Abstract
The aggregation of imaging, clinical, and behavioral data from multiple independent institutions and researchers presents both a great opportunity for biomedical research as well as a formidable challenge. Many research groups have wellestablished data collection and analysis procedures, as well as data and metadata format requirements that are particular to that group. Moreover, the types of data and metadata collected are quite diverse, including image, physiological, and behavioral data, as well as descriptions of experimental design, and preprocessing and analysis methods. Each of these types of data utilizes a variety of software tools for collection, storage, and processing. Furthermore sites are reluctant to release control over the distribution and access to the data and the tools. To address these needs, the Biomedical Informatics Research Network (BIRN) has developed a federated and distributed infrastructure for the storage, retrieval, analysis, and documentation of biomedical imaging data. The infrastructure consists of distributed data collections hosted on dedicated storage and computational resources located at each participating site, a federated data management system and data integration environment, an Extensible Markup Language (XML) schema for data exchange, and analysis pipelines, designed to leverage both the distributed data management environment and the available grid computing resources.
Index Terms: Biomedical Informatics Research Network (BIRN), cyberinfrastructure, database, data grid, Extensible Markup Language (XML), Extensible Markup Language (XML)-based clinical experiment data exchange schema (XCEDE), Extensible Neuroimaging Archive Toolkit (XNAT), globus, high intensity discharge (HID), informatics, magnetic resonance imaging (MRI), neuroimage, storage resources broker (SRB)
I. INTRODUCTION
The field of biomedical imaging has undergone explosive growth in the last 50 years, driven in part by emergent imaging technologies and the information superhighway. The ability to collect vast amounts of complimentary data has empowered scientists to ask multifaceted questions of the data that was difficult at best in earlier years. In recent years, this data explosion has grown, as more publicly accessible data have become available. Due to the size and specialized nature of certain individual datasets, multidisciplinary teams of domain scientists are required to work together toward a common goal of understanding the illness and the disease. Complicating this endeavor is the reality that these scientists are geographically dispersed, necessitating effective means for domain scientists to collaborate on such multidisciplinary research problems. Improvements in critical information technologies and internet speeds have made the construction of a national collaboratory of federated and distributed data resources connected through a common semantic fabric viable.
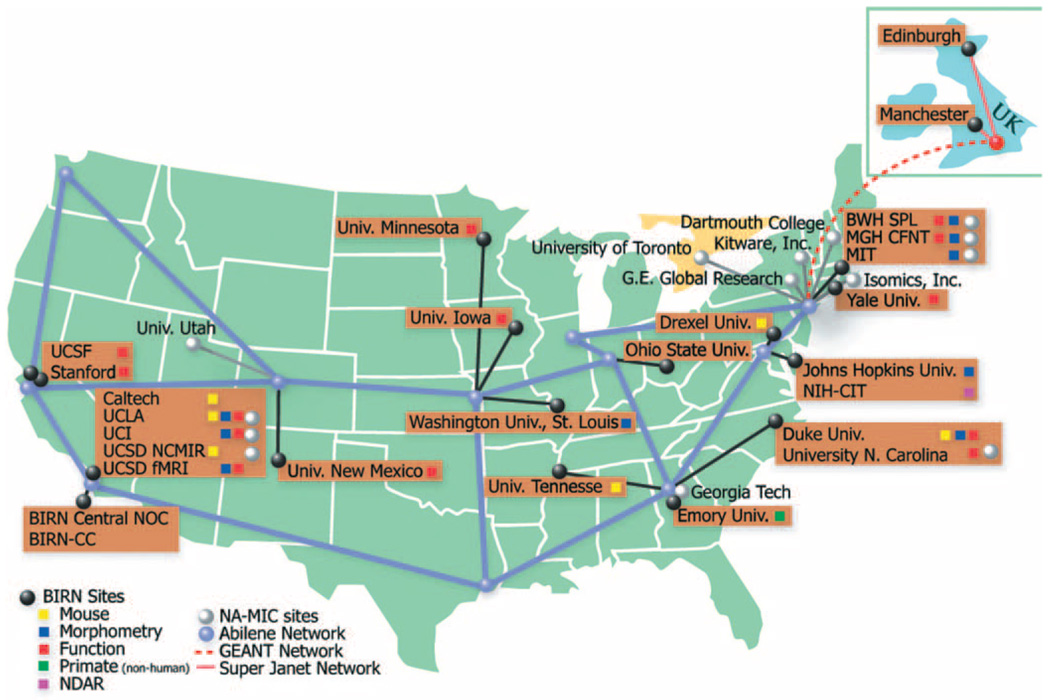
The aggregation of imaging, clinical, and behavioral data from multiple independent institutions and researchers presents both a great opportunity as well as a formidable challenge. Many research groups have well-established data collection and analysis procedures, as well as data and metadata format requirements that are particular to that group. Building a large-scale infrastructure to support these scientists and requirements has been accomplished by the Biomedical Informatics Research Network (BIRN; http://www.nbirn.net). BIRN has developed and deployed a federated and distributed infrastructure for the storage, retrieval, analysis, and documentation of biomedical data using state-of-the-art open source toolkits and grid technologies built on top of the high-speed internet-2/Abilene backbone (http://www.nbirn.net). The BIRN is a virtual community of shared resources consisting of a central coordinating center, four test beds, and other associated collaborative projects [e.g., the National Alliance for Medical Imaging Computing (NAMIC); http://www.na-mic.org/]. The BIRN coordinating center (BIRN-CC) is tasked with defining, integrating, packaging, fielding, and updating a complete end-to-end infrastructure in support of the biomedical test beds. More specifically, the BIRN-CC is responsible for hardware, the grid middleware for security, data, and computation, the data mediation and integration environment, and the BIRN portal, as well as the underlying system software, and the BIRN packaging of scientific tools and biomedical applications that have been defined by the BIRN scientific test beds. The three test beds focus on different imaging technologies in animal models or in human populations (Fig. 1).
Fig. 1.
Map of BIRN participating sites and BIRN nodes.
The Function BIRN test bed is working to understand the underlying causes of schizophrenia and to develop new treatments for the disease. The effort brings together researchers in different aspects of functional neuroimaging to apply recently developed multimodal and interdisciplinary techniques to investigate the neural substrates of schizophrenia. The scientific goals of the project are to determine the role of frontal and temporal lobe dysfunction in schizophrenia, and to assess the impact of treatments on functional brain abnormalities.
Morphometry BIRN participants are examining neuroanatomical correlates of neuropsychiatric illnesses in disorders such as unipolar depression, mild Alzheimer’s disease, and mild cognitive impairment. Through large-scale analyses of patient population data acquired and pooled across sites, these scientists are investigating whether brain structural differences correlate to symptoms such as memory dysfunction or depression, and whether specific structural differences distinguish diagnostic categories.
