Abstract
Growing tumors face two major metabolic challenges—how to meet the bioenergetic and biosynthetic demands of increased cell proliferation, and how to survive environmental fluctuations in external nutrient and oxygen availability when tumor growth outpaces the delivery capabilities of the existing vasculature. Cancer cells display dramatically altered metabolic circuitry that appears to directly result from the oncogenic mutations selected during the tumorigenic process. An emerging theme in cancer biology is that many of the genes that can initiate tumorigenesis are intricately linked to metabolic regulation. In turn, it appears that a number of well-established tumor suppressors play critical roles in suppressing growth and/or proliferation when intracellular supplies of essential metabolites become reduced. In this review, we consider the potential role of tumor suppressors as metabolic regulators.
Keywords: Cancer biology, cellular metabolism, tumor suppressors, oncogenes, proliferation, apoptosis
During proliferation a cell must increase its biomass and replicate its genome prior to dividing to create two daughter cells. Thus, the cell must generate enough energy and acquire or synthesize biomolecules at a sufficient rate to meet the demands of proliferation. Cancer is essentially a disease in which cells have lost their normal checks on cell proliferation. By extension, it may not be surprising that tumor cells, in order to meet the increased requirements of proliferation, often display fundamental changes in pathways of energy metabolism and nutrient uptake (Garber 2006). Otto Warburg first proposed the theory in the 1920s that defects in energy metabolism, particularly in mitochondrial function, may be the root cause of cancer. This theory was largely based on his finding that tumor cells preferentially use glycolysis over mitochondrial oxidative phosphorylation (OXPHOS) for glucose-dependent ATP production even in the presence of ample oxygen to fuel mitochondrial respiration, a phenomenon known as the “Warburg effect” (Warburg 1956). This principle is exploited in clinical settings today through the use of 18F-deoxyglucose positron emission tomography (FDG-PET) to image tumors with increased glucose uptake. Enhanced glucose uptake visualized by FDG-PET correlates with poor prognosis and higher metabolic potential in many tumor types (Gatenby and Gillies 2004; Kroemer and Pouyssegur 2008), supporting the theory that alterations in cellular metabolism may contribute to the malignant phenotype.
The revolution in molecular biology over the past 30 yr has led to the theory that genetic lesions, through either oncogene activation or loss of tumor suppressors, drives cancer progression. As we discuss here, several of the mutations that lead to cancer also drive the altered metabolism of tumor cells (Vogelstein and Kinzler 2004). This fundamental metabolic switch (“metabolic transformation”) may confer a selective growth advantage and/or resistance to apoptosis to allow cancer cells to maintain mitochondrial bioenergetics and integrity during cell growth and proliferation (DeBerardinis et al. 2008). However, increased metabolic potential, while beneficial for unchecked growth, poses an inherent challenge for malignant cells. As a growing tumor with increased metabolic demands outstrips nutrient supply, it must enact a strategy to maintain cellular bioenergetics lest cell growth, proliferation, or viability be affected. Without such a strategy, cells undergo apoptosis. Thus, tumors face two distinct metabolic challenges: (1) how to modify their cellular metabolism to support enhanced cell growth and proliferation, and (2) how to engage strategies of metabolic adaptation to survive periods of metabolic stress and maintain viability as the cells accumulate. In this review, we discuss how oncogene and tumor suppressor networks influence cellular metabolism and bioenergetics to support growth and proliferation and survive metabolic stress. We also discuss the implications of metabolic reprogramming for tumor growth, and the potential to exploit changes in tumor metabolism for cancer therapy.
Glycolysis—metabolic reprogramming fuels cell growth and proliferation
The decision to proliferate presents a significant bioenergetic challenge for a cell. In nonproliferating tissues such as heart or muscle, cellular bioenergetics are directed toward OXPHOS. Enzymes of the tricarboxylic acid (TCA) cycle resident in the mitochondrion facilitate the oxidation of pyruvate and other substrates for maximal ATP production through electron transport-coupled OXPHOS. However, cell proliferation requires that bioenergetic resources and biosynthetic activity be redirected toward the duplication of all macromolecular components (DNA, membranes, and proteins) to ensure successful passage through the cell cycle. To meet this challenge, metabolic activities are reorganized in proliferating cells resulting in an anabolic shift in cellular metabolism.
