing 3C interactions by genotypic status may be informative.
Figure 2.
Long-range physical interaction of the CRC risk variant rs6983267 with MYC in colon cancer cell lines. a) Physical map of the region interrogated by 3C, spanning a 395-kb distance with rs6983267 at one end and MYC at the other. The position of the constant fragment containing rs6983267 is marked by a red bar; positions of target fragments are marked by black bars, with an expanded view of MYC target fragments 5–9. b) Graph showing 3C interaction frequency of the constant fragment containing rs6983267 with each target fragment. The results demonstrate decreased interaction at 12 kb, 95 kb (fragments 2 and 3) and 395 kb (fragment 10), and increased interaction frequency in CRC cell lines at the MYC promoter (fragment 4) and the first half of the MYC structural gene (fragments 5 and 6), located ~330 kb away. Distance from rs6983267 (x-axis) is not represented to scale. The y-axis refers to number of molecules in 3C libraries from each interrogated ligation product. Labels above each data point in the graph refer to the name of each target fragment 1–10.
We have demonstrated that the CRC risk locus containing rs6983267 has in vitro and in vivo properties of an enhancer, and demonstrates long-range physical interaction with MYC. We propose that this risk locus acts as part of a cis-regulatory enhancer element for the MYC proto-oncogene in CRC, mediating colorectal cancer risk in part through its differential binding with TCF7L2. Since 8q24 is known to be associated with other cancers 15, it will be informative to evaluate whether these other 8q24 risk loci also interact with MYC. More broadly, this integrative approach offers a framework for understanding the molecular consequences of non-protein coding risk alleles.
METHODS
Chromosome 8q24 tiling array
A Roche NimbleGen tiled microarray (385,000 features) was designed to interrogate the 8q24 region of the human genome (chr8: 125M to 130M) with a median probe spacing of 5bp. RNA and genomic DNA were derived from histologically normal colon tissue in five human subjects who underwent surgery and treatment at the University of Southern California/ Norris Comprehensive Cancer Center (USC/NCCC) or the Los Angeles County/University of Southern California Medical Center (LAC/USCMC) between 2001 and 2008. Five arrays were each co-hybridized under standard conditions with 5ug Cy3-labeled cDNA and 5ug Cy5-labeled genomic DNA. (Roche NimbleGen, Madison, WI). The genomic DNA was hybridized to help identify spuriously cross-hybridizing probes which could erroneously be interpreted as novel transcription (see below). After hybridization for 16 to 20 hours at 42°C, the arrays were washed and scanned using standard NimbleGen procedures.
Initially, data generated from each group of microarray scans (cDNA and genomic DNA) were normalized separately. Quantile normalization of probe intensities was carried out using the Multi-Array quantile normalization module, part of the NMPP 1.01 package 16 with the two-step normalization option. First, replicate slides within each set were normalized, followed by a global normalization to adjust both sets to an average baseline. Next, background subtraction was done by subtracting the top 3% of random probe intensity values from individual experimental probe values. These random probes (which represent 4496 of the 385,000 features on the array, generated by Roche NimbleGen) were included on the array to help define background probe intensity levels, since virtually all of the sequences represented by these probes are not found in the human genome. Values obtained were summed, resulting in a single intensity value per probe for each group. A probe was considered positive in the cDNA array when its intensity was higher (> 1) than its genomic DNA probe value. This strategy is an improvement over most of the microarray-based strategies which empirically seek to assess transcription in a genome by cDNA-only hybridizations. Probe behavior is difficult to assess without empirically determining hybridization specificity. In fact, some probes are prone to spurious cross-hybridization; this can only be revealed by hybridization of genomic DNA, which should hybridize consistently across a genomic region provided that repeat regions are masked. Thus, any probe intensities significantly higher than the median probe intensity in the genomic DNA hybridization would indicate spurious cross-hybridization behavior of those particular probes. Consistent with this reasoning, elevated probe intensity levels in the same genomic regions for both the cDNA and genomic DNA hybridizations are often observed, indicating that these probes are not accurately assessing transcription.
After the above analysis of the 385K features on the array, 9381 probes were considered as expressed in the cDNA set, and were used for generating Transcriptionally Active Regions (TARs). For this, a minimum run of 3 probes and a maximum gap of 2 probes 17 were applied to the above dataset which resulted in the filtering out of 2537 probes. Of the remaining probes, 5029 were mapped to the mRNA regions of 14 of the genes in this genomic interval, while 1803 were assigned to the intergenic regions. Log2 Expression values for these 14 genes were calculated by taking the median intensity value of the CDS and UTR probes (1733) from each mRNA (Supplementary Table 1).