Researchers in Mouse BIRN are studying animal models of disease at different anatomical scales to test hypotheses associated with human neurological disorders. The aim is to share and analyze multiscale structural and functional data, and ultimately to integrate them with genomic and gene expression data on the mouse brain. The ongoing collaborations in basic mouse models of neurological disorders include animal models of relevance to schizophrenia, Parkinson’s disease, brain cancer, substance abuse, and multiple sclerosis. Correlated multiscale analyses of data from the Mouse BIRN projects promise to provide a comprehensive basis upon which to interpret signals from the whole brain relative to the tissue and cellular alterations characteristic of the modeled disorder.
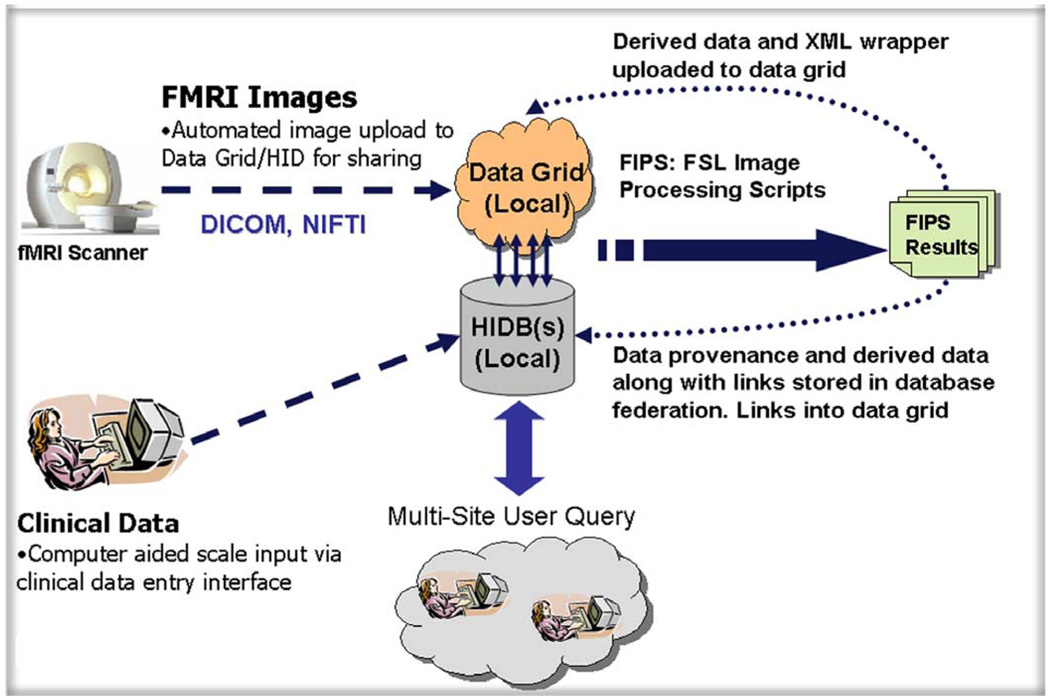
The development and deployment of a distributed end-to-end infrastructure to support the collaborative biomedical imaging research must integrate tools for data acquisition, data analysis workflows, and data retrieval/interpretation systems with the core enabling infrastructure components and middleware. Common workflows in support of the biomedical imaging consist of data import from scanning systems, storage of clinically relevant information in community, or laboratory databases, preprocessing and analysis of imaging and clinical information, and storage of the derived data back into a data management system (Fig. 2). BIRN has focused its efforts on supporting each of these steps in the context of the distributed infrastructure. In addition, BIRN has provided access to software systems and computational resources that sites could not realistically support or gain access to in the past.
Fig. 2.
Dataflow diagram showing clinical data being entered into a local database, images uploaded to data grid. Multiqueries run against the federated databases to identify interesting datasets. Datasets are downloaded and analyzed. Analysis results are uploaded to the data grid and linked back to local database.
II. NATIONAL GRID INFRASTRUCTURE
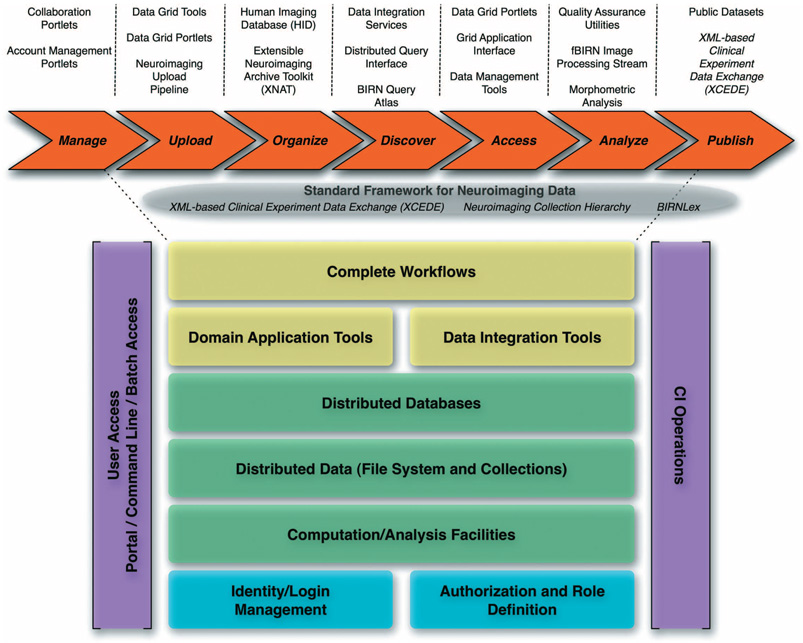
This overall infrastructure consists of distributed data collections hosted on dedicated storage and computational resources located at each participating site, a federated data management system, and data integration environment, an extensible Markup Language (XML) schema for data exchange, and analysis pipelines, designed to leverage both the distributed data management environment and the available grid computing resources, and visualization environments to enable data discovery and interpretation by domain scientists (Fig. 3).
Fig. 3.
BIRN integrated cyber-infrastructure showing the tools developed to facilitate data management, uploading, organization, discovery, access, analysis, and publications.