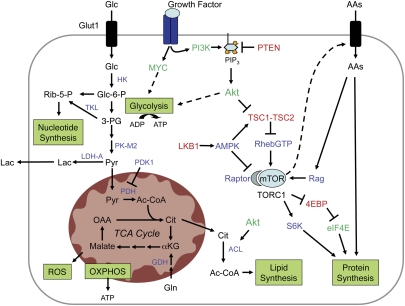
One of the primary metabolic changes associated with proliferating tumor cells is induction of aerobic glycolysis (Fig. 1). Glucose is a critical nutrient for proliferating cells (Holley and Kiernan 1974; Pardee 1974). It serves as the primary substrate for ATP generation and is an essential carbon source for biosynthesis of other macromolecules. In proliferating cancer cells, the majority of the pyruvate generated from glucose (>90%) is converted to lactate by lactate dehydrogenase (LDH-A), where it is readily secreted into the extracellular environment rather than oxidized to completion. By converting pyruvate to lactate, LDH-A recovers the NAD+ needed to maintain glycolysis. This step is critical for the maintenance of tumor proliferation in vivo (Fantin et al. 2006). At face value, the preferential use of glycolysis for ATP production appears to be wasteful form of metabolism. Although the energetic yield per molecule of glucose is much lower for aerobic glycolysis compared with OXPHOS, when glucose is in excess and flux through the pathway high, glycolysis has the potential to produce ATP in greater quantities and at a faster rate (Guppy et al. 1993).
Figure 1.
Metabolic pathways of growth and proliferation. Growth factor-independent activation of the PI3K/Akt and c-Myc pathways drives changes in cellular metabolism to promote cancer cell growth and proliferation. PI3K/Akt and c-Myc facilitate increased rates of glucose uptake and glycolysis. Lactate dehydrogenase (LDH-A) maintains glycolytic flux by converting excess pyruvate to lactate and regenerating NAD+ in the process. Akt signaling promotes increased surface expression of the glucose transporter Glut1 and enhances the activity of glycolytic enzymes. Glucose (Glc) has multiple catabolic and biosynthetic fates, including glycolytic processing for production of ATP and pyruvate, processing through the pentose phosphate shunt to generate ribose 5-phosphate (Rib-5-P) and NADPH for nucleotide biosynthesis, or entry into the mitochondrion for biosynthesis or ATP generation by the TCA cycle and electron transport chain (ETC). Glucose-derived citrate (Cit) is exported to the cytosol and processed by ATP citrate lyase (ACL) to acetyl-CoA (Ac-CoA), which is channeled into lipid production. Glutamine (Gln) is deaminated to form glutamate, which can be processed further in the mitochondria by glutamate dehydrogenase (GDH) to generate α-ketoglutarate (αKG) and maintain TCA cycle function. Reactive oxygen species (ROS) are the result of mitochondrial OXPHOS. The TORC1 complex, which contains mTOR and its binding partner Raptor, regulates protein translation. mTOR coordinates surface expression of amino acid (AA) transporters, and promotes translation initiation by stimulating ribosomal S6 kinase activity and relieving 4E-BP-mediated inhibition of eIF4E. Amino acids stimulate mTOR activity through the activity of the Rag GTPases. PI3K/Akt signals activate mTOR primarily through inhibition of the TSC1–TSC2 complex, thus releasing inhibition of RhebGTP and activating mTOR. Bioenergetic signals antagonize the TORC1 complex through the LKB1–AMPK pathway. Known oncogenes are shown in green, tumor suppressors in red, and effectors with undefined roles in tumorigenesis are shown in blue.
In addition to energy, glycolysis generates metabolic intermediates important for cell growth. Metabolism of glucose through the oxidative or nonoxidative arms of the pentose phosphate pathway (PPP) generates ribose-5-phosphate (Rib-5-P), a key intermediate in nucleotide biosynthesis. The oxidative arm of the PPP can also produce NADPH, which supplies a pool of reducing equivalents for both nucleotide and fatty acid biosynthesis. Recent data suggest that pyruvate kinase-M2 (PK-M2), an isoform of the rate-limiting enzyme for pyruvate generation that is preferentially expressed in tumor cells, regulates the flux of carbon into Rib-5-P and nucleotide synthesis (Christofk et al. 2008a). Interestingly, the enzymatic activity of PK-M2 is negatively regulated by phosphotyrosine signaling downstream from growth receptors (Christofk et al. 2008b). By regulating the amount of pyruvate generated by glycolysis, PK-M2 functions to direct the flow of carbon into pathways of nucleotide and fatty acid biosynthesis that support the proliferation of tumor cells. Glycolytic intermediates including 3-phosphoglycerate (3-PG) serve as carbon sources for amino acid and lipid synthesis. Finally, pyruvate can be imported into the mitochondria and converted into substrates for the production of additional amino acids or fatty acid synthesis, and used to maintain mitochondrial membrane potential. Thus, maintaining high rates of glycolysis allows cells to meet the energetic and biosynthetic demands of anabolic growth.
The mitochondrion—from engine to biosynthetic hub
The majority of macromolecules required for proliferation (i.e., lipids, proteins, and nucleic acids) are generated de novo from glucose (Bauer et al. 2005). The remaining pyruvate from aerobic glycolysis that is not converted to lactate (∼10% of total) enters the mitochondrion and is extruded from the TCA cycle at various steps for use in biosynthetic pathways. Thus, in addition to its role as cellular energy center, the mitochondrion takes up a new role as a biosynthetic hub to meet the increased biosynthetic demand during proliferation, converting metabolites such as pyruvate and glutamine into intermediates utilized by other biosynthetic pathways (Fig. 1). Despite the low overall percentage of pyruvate available for mitochondrial synthesis, the high rate of glycolytic flux provides sufficient amounts for biosynthesis.