The 1803 positive intergenic probes could be clustered into 385 TARs, which were scattered across the chromosome and when totaled, measured 34.8 kb of genomic space. A vast majority of these (334 of 385; 87%) were 100 bp in length or less (total, 26.8 Kb), smaller than the mean and median human exon size of 145 and 122 bp, respectively 18; and given their small size and random occurrence in this genomic region they are thus unlikely to represent novel transcripts. Of the remaining TARs, a cluster of 10 are located between the genomic coordinates 12508000-125127143 (>3 Mb centromeric to the risk region), are more than 100 bp in size, and based on their clustering appear to represent a novel transcript or gene extension of C8orf78 (an in silico ORF prediction). Only two TARs in the risk region are greater than 100 bp; one is located at the position spanning 128497309 to 128497645 (336 bp, Supplementary Figure 1) whereas the other is only 102 bp spanning position 128816692 to 128816794 and still smaller than the mean/median exon size of the genome. The 336 bp TAR overlaps part of a pseudogene called POU5F1P1 (located between 128497039 and 128498621). However, of the probes interrogating the pseudogene, 85% did not pass our thresholding strategy. Thus, the probes that did make it to the TAR stage likely represent probes exhibiting spurious cross-hybridization. Consistent with this idea, the pseudogene region was partially repeat masked during design of the array due to repeats within this region. Finally, qPCR of a unique portion of the pseudogene confirmed that expression from this region is negligible (primers and probes available upon request).
Of note, RNA isolation was achieved using Trizol (Invitrogen), which captures both large and small RNA species. The tiling array is reliably able to detect all RNA species greater than 200 bp (e.g., unprocessed miRNAs). The labeling and hybridization of shorter RNA species to tiling arrays, however, are less well studied. It is conceivable that processed miRNAs may not be detected. Of note, high throughput sequencing for small RNAs using Solexa/Illumina in prostate tissue did not demonstrate any evidence for miRNAs in this region 19. Importantly, the colon cancer risk allele, rs6983267, is exactly the same allele that is implicated in prostate cancer 20–22.
ChIP qPCR for markers of enhancers
ChIP analyses of cultured Colo205 cells were performed as described previously 23. Briefly, chromatin from 1×107 fixed cells was sonicated to a size range of 200–1500bp. Solubilized chromatin was subjected to immunoprecipitation with antibody against p300 (sc-585, Santa Cruz), RNAPII (sc-9001, Santa Cruz), H3K4me1 (ab8895, Abcam), or H3K4me3 (ab8580, Abcam). DNA from ChIP preparation was quantified by qPCR using TaqMan PCR Master Mix (Applied Biosystems). Triplicate qPCR results were averaged. Enrichment of each ChIP at rs6983267 site was normalized against a neighboring 8q24 control region (defined as 1). The control region was chosen based on the absence of histone 3 acetylation as assessed by ChIP on an 8q24 NimbleGen tiling array (data not shown). The primers and probes are listed in Supplementary Table 2.
Luciferase reporter assays
The cloned enhancer candidate region (chr8:128482317-128483854) spanned 1538 bp and contained either the T or the G allele the risk variant rs6983267. The insert was amplified from genomic DNA using High Fidelity Platinum Tag DNA polymerase (Invitrogen). All clones were confirmed by sequencing. The amplified sequences were then subcloned in either the KpnI or Sac II restriction sites upstream of a thymidine kinase (TK) minimal promoter-luciferase vector in both directions. All clones were confirmed by sequencing. COLO 205 cells were transfected with reporter plasmids along with constitutively active pRL-TK Renilla luciferase plasmid (Promega) using Lipofectamine LTX Reagent (Invitrogen) according to the manufacturer’s protocol. Dual luciferase activities were measured as previously described 23. To assess allele-specific effects, point mutations were introduced to create enhancer reporter constructs with specific SNP alleles- the risk allele (G) or the wild-type allele (T). In this assay, nine independent clones of each construct were made, and confirmed by sequencing. Six clones (3 G and 3 T) were assayed on Day 1. Twelve clones (6 G and 6 T) were assayed twice, on Day 2 and Day 3. Each individual clone was tested in triplicate. The mean luciferase activity of each triplicate was calculated. Luciferase activity was normalized to Renilla activity. A linear mixed model was used for analysis with random effect for clone and fixed effects for allele and experiment.