A. Hardware Infrastructure
The BIRN hardware infrastructure consists of physical racks deployed at each participating site (see Fig. 1), and a collection of services and servers hosted by the BIRN-CC. Currently, there are over 25 BIRN racks installed. Each rack supports a standardized configuration of BIRN applications and data storage. The physical racks consist of a gigabit Ethernet switch managing separate internal and external networks, a grid point of presence node (GPOP), a general compute node, a network statistics system, a minimum of 1 TB network attached storage, a uninterruptible power system (UPS), and a direct power redundancy. All systems are dual processor with Linux® operating system installations. The GPOP is the main externally visible node, and contains the data grid and middleware services. As systems age, they are replaced with current technologies. There are over 45 TB of available storage attached to BIRN racks across the consortium with more than 30 TB to be brought online in the near future. Network and data grid monitoring tools provide real-time system performance and availability statistics. In order for the BIRN-CC to efficiently deploy this integrated software stack across the entire BIRN infrastructure, the BIRN-CC has adopted the National Partnership for Advanced Computational Infrastructure (NPACI) Rocks Cluster Toolkit [1], a set of open-source enhancements for building and managing Linux®-based clusters. While BIRN racks are not traditional high-performance computing (HPC) clusters (each node in a BIRN rack has a specialized function), Rocks has enabled the BIRN-CC to automate the loading of its software stack, so that, after a few configuration parameters are provided, each resource is then deployed in an automated fashion. This ensures consistent configuration across all grid sites, provides greater reliability, simplifies the diagnostic efforts to correct problems, and thus, increases the availability of the BIRN resources. As the BIRN continues to grow, the Rocks Cluster Toolkit will continue to evolve to support more platforms and a greater heterogeneity of site configurations.
B. Middleware Stack
The grid middleware enables the assembly of geographically disparate resources into an application-specific virtual resource. The low-level grid software layer for the BIRN is built upon a collection of community-accepted software systems and services distributed as part of the National Science Foundation (NSF) Middleware Initiative (NMI; http://archive.nsfmiddleware.org). A key component of the NMI distribution, the Globus Toolkit [2], [3] supplies the essential grid services layer that includes authentication, encryption, resource management, and resource reporting. In addition, Condor [4], [5], a specialized workload management system for compute-intensive jobs is utilized. The Globus combined with Condor provide the computational and security components that allow test bed researchers to launch long running jobs to any available computing resource on the BIRN grid, without having to worry about where the job runs, which communications protocols are supported, and how to sign on to different computing systems.
C. Data Grid
The technological challenges of accessing data across geographically distributed sites are formidable. Storing and accessing the data can be achieved in a centralized data warehouse or, as the BIRN has chosen, in a distributed data grid (Fig. 1). The storage resources broker (SRB) [6] was chosen as the distributed data collections system because of its maturity and ability to provide a uniform access interface to different types of storage devices (http://www.sdsc.edu/srb/index.php/Main_Page). The SRB provides a uniform application programmer interface that can be used to connect to heterogeneous and distributed resources, allowing users to seamlessly access and manage these data sets. Also, the physical location of files is abstracted from the users and application programs, using a logical uniform resource identifier (URI) string to uniquely identify image files across the distributed sites. The hallmark of the SRB system is the ability for investigators to establish a hierarchy, and the appropriate access permissions, for multiple institutions and their researchers to contribute heterogeneous data. Each participating institution has a sufficient read and write access to the formal data hierarchy, regardless of their location and environment. Regardless of which site and what type of environment, the hierarchy looks and acts the same. While technologies such as iSCSI might perform faster than the SRB, configuring mount points, access permissions, firewalls rules for hundreds of users across 20 institutions would not be feasible for the types of projects that the BIRN infrastructure can handle today. In addition, the SRB supports file and directory metadata in the form of key-value pairs, allowing any relevant metadata to be attached and searched within the data grid.
D. Security
A key-enabling component of the middleware is the common security model. The de facto standard for grid security is the grid security infrastructure (GSI) [3], [7]. GSI is a public key-based X.509 conforming system that relies on trusted third parties for signing user and host certificates. Typical usage models require that each user be assigned a user credential consisting of a public and a private key. Users generate delegated proxy certificates with short life spans that get passed from one component to another, and form the basis of authentication, access control, and logging. Users can typically manage the credentials (and proxy) manually or use a number of command line-based utilities to manage their credentials.
Despite the general acceptance of the GSI and its use over the course of many years, GSI-based security systems are known to be difficult for administrators to deploy and for users to use. In order to provide a suitable solution for researchers, the grid accounts management architecture (GAMA) [8] system was adapted for use in the BIRN infrastructure. In addition to the core GAMA services, the GAMA server contains a collection of third party components: the BIRN certificate authority and MyProxy [9], [10] for credential management.
With GAMA, end users do not need to know anything about the grid security, credentials, proxies, or other technical matters. They simply request an account using a typical Web form interface, and after the account is created, they log in to the portal using a familiar username/password combination. Once logged into the portal, the portal has a proxy certificate that can be used for a secure access to the SRB server to retrieve data files or submit jobs to the Condor or Globus gatekeepers deployed throughout the BIRN infrastructure.
However, many applications, in particular, visualization packages, run on the client side. To provide a bridge between the BIRN infrastructure and this collection of domain-specific biomedical applications, BIRN has created a portal compliant Java grid interface. This platform independent “grid wrapper” brokers communications and information/data transfer between the application (running on the users desktop) and BIRN resources managed via the portal. Using the Java Webstart, the portal is able to delegate the user’s proxy credentials to the user’s workstation, where the grid interface is able to transparently access the data grid for the user.
III. DATA ACQUISITION AND MANAGEMENT
Building on top of the BIRN hardware and data grid infrastructures are a number of data management, analysis, information, and visualization systems (Fig. 3). Each system is an open source, and is provided free to the community. The tools discussed next are not an exhaustive list, but feature two data management systems developed with different data models and access patterns, both functioning on the BIRN distributed infrastructure, and an image quality assurance tool. For more information regarding the large number of open source tools built upon the BIRN infrastructure, please visit http://www.nbirn.net/tools/index.shtm.