Shunting of pyruvate into the TCA cycle generates a source of acetyl-CoA for the biosynthesis of lipids and amino acids. Acetyl-CoA condenses with oxaloacetate (OAA) to generate citrate, which is exported from the mitochondrion to the cytosol and converted back to acetyl-CoA by ATP citrate lyase (ACL). Inhibition of ACL results in decreased glucose-dependent lipid synthesis and impaired cell proliferation (Hatzivassiliou et al. 2005). This cytosolic pool of citrate-derived acetyl-CoA is essential for the synthesis of lipids (fatty acids, sphingomyelin, cholesterol, and isoprenoids). One risk of converting mitochondrial activity to biosynthesis is the loss of mitochondrial integrity due to depletion of TCA cycle intermediates. The export of citrate for lipid synthesis results in a net loss of OAA, the acceptor for pyruvate-derived acetyl-CoA, which must be regenerated to maintain the integrity of the TCA cycle. This can be achieved through the stepwise oxidation of glutamine in proliferating cells. Metabolic flux analysis has revealed that glutamine is used by proliferating cells to generate a pool of α-ketoglutarate, which can be metabolized through the TCA cycle to regenerate the OAA consumed by biosynthesis (DeBerardinis et al. 2007). This may explain why glutamine is an essential metabolite for proliferation (Griffiths and Keast 1990). Given the importance of glutamine-mediated anapleurosis to TCA function and cell proliferation, regulators of this process are likely to possess oncogenic properties.
Tumor suppressors and oncogenes constitute metabolic signaling networks
Many of the predominant mutations observed in cancer also control tumor cell metabolism (Vogelstein and Kinzler 2004), leading to the theory that oncogene and tumor suppressor networks influence the metabolic shift in cancer. A central regulator of metabolism in both nontransformed and transformed cells is the phosphatidylinositol 3-kinase (PI3K), a lipid kinase that regulates the levels of phosphorylated phosphatidylinositol (PIP3) at the plasma membrane. Growth factor-dependent activation of PI3K leads to the activation of downstream effectors including Akt and the mammalian target of rapamycin (mTOR) that coordinate metabolic activities that support cellular biosynthesis. In normal cells, PI3K activation is tightly controlled by dephosphorylation of PIP3 by the phosphatase PTEN, a potent tumor suppressor. Activity of the pathway is deregulated in cancer through a variety of mechanisms, including activating mutations in PI3K (Samuels et al. 2004, 2005; Jia et al. 2008) or PTEN loss (Cairns et al. 1997). Together, mutations in PI3K signaling constitute one of the most common sets of mutations in human tumors (Shaw and Cantley 2006). Enhanced PI3K/Akt signaling promotes metabolic transformation through multiple pathways, including (1) increased surface expression of nutrient transporters enabling increased uptake of glucose, amino acids, and other nutrients; (2) Akt-dependent stimulation of hexokinase and phosphofructokinase to drive glycolysis; (3) enhanced transcription of genes involved in glycolysis and lipogenesis; and (4) enhanced protein translation through Akt-dependent mTOR activation (Edinger and Thompson 2002; Bauer et al. 2004; Wullschleger et al. 2006).
Protein translation is essential for tumor growth, and deregulated translation itself can be tumorigenic (Mamane et al. 2004). mTOR coordinates protein synthesis by regulating amino acid uptake, tRNA charging, and translation initiation. mTOR functions as the central kinase in two distinct multiprotein complexes—known as TOR complex 1 (TORC1) and TOR complex 2 (TORC2), respectively—that vary slightly in composition and physiological function. mTOR enhances the initiation of cap-dependent translation by altering the activity of components of the translational machinery including inhibition of the translational inhibitor 4E-BP1, which in turn promotes translation initiation by the eIF4F complex, and stimulation of ribosomal S6 kinase. TORC1 activity is kept in check by the tumor suppressors TSC1/TSC2 and LKB1, which function to block mTOR activation through inhibition of the small GTPase Rheb; mutation of these genes leads to elevated mTOR activity and the development of hamartomas (Inoki et al. 2003; Corradetti et al. 2004; Shaw et al. 2004a). Akt regulates mTOR function by phosphorylating and inhibiting TSC2, which in turn disrupts the function of the TSC1–TSC2 complex, promoting mTOR activation (Inoki et al. 2002). In contrast, the bioenergetic sensor AMP-activated protein kinase (AMPK) phosphorylates the TSC1–TSC2 complex in an LKB-dependent fashion to promote mTOR inhibition (Inoki et al. 2003). Regulation of translation initiation by the abundance of tRNAs may represent an additional control point for transformation. Increased expression of RNA polymerase III-dependent initiator methionine tRNA results in increased cellular transformation (Marshall et al. 2008). Amino acid availability represents an additional regulatory step for protein translation. Signal transduction by mTOR promotes increased amino acid uptake by up-regulating