TCF7L2 binding ChIP-chip
TCF7L2 ChIP DNA from LS174T cells was linearly amplified and hybridized to Affymetrix GeneChip Human Tiling 2.0RF using the same techniques described previously 24. Data were analyzed using MAT 25. Data were visualized using the Affymetrix Integrated Genome Browser, version: 5.12. Probes are tiled at a resolution of 25bp.
Allele-Specific ChIP using Sequenom platform
The Sequenom (Carlsbad, CA) platform is a genotyping platform using Matrix Assisted Laser Desorption /Ionization- Time of Flight (MALDI-TOF) mass spectrometry and has the capacity to be quantitative (protocols available at www.sequenom.com). Primers were designed to amplify a region containing rs6983267. A probe was designed to hybridize to ~10 bases immediately adjacent to the SNP in the amplified product. A single base extension step with mass-modified alleles followed. The genotype of rs6983267 was identified by expected mass when run through the spectrophotometer. While the absolute peak heights of each allele of a heterozygous SNP on the spectral display vary from run to run, the peak heights of samples relative to each other is maintained. To determine allele-specificity or allelic imbalance, deviation away from the 1:1 allelic ratio typically observed in a heterozygote genotype was measured. A similar approach has been used previously by Knight et al 26.
To determine allele-specific binding to TCF7L2, control input DNA and ChIP DNA immunoprecipitated with TCF7L2 (sc-8631, Santa Cruz) antibody from 6 independent ChIP experiments were tested separately. Each sample within each experiment was run in quadruplicate, and the peak height ratio of allele A to allele B measured in each replicate. The quadruplicates were averaged, and the mean peak-height ratios for control input DNA was compared with the mean peak height ratios for DNA immunoprecipitated with TCF7L2 in a paired t-test, p <0.05 taken as significant.
Quantitative Gene Expression of MYC
RNA was isolated from fresh frozen colorectal cancer tissue. Five micron sections were reviewed by a pathologist (J.C.) to confirm colonic adenocarcinoma and benign tissue. Areas of tumor were selected where >40% of cells consisted of tumor cells. Areas of benign tissue were selected where 100% of cells consisted of non-neoplastic epithelium. Two 2 mm punch biopsy cores of frozen tissue were processed for RNA extraction using a modified Qiagen Allprep DNA/RNA protocol. cDNA was prepared for QGE from fresh frozen RP tissue using Invitrogen SuperScript III Reverse Transcription kit, incubating approximately 2 micrograms of RNA with random hexamers followed by PCR.
In addition to MYC, seven normalization genes (B2M, HMBS, HPRT1, SDHA, TBP, UBIQ, YWHAZ) were included. MassARRAY QGE Assay Design software from Sequenom was used to design all primers. One of the two primers in each assay spanned an exon/exon boundry. Competitor oligos were designed, 80–100 base pairs in length and identical to each assay’s PCR product except for a one base pair change. All assays were plexed into a single reaction mix. Sequences are available upon request.
Competitor oligos and 10 ng cDNA were co-amplified in a final volume of 5 µL. Reactions were performed in quadruplicate using 8 serial dilutions of competitor, following the standard protocol 27. Probes hybridized to the amplified product and a single base extension distinguished cDNA from competitor.
The EC50- the point at which cDNA and competitor concentrations are equal-was calculated for each gene in each sample using QGE Analyzer software (Sequenom at www.sequenom.com). The geometric mean of the expression of the seven housekeeping genes for each sample was taken as the normalization factor for that sample. MYC expression in each sample was then normalized to this normalization factor. Normal and tumor tissues were analyzed separately by genotype. Two-tailed Kruskal-Wallis test was employed to determine significant differences between expression levels of normalized MYC among each genotype class. R-software was used for statistical analysis (http://www.R-project.org).
Chromosome Conformation Capture (3C)
3C Library preparation
Colo205 and LS174T colon cancer cell lines and the LL24 lung fibroblast cell line were grown to 80% confluence, washed with 1%PBS and trypsinized with Trypsin EDTA 0.25%. Trypsin was neutralized with 1XPBS and 10% FBS. Cultured cells were collected into a 50ml tube, passed through a cell strainer, and fixed with formaldehyde to a final concentration of 1% for 10 minutes. Fixation was quenched with 2M glycine to a final concentration of 0.125mM for 5 minutes on ice. Cells were counted and placed into aliquots 10×106 cells, snap frozen and stored in −80oC in 1XPBS 0.125mM glycine. A biologic replicate (processed independently on separate days) was performed on LS174T.