A. Human Imaging Database (HID)
The HID [11] is an open-source, extensible database schema implemented in Oracle (http://www.oracle.com) 10g, 9i, and PostgreSQL (http://www.postgresql.org/) 7.x, 8.x, and associated three-tier J2EE application environment for the storage and retrieval of biomedical data designed to operate in a federated database environment. The HID at a particular site can be extended to contain relevant information concerning the research subjects used in an experiment, subject assessments, the experimental data collected, the experimental protocols used, and any annotations or statistics normally included with an experiment. The database is composed of an extensible schema and structured core. The core database contains a hierarchical description of an experiment and how experimental protocols relate to this hierarchy. Each descriptor in the database consists of a “base tuple” that defines the minimum informational requirements of that descriptor. For example, the base description of an experimental event (i.e., stimulus) contains the base information required to describe when that event occurs during an experimental protocol; the actual information regarding the specifics of various stimuli (e.g., the frequency of a tone stimulus) are stored as extended information in the database. Therefore, the database can be extended for various experiments utilizing these extended tuples that can be reused and/or modified for other experiments. In addition, the database contains an extensible framework for the definition and storage of clinical assessment and demographic data. This dedicated section of the schema handles the storage of assessment data from a wide variety of assessments and their modifications (i.e., assessment version control). The information is stored that allows for the annotation of the status or quality of assessment data. All missing data are coded to differentiate between data that were not entered for unknown reasons and those that the subject declined to answer or other possibilities that can affect the interpretation of resulting analyses. Some of the data quality measures put in place for clinical data includes double-entry, data type and range validation, and manual validation and logging for those data that fail the double-entry, prior to being exposed to the mediated query services. There are currently 11 federated HID databases, ten Oracle, and one PostgreSQL versions, storing clinical information on more than 419 subject imaging visits and 3174 subject assessments. The image and derived data are physically stored in the data grid and linked back to the HID located at the site that imported the data into the system. The HID environment also contains: 1) an intuitive web-based user interface that can be used for the entry and management of the subject’s data. A core component of this interface involves the management of behavioral and/or clinical data that uses modules that streamline the development of on-line forms for entry and maintenance of large numbers of measures and 2) a data integration engine that builds on top of the BIRN data integration environment, allowing multiple sites running the HID to create a federated database, so that these sites can be queried as a single database resources from the web-based user interface.
B. Extensible Neuroimaging Archive Toolkit (XNAT)
XNAT is a software platform designed to facilitate common management and productivity tasks for neuroimaging and associated data [12]. XNAT is an open source Java-based application. It follows a three-tiered architecture that includes a data archive, user interface, and middleware engine. XNAT uses an XML data model from which a relational database is generated.
XNAT implements a workflow to support the quality, integrity, and security of data from acquisition, and storage of analysis and public sharing. Imaging data from the scanner enter the workflow using one of several transfer mechanisms, including the digital imaging and communications in medicine (DICOM) (http://medical.nema.org/) “pushes,” secure file transfer protocol (SFTP), or portable hard media. Nonimaging data (e.g., clinical assessments, demographics, genetic measures) are entered via web-based forms, spreadsheet uploads, or XML. Newly entered data are placed in a “virtual quarantine” until an authorized user validates that the integrity of the data is intact. Once the data have been validated, they are moved into a secure archive. The archive unifies the data acquired from various sources associated with a study into a single-integrated resource. Archived data are made available to additional authorized users and project-specific automated processing and analysis pipelines. XNAT’s web-based user interface provides tools for monitoring the XNAT workflow and for exploring the resulting archive. As XNAT-managed studies progress, the data can be made available to successively broader groups of users, from collaborators to reviewers to the general scientific community.
A more complete description of the HID and XNAT’s features, tools, and design, as well as source code and installation packages, are available at http://www.nbirn.net/tools/index.shtm and http://www.xnat.org.
C. XML-Based Clinical Experiment Data Exchange Schema (XCEDE)
The BIRN test beds generate many types of data, including imaging data, clinical assessments, results from statistical analysis, behavioral data, and many others. One of the major goals of the BIRN is to share this data with the public. XCEDE is an XML-based data exchange schema designed to facilitate metadata transfer between databases (http://www.nbirn.net/tools/index.shtm), within and between software tools, and between the BIRN and external sites. XCEDE consists of several broadly scoped modules dedicated to representing metadata of various types, a few of which include the following.
1) Experiment hierarchy
XCEDE supports a multilevel hierarchy corresponding to the project, subject, visit, study, and series levels in a clinical experiment. These generic levels can be “subclassed” using standard XML schema type derivation to extend or restrict the level to support modality-specific metadata, such as that in a clinical screening visit, or an MRI study.
2) Event data
An “event” specification allows for the representation of time intervals annotated with arbitrary name-value pairs that help to characterize the intervals. This module is used to store many types of time-tagged data, such as button responses to stimuli during a functional NRI (fMRI) scan, metadata for the stimuli themselves, or image quality assurance (QA) measures assigned to each acquired volume in the same fMRI scan.
3) Binary data interface
XCEDE provides an abstract data reading interface intended to abstract away the particulars of reading any particular data format and provide a common interface to the binary data within. As this interface is formatagnostic, it can be used, for example, to “wrap” data in most MRI data format [NIfTI-1 (http://nifti.nimh.nih.gov/), DICOM, management interactive network connection (MINC) (http://www.bic.mni.mcgill.ca/software/minc/), and several others].
4) Data provenance
XCEDE supports the documentation of the processing workflow steps that brought the data to its current state. The goal is to allow one to replicate, as closely as possible, the same pipeline (with the same versions of software) at some future time for data verification or recreation.
The XCEDE XML schema has been used to support the extraction of activation maps from a common brain image analysis tool, the statistical parametric mapping software (SPM, http://www.fil.ion.ucl.ac.uk) [13]. Members of several BIRN test beds and related groups [functional magnetic resonance imaging data center (fMRIDC); http://www.fmridc.org/f/fmridc, NIfTI; http://nifti.nimh.nih.gov/] are currently developing XCEDE version 2, the goal of which is to streamline and extend the schema based on our experience implementing and deploying the current version in and outside the BIRN.
D. Quality Assurance Utilities
A critical activity in the BIRN collaboratory that builds on the data management strategies described earlier is ensuring the quality of the data shared within the collaboratory and with the larger research community. To that end, the BIRN has released to the public several QA measurement tools that are in active use within the BIRN. One is a tool designed to measure characteristics of the fMRI signal, such as signal-to-fluctuation-noise ratio (SFNR), generated by a scanner on an agar phantom [14]. Another is a tool designed to collect many metrics on fMRI scans of human subjects, such as center-of-mass, mean intensity, per-slice spikiness, and put them both in human-readable HyperText Markup Language (HTML) pages (Fig. 4) for viewing in a standard web browser and in computer-readable XML files (using the XCEDE events module). The XML files can then be used, for example, by subsequent analysis tools in a pipeline to automatically recognize outlier time points and deal with them appropriately. The phantom QA tools are being run by all Function BIRN sites regularly on upload of their QA scans, and the results are placed in an online, federated database. The human fMRI QA tools have been expanded significantly since their introduction, based on the feedback from BIRN researchers, and are now being used by Function BIRN sites, and will soon be integrated into the fBIRN image processing stream (FIPS).
Fig. 4.
Example of image QA report for multiple auditory oddball EPI data collected during an fMRI experiment.
IV. DATA ANALYSIS
Once data have been collected, checked for quality, and made available to the BIRN community, it is now available for analysis by any researcher within the collaboratory. This enables many analyses to be completed, simultaneously accelerating the scientific discovery process. However, in order to enable easier construction and sharing of these analyses, various tools are being developed.
A. FBIRN Image Processing Stream (FIPS)
The purpose of the BIRN infrastructure is not only to store data in a reliable and retrievable way, but also to provide project management for the analysis of the data. To this end, we have developed FIPS, a package for the comprehensive management of large-scale multisite fMRI projects, including data storage, retrieval, calibration, analysis, multimodal integration, and quality control. There are many challenges associated with the distributed analysis of large-scale, multisite fMRI data. These include: 1) assuring identical analyses across the member sites; 2) maintaining flexibility in the setting of analysis pipeline components and parameters; 3) unburdening the users from having to know the details about a particular data set; and 4) automatically checking the quality of the data. The FIPS allows the user to analyze data in a query-driven fashion, i.e., the user specifies match criteria (e.g., subject diagnosis and age), and the FIPS finds the matching data in the BIRN hierarchy, analyzes all of it, and places the results back into the hierarchy. The configuration of the analyses is divided into data-specific and data-independent parts. The data-specific parameters are specified at the time the data are uploaded and are stored with the data. For example, these parameters include the slice timing and number of dummy scans (which can change from site-to-site) and stimulus schedule (which can change from run-to-run). The data-independent parameters include the amount of spatial smoothing to use, whether to include the slice-timing correction, the shape of the hemodynamic response, and hypotheses to test. These are specified only once, regardless of how many data sets are to be analyzed. These parameters are stored outside of the hierarchy in one location that is available to all member sites. When a particular fMRI data set is analyzed, the dataindependent and the data-specific parameters are automatically retrieved and combined to tailor the analysis to the given data set. Currently, the analysis is accomplished using the Functional Magnetic Resonance Imaging of the Brain (FMRIB) Software Library (FSL, http://www.fmrib.ox.ac.uk/fsl) and FreeSurfer (http://surfer.nmr.mgh.harvard.edu), but is general enough to incorporate other software packages.
B. FIPS Preprocessing Modules
The FIPS architecture includes modules that allow fine grain control over fMRI preprocessing steps, allowing researchers to explicitly specify both the order and specific tool used for each preprocessing step. These modules are typically wrappers, which call specific tools from FSL, analysis of functional neuro imaging (ANFI), and/or other image processing packages to perform the bulk of the preprocessing operations, with these XCEDE aware modules recording data provenance information, handling file conversion issues, and restructuring the data as required for each specific tool. To date, FIPS preprocessing modules are available for motion correction, B0 field inhomogeneity correction, slice timing correction, and spatial smoothing. These modules can be automatically invoked during an FIPS analysis to perform preprocessing on the fly or can be explicitly run prior to any FIPS or non-FIPS analysis, where a traditional fMRI preprocessing is desired. At the moment, the FIPS preprocessing modules run on data extracted from the SRB (either on a local laboratory resource or on a large scale distributed computational resource), and produce an output dataset, which is then uploaded to the data grid and linked into the federated database (Fig. 2).
V. BIRN PORTAL AND WORKFLOW MANAGEMENT SYSTEM
The ability to send data through a succession of software programs is critical for the successful analysis of complex images. Over the years, various BIRN sites have developed applications that are “data pipelines,” many of which are simple scripts, but some of which are complex applications to handle a series of processes. One such application is the Louisiana Optical Network Initiative (LONI) pipeline [15] from the Laboratory of NeuroImaging at the University of California at Los Angeles. Others in use include the Kepler pipeline [16] from the University of California at San Diego and the j-business process management (BPM) workflow engine from JBoss (http://jboss.com). Although the pipelines are effective in their various local environments, it is much more difficult to export these environments to other researchers, not familiar with these tools, and they are not designed to operate in environments where a high degree of collaboration is required in a calculation. In order to allow for the successful sharing of such data pipelines, a system was envisioned that could consume the existing pipeline applications and achieve the following goals: 1) allow data pipelines produced by the BIRN to be made available to domain scientists inside and outside of the BIRN collaboratory; 2) allow a consistent environment for these pipelines, regardless of pipeline application software, to be maintained with special attention to metadata and data provenance; 3) allow study metadata to be tightly organized across groups to allow for collaboration and comparison of results; and 4) be made available through the BIRN portal environment.
A. BIRN Portal
A critical activity area for the BIRN has been the development of an effective and intuitive interface to the BIRN cyber-infrastructure that facilitates and enhances the process of collaborative scientific discovery for domain scientists in the BIRN test beds. Ubiquitous access for all users has been accomplished by deploying the BIRN portal, which:
provides transparent and pervasive access to the BIRN cyber-infrastructure requiring only a single username and password;
provides a scalable interface for users of all backgrounds and levels of expertise;
provides customized “work areas” that address the common and unique requirements of test bed groups and individual users;
has a flexible architecture built on emerging software standards, allowing for a transparent access to sophisticated computational and data service; and
requires a minimum amount of administrative complexity.
More than a simple web interface, the BIRN portal environment is designed to provide the integrated collection of tools, infrastructure, and services that BIRN test-bed researchers and databases users need access to in order to perform comprehensive and collaborative studies from any location with an Internet access. The BIRN portal environment is built on top of GridSphere (http://www.gridsphere.org; Novotny et al., 2004), a leading open-source portlet environment.
B. Workflow Management Portlet
The main system software is divided amongst a portal interface and an execution server. The system relies on uploads and downloads of images and other accompanying data to and from the data grid, which provides a way to access data sets and resources based on their attributes and/or logical names rather than their names or physical locations, and allows file security to be managed on a network-shared resource. The execution server has access to multiple execution environments (e.g., Condor, jBPM, LONI pipeline). The execution sever utilizes jBPM as the principle engine for scheduling and executing domain science and community developed data pipelines within the portal, because it is a reliable, open-source workflow engine that is particularly geared toward making human handoffs in a workflow. Its “out of the box” functionality includes a set of services that allows breakpoints in a workflow to be defined where the workflow will enter a “wait” state until human intervention occurs. This gives the chance for handoffs between groups to occur and intermediate calculations to be checked.