Frozen aliquots were pelleted at 4oC and storing media aspirated. Lysis buffer was added (500ul of 10mM TrisHClph8, 10mM NaCl, 0.2%NP40) with protease inhibitor and incubated for 15 min on ice. Cell nuclei were pelleted and washed with 1X restriction enzyme buffer B. Nuclei were pelleted and resuspended in 200ul of 1X RE buffer B and distributed into four tubes. 337ul of 1X RE buffer B were added to each tube. SDS was added to each aliquot, vortexed and incubated for 10 minutes at 65oC at a final concentration of 0.1%. TritonX-100 was added to a final concentration of 1.8% followed by 400 units of Csp6I restriction enzyme (Fermentas) and incubated for 24hr at 37oC. Each aliquot received 10%SDS and was incubated for 30 minutes at 65oC. Ligation mixes were prepared on 15ml tubes (745ul 10X T4 ligase Buffer, 10% TritonX100, 80ul 10mg/ml BSA, 6ml water, 575ul of cell lysate, 4000 units of T4 ligase) and incubated for four full days at 16oC. Aliquots of the ligated samples were QC’ed by taking 350ul, digesting with Proteinase K for 2 hours, ethanol precipitated, and run on 0.8% 1X TAE agarose gel. Ligated 3C libraries were incubated with Proteinase K for 24 hours. Samples were cleansed with phenol then phenol-chloroform. Aqueous phase was ethanol precipitated overnight (8ml of water, 0.1% of 3M Sodium Acetate pH 5.5 and 2.5 volumes of ethanol). DNA was extracted by centrifugation at max speed at 4oC and washed 1X with 70% ethanol. The DNA pellet was re-hydrated with water and passed through a Microcon Centrifugal Filter device Y-100 (Millipore) for further desalting, and re-hydrated in 25ul of water. Each DNA aliquot was analyzed on 1X TAE agarose gel. Samples that passed QC were combined and final concentration was estimated on 0.8% 1XTAE agarose gel.
3C fragment quantification
The Sequenom quantitative gene expression (QGE) platform was first described for quantifying gene expression products using competitive PCR 28. This platform possesses the properties necessary for rigorous quantification and has been shown to be sensitive, accurate, and precise in the detection of nucleic acids 28. We adapted this competitive PCR approach to the determination of the abundance of each ligated product of interest in the 3C library, and initially confirmed the identity of our expected ligation product by sequence verification.
Restriction enzyme Csp6I cuts at predictable sites, and the region of interest (chromosome 8:128482237-128877490) contains numerous digestion fragments that could potentially ligate with the constant fragment containing rs6983267. These ligation events will occur at varying frequencies depending on the proximity of the constant and target fragments. We interrogated 10 target fragments at varying distances from the constant fragment at short (0–12kb), intermediate (95kb), and long distances (>300kb) from the constant fragment, including several fragments within MYC (fragments 5–9).
Competitor oligonucleotides of known quantity were synthesized with sequence complementary to each of the 10 ligation products of interest. The competitors were used to determine the quantity of amplified product present in the 3C library. Competitor oligonucleotides differed from 3C template only at the G/T SNP rs6983267, where a C allele (G in the reverse complement) was created. Competitor oligonucleotides were from Integrated DNA Technologies (www.idtdna.com). Primers and probes were from Invitrogen. Competitor was co-amplified with 3C template in a PCR reaction using primers for the predicted ligation product of interest. Primer and competitor oligonucleotide sequences are listed in Supplementary Table 2.
Each PCR reaction was a 5ul mixture containing the following- 3C template DNA (33ng/ul) 0.625ul, Constant primer (1uM) 0.625 ul, Target primer (1uM) 0.625 ul, Competitor for the specific product of interest (1 ul), H20 0.4375ul, 10X PCR buffer 0.78125 ul, 25mM MgCl2 0.40625 ul, 25mM dNTP mix 0.125ul ul, and 5U/ul Hotstart Taq Plus 0.125 ul. PCR cycling conditions: 94 deg 5min; 40 cycles of 94 deg 20 sec, 56 deg 30 sec, 72 deg 30 sec; 72 deg 10 min; 4 deg hold. Competitors were serially diluted 8 times and each dilution was used in a separate PCR reaction. Competitor concentrations in the 5ul PCR reaction volume varied as follows: 1.00E-16 M; 3.73E-17 M; 1.39E-17 M; 5.16E-18 M; 1.93E-18 M; 7.20E-19 M; 2.68E-19 M, and 0 M. Each PCR reaction was performed in quadruplicate. Therefore, a total of 320 PCR reactions were performed per cell line (10 ligation products of interest × 8 competitor dilutions × 4 replicates).