The development of this portal-based workflow application framework allows BIRN applications that previously were not generally available to become accessible to the clinical researcher, without them having to worry about the environment in which the data pipeline was developed. This expands the impact that the BIRN can make on the clinical research, and allows efficient sharing of available hardware resources. Additionally, the workflow application allows for a stabilization of the BIRN calculation process and the ability of more efficient collaborations. Setting up the BIRN analysis portal allows general use of BIRN resources, and enables effective collaborations between sites. It allows greater exploration of recalculated experiments and the ability to routinely explore complex parameter spaces.
VI. LARGE-SCALE COMPUTATION WITHIN BIRN
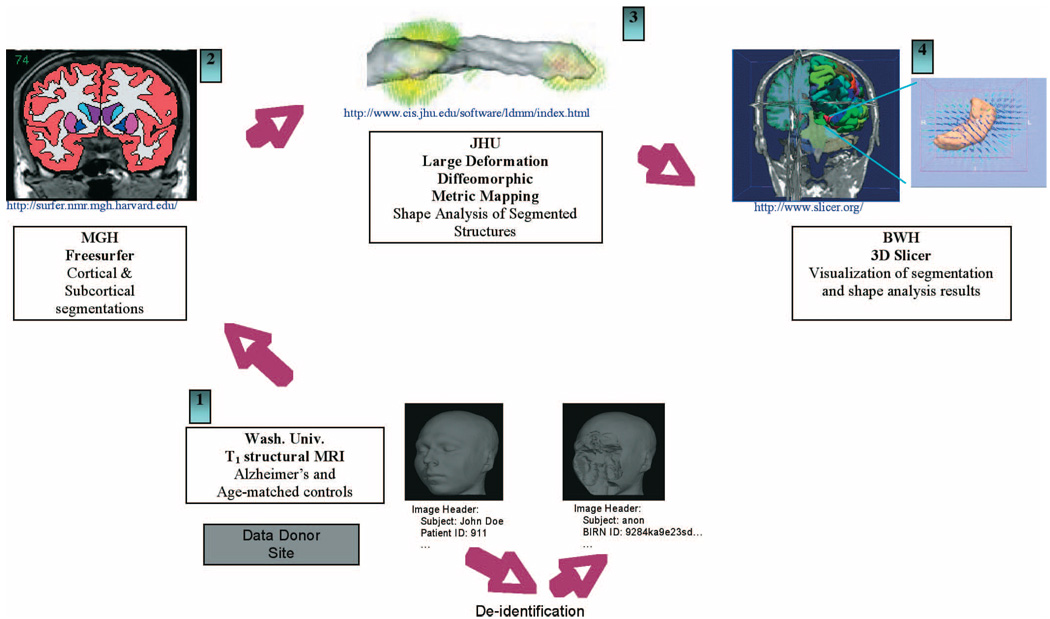
In addition to the construction, management, and sharing of analyses, BIRN researchers also require access to large-scale computational resources to be able to perform certain analyses. An example of such a workflow process is the semiautomated shape analysis pipeline (SASHA), as shown in Fig. 5. First, 3-D structural MRI data of the brain with good gray–white matter contrast-to-noise ratio is acquired at a participating site. In order to be shared, the image data has to be deidentified within the site’s firewall: patient information is removed from the image headers and face information is stripped from the images while leaving the brain intact. The defacing tool was developed to minimize the impact of the defacing procedure on the brain tissue [17]. A behavioral study was then conducted to validate the effectiveness of the defacing procedure by asking subjects to match photographic images with 3-D reconstructed views of the defaced MRI images [18]. The images were found to be sufficiently unidentifiable even for persons familiar with those depicted in the rendered image. The deidentified data then needs to be uploaded to the data grid where it can be accessed by other participating sites. Second, the deidentified structural brain MRI data is automatically segmented using Massachusetts General Hospital’s (MGH) FreeSurfer morphometry tools. The derived segmented data (e.g., the hippocampal surfaces) was consumed by the Johns Hopkins University’s (JHU) shape analysis using their large deformation diffeomorphic metric mapping tool (LDDMM). LDDMM because of its computational demands requires numerous processor hours to complete benefiting from access to the BIRN computational grid and support from the TeraGridTM project (http://www.teragrid.org/). The combined morphometric results (surfaces, volumes, labels, deformation fields) can be viewed from the database using Brigham and Woman Hospital’s (BWH) 3-D Slicer as the common visualization platform.
Fig. 5.
Integration of segmentation, metric mapping, and visualization software located at distributed sites working together through integrative workflows built on the BIRN distributed infrastructure.
The preliminary study, which involved only 45 subjects, used more than 30 000 processor hours and generated more than four terabytes of data. The resulting data were uploaded into the BIRN data grid for sharing and further analysis. From this analysis, it is evident that the use of these advanced analyses provides a powerful means of distinguishing shapes and providing the neuroanatomist an increased understanding of diseases and disorders with greater statistical power. More specifically, this initial study details a potential mechanism for diagnosing subjects through the utilization of noninvasive imaging methodologies and advanced computational methods. This initial study is now being followed by a much larger study involving data from more than 100 subjects.
VII. DATA INTEGRATION AND VISUALIZATION
The neuroscience research community deals not only with large distributed databases, but also with highly heterogeneous sets of data. A query may need to span several relational databases, ontology references, spatial atlases, and collections of information extracted from image files. To that end, the BIRN-CC has deployed a data source mediator that enables researchers to submit multisource queries and to navigate freely between distributed databases. This data integration architecture for BIRN builds upon work in the knowledge-guided mediation for integration across heterogeneous data sources [19]–[21]. In this approach, the integration environment uses additional knowledge captured in the form of ontologies, spatial atlases, and thesauri to provide the necessary bridges between heterogeneous data. Unlike a data warehouse, which copies (and periodically updates) all local data to a central repository and integrates local schemas through the repository’s central schema, this mediator approach creates the illusion of a singleintegrated database while maintaining the original set of distributed databases. This is achieved via so-called integrated (or virtual) views—in a sense the “recipes” describing how the local source databases can be combined to form the (virtual) integrated database. It is the task of the data integration system to accept queries against the virtual views and create query plans against the actual sources whose answers, after some postprocessing, are equivalent to what a data warehouse would have produced. However, querying and retrieving data distributed within the collaboratory is only the first step, the scientist must then also be able to utilize this information in an easy and intuitive fashion.
A. BIRN Query Atlas
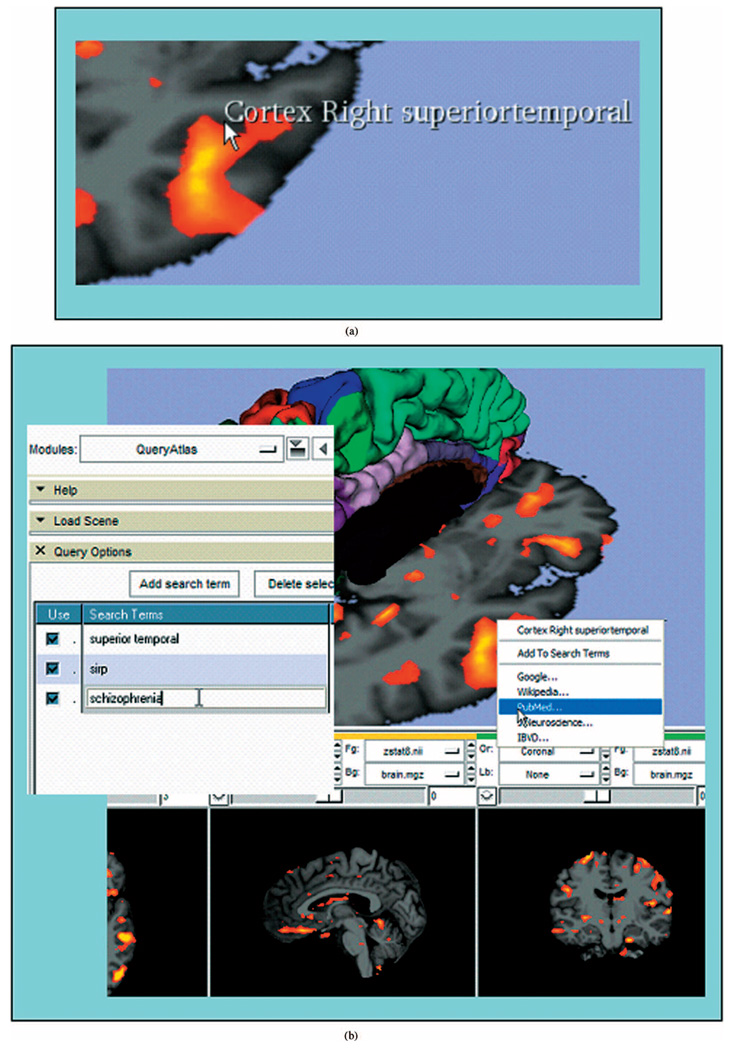
The extraordinarily rich combination of data resources provided by the Function BIRN is a great benefit to researchers, but it presents a challenge when seeking to develop a cohesive scientific interpretation of issues that span a variety of knowledge domains and scientific disciplines. We are addressing this issue of “information overload” by developing an intuitive, interactive graphics application that allows users to search databases, the internet, and the scientific literature for information that is specifically relevant to the image data they are viewing. This system, called the BIRN query atlas, is tightly integrated with the rest of the informatics infrastructure, and builds on the notion that the functional and structural MRI information has been ‘tagged’ with metadata defining not only an anatomical segmentation of the images, but also clinical and demographic information about the subject and protocol information about the image acquisition. The BIRN query atlas organizes this information for easy access when reviewing a particular analysis result. Fig. 6(a), for example, is a screen shot from an interactive session showing a structural MRI image (grayscale) with superimposed fMRI activation map (orange). As the user moves the cursor over the structural image, the name of the anatomical structure is shown as a text overlay. By pressing the right mouse button, the user can access a context menu of search options for that structure; complex queries can be constructed by combining interactively selected anatomical search terms with additional terms drawn from the image metadata, as illustrated in Fig. 6(b). The BIRN query atlas is built atop the 3-D Slicer application software (http://www.slicer.org) and relies on FreeSurfer cortical and subcortical structural analyses (http://surfer.nmr.mgh.harvard.edu) in addition to directly loading the output of the FIPS pipeline. The design and implementation of the BIRN query atlas have been driven by requests from the BIRN clinical user community for more powerful tools to interpret complex functional/structural data in anatomical context, and to draw upon the growing body of scientific literature and other databases that may bear upon the BIRN areas of research. The Function BIRN informatics group is looking to improve the accuracy of user searches through the addition of machine learning techniques that adapt the search results based on the feedback from previous searches.
Fig. 6.
(a) BIRN query atlas interactive anatomical exploration of a functional/structural imaging study. (b) Formulation of a multiterm query using anatomical information collected interactively and supplemental query terms derived from metadata about the images.
VIII. CONCLUSION
The availability of multimodal data and large human imaging datasets have become more prevalent in recent years, but combining these datasets across sites and providing researchers with an infrastructure to both share and query across species has been largely inaccessible to the majority of research centers. The BIRN consortium and its associated test beds have been focused on building a distributed infrastructure for sharing and combining data, both within a modality and across modalities and species. The BIRN will provide a central data repository for scientists wanting to share their data. The BIRN is committed to building open-source tools leveraging on the state-of-the-art cyber-infrastructure, and providing these tools and infrastructure to the larger research community. The BIRN will continue to release new tools and updates available through the website www.nbirn.net.
ACKNOWLEDGMENT
The authors would like to thank all the numerous individuals across the BIRN test beds that made significant intellectual contributions to the ongoing research of the consortium. They further thank the Principle Investigators (PIs) from each of the test beds for their leadership and vision in this project: M. Ellisman, BIRN Coordinating Center, S. Potkin, Function BIRN, B. Rosen, Morphometry BIRN, and A. Toga, Mouse BIRN.
Biographies
David B. Keator is the Director of Scientific Computing at the University of California, Irvine Brain Imaging Center in the Department of Psychiatry and Human Behavior and the Chair of the Neuroinformatics Working Group of the Function Biomedical Informatics Research Network. His research areas include neuroinformatics, multi-modality data visualization, and computational problems in functional neuroimaging.
J. S. Grethe, photograph and biography not available at the time of publication.
D. Marcus, photograph and biography not available at the time of publication.
B. Ozyurt, photograph and biography not available at the time of publication.
S. Gadde, photograph and biography not available at the time of publication.
Sean Murphy is Medical Director of Research Computing at Partners Healthcare, overseeing the Research Patient Data Registry, a data warehouse used for repurposing clinical data for research. He is also a Principal Investigator of the “Informatics for Integrating Biology to the Bedside” NIH Roadmap project, and the Biomedical Informatics Research Network.
S. Pieper, photograph and biography not available at the time of publication.
D. Greve, photograph and biography not available at the time of publication.
R. Notestine, photograph and biography not available at the time of publication.
H. J. Bockholt, photograph and biography not available at the time of publication.
P. Papadopoulos, photograph and biography not available at the time of publication.
Contributor Information
David B. Keator, University of California, Irvine, CA 92697 USA (e-mail: dbkeator@uci.edu).
J. S. Grethe, University of California, San Diego, CA 92093 USA (e-mail: jgrethe@ncmir.ucsd.edu).
D. Marcus, Washington University School of Medicine, St. Louis, MO 63110 USA (e-mail: dmarcus@npg.wustl.edu).
B. Ozyurt, University of California, San Diego, CA 92093 USA (e-mail: iozyurt@ucsd.edu).
S. Gadde, Duke University, Durham, NC 27708 USA (e-mail: gadde@biac.duke.edu).
Sean Murphy, Laboratory of Computer Science, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02115 USA..
S. Pieper, Surgical Planning Laboratory, Brigham and Women’s Hospital, Boston, MA 02115 USA (e-mail: pieper@bwh.harvard.edu).
D. Greve, Center for Magnetic Resonance Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA 02115 USA (e-mail: greve@nmr.mgh.harvard.edu).
R. Notestine, University of California, San Diego, CA 92093 USA (e-mail: RNotestine@ucsd.edu).
H. J. Bockholt, Medical Investigation of Neurodevelopmental Disorders (MIND) Institute, Albuquerque, NM 87131 USA (e-mail: jbockholt@themindinstitute.org).
P. Papadopoulos, University of California, San Diego, CA 92093 USA (e-mail: phil@sdsc.edu)..
REFERENCES
- 1.Papadopoulos PM, Katz MJ, Bruno G. NPACI Rocks: Tools and techniques for easily deploying manageable Linux clusters. Proc. IEEE Int. Conf. Cluster Comput. 2001:258–267. [Google Scholar]
- 2.Foster I, Kesselman C. The Grid: Blueprint for a New Computing Infrastructure. San Mateo, CA: Morgan-Kaufman; 1999. Computational Grids. ch. 2. [Google Scholar]
- 3.Foster I, Kesselman C. The globus project: A status report. Proc. IPPS/SPDP’1998 Heterogeneous Comput. Workshop. :4–18. [Google Scholar]
- 4.Thain D, Tannenbaum T, Livny M. Condor and the grid. In: Berman F, Hey A, Fox G, editors. Grid Computing: Making the Global Infrastructure a Reality. New York: Wiley; 2003. [Google Scholar]
- 5.Tannenbaum T, Wright D, Miller K, Livny M. Condor—A distributed job scheduler. In: Sterling T, editor. Beowulf Cluster Computing With Linux. Cambridge, MA: MIT Press; 2002. [Google Scholar]
- 6.Rajasekar A, Wan M, Moore R, Schroeder W, Kremenek G, Jagatheesan A, Cowart C, Zhu B, Chen S, Olschanowsky R. Storage resource broker—Managing distributed data in a grid. Comput. Soc. India J., Spec. Issue SAN. 2003;vol. 33(no 4):42–54. [Google Scholar]
- 7.Lorch M, Kafura D. Supporting secure ad-hoc user collaboration in grid environments. presented at the 3rd Int. Workshop Grid Comput; Baltimore, MD. 2002. Nov., [Google Scholar]
- 8.Bhatia K, Muller K, Chandra S. GAMA: Grid account management architecture. presented at the IEEE Int. Conf. ESci. Grid Comput.,; Los Alamitos, CA. 2005. Dec, [Google Scholar]
- 9.Basney J, Humphrey M, Welch V. The MyProxy online credential repository. Softw.: Practice Exp. 2005;vol. 35(no 9):801–816. [Google Scholar]
- 10.Novotny J, Tuecke S, Welch V. An online credential repository for the grid: MyProxy. Proc. 10th Int. Symp. High Perform. Distrib. Comput. (HPDC-10) 2001:104–111. [Google Scholar]
- 11.Keator DB, Ozyurt B, Wei D, Gadde S, Potkin SG, Brown G, Grethe J Morphometry BIRN, Function BIRN. A general and extensible multi-site database and XML based informatics system for the storage, retrieval, transport, and maintenance of human brain imaging and clinical data. presented at the Annu. Meeting Organ. Hum. Brain Mapping, Florence; Italy. 2006. [Google Scholar]
- 12.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit (XNAT): An informatics platform for managing, exploring, and sharing neuroimaging data. NeuroInformatics. 2007;vol. 5(no 1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 13.Keator DB, Gadde S, Grethe JS, Taylor DV, Potkin SG FIRST BIRN. A general XML schema and SPM toolbox for storage of neuro-imaging results and anatomical labels. Neuroinformatics. 2006;vol. 4(no 2):199–212. doi: 10.1385/ni:4:2:199. [DOI] [PubMed] [Google Scholar]
- 14.Friedman L, Glover GH. Reducing interscanner variability of activation in a multicenter fMRI study: Controlling for signal-to-fluctuation-noise-ratio (SFNR) differences. Neuroimage. 2006;vol. 33(no 2):471–481. doi: 10.1016/j.neuroimage.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Rex DE, Ma JQ, Toga AW. The LONI pipeline processing environment. Neuroimage. 2003;vol. 19(no 3):1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- 16.Ludäscher B, Altintas I, Berkley C, Higgins D, Jaeger-Frank E, Jones M, Lee E, Tao J, Zhao Y. Scientific workflow management and the Kepler system. Concurrency Computat.: Practice Exp. 2005;vol. 18(no 10):1039–1065. [Google Scholar]
- 17.Bischoff-Grethe A, Ozyurt IB, Busa E, Quinn BT, FennemaNotestine C, Clark CP, Morris S, Bondi MW, Jernigan TL, Dale AM, Brown GG, Fischl B. A technique for the deidentification of structural brain MR images. Hum. Brain Mapping. 2007 Sep.vol. 28(no 9):892–903. doi: 10.1002/hbm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bischoff-Grethe A, Fennema-Notestine C, Kaelberer M, Brown G, Fischl B. The anonymization of cranial MR images: A behavioral study. presented at the Soc. Neurosci. Annu. Meeting; Atlanta, GA: Georgia World Congress Center; 2006. [Google Scholar]
- 19.Martone ME, Gupta A, Ellisman MH. E-neuroscience: Challenges and triumphs in integrating distributed data from molecules to brains. Nat. Neurosci. 2004;vol. 7:467–472. doi: 10.1038/nn1229. [DOI] [PubMed] [Google Scholar]
- 20.Ludascher B, Gupta A, Martone ME. Model-based information integration in a neuroscience mediator system. presented at the 26th Int. Conf. Very Large Databases (VLDB); Egypt: Cairo; 2000. [Google Scholar]
- 21.Gupta A, Ludascher B, Martone ME. An extensible model-based mediator system with domain maps. presented at the Proc. 17th Int. Conf. Data Eng. (ICDE); Germany: Heidelberg; 2001. [Google Scholar]