A probe was hybridized immediately adjacent to the rs6983267 polymorphism (Supplementary Table 2). After desalting, single base probe extension was performed. A single base extension step distinguished the 3C template (G/T) from competitor (C). Based on the relative abundance of competitor-to-product in the final PCR reaction, an EC50 (concentration) was calculated for each ligation product of interest, where the concentration of ligation product (unknown) equals concentration of competitor (known) (Supplementary Figure 6). The EC50 was determined from a non-linear regression fitting procedure calculated by the Sequenom software. The number of molecules in the reaction was calculated: EC50 × 6.02 × 1023 molecules/mole × 1 × 10−6L/ul × 5ul PCR reaction and served as a proxy for 3C interaction frequency.
To calculate standard errors for the number of molecules, the following formula was used: Number of molecules × SE(logEC50) × ln(10). The SE(logEC50) is a value that is determined by the Sequenom software. Because Colo205 and LS174T are heterozygous for rs6983267, we needed to have a summary SE estimate for both alleles.
To establish tissue-specificity of the fragment containing rs6983267 and MYC, interaction frequencies across the LS174T line were compared to those across the LL24 lung fibroblast line. 3C interactions were normalized using a housekeeping gene, FAM32A on chromosome 19 (Supplementary Figure 4). The fragments interrogated at FAM32A were located at chromosome19: 16156175-16157698 and 16159199-16161288. Primer, competitor and probe sequence for FAM32A are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
Matthew Freedman is a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee and is a recipient of a 2006 Doris Duke Clinical Scientist Development Award. Grant support from the NIH (R01 CA129435 to MLF) and (R01 CA109147 to GAC), the Mayer Foundation (to MLF) the H. L. Snyder Medical Foundation (to MLF), the Dana-Farber/Harvard Cancer Center Prostate Cancer SPORE (National Cancer Institute Grant No. 5P50CA90381), the Prostate Cancer Foundation (to GAC and to MMP), the Whittier Foundation (to GAC), the American Cancer Society Institutional Research Grant (IRG – 58-007-48 to LJ), American Society of Clinical Oncology (to MMP).
The authors acknowledge Dr. Myles A. Brown for his support and guidance.
Footnotes
URLs:
Expression array data will be made available through the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database.
REFERENCES
- 1.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 2.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 3.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeager M, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum Genet. 2008;124:161–170. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 6.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009 doi: 10.1038/ng.406. in press. [DOI] [PubMed] [Google Scholar]
- 8.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 9.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 10.Barker N, Morin PJ, Clevers H. The Yin-Yang of TCF/beta-catenin signaling. Adv Cancer Res. 2000;77:1–24. doi: 10.1016/s0065-230x(08)60783-6. [DOI] [PubMed] [Google Scholar]
- 11.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 12.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 13.Sansom OJ, et al. MYC deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 14.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by chromosome conformation capture. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb2111s74. Chapter 21, Unit 21 11. [DOI] [PubMed] [Google Scholar]
- 15.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, et al. NMPP: a user-customized NimbleGen microarray data processing pipeline. Bioinformatics. 2006;22:2955–2957. doi: 10.1093/bioinformatics/btl525. [DOI] [PubMed] [Google Scholar]
- 17.Manak JR, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- 18.Pevsner J, editor. Bioinformatics and functional genomics. John Wiley and Sons; 2003. p. 636. [Google Scholar]
- 19.Pomerantz MM, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-0387. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wokolorczyk D, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 21.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 22.Haiman C, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia L, et al. Androgen receptor activity at the prostate specific antigen locus: steroidal and non-steroidal mechanisms. Mol Cancer Res. 2003;1:385–392. [PubMed] [Google Scholar]
- 24.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 27.Elvidge GP, Price TS, Glenny L, Ragoussis J. Development and evaluation of real competitive PCR for high-throughput quantitative applications. Anal Biochem. 2005;339:231–241. doi: 10.1016/j.ab.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 28.Ding C, et al. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc Natl Acad Sci U S A. 2003;18:3059–3064. doi: 10.1073/pnas.0630494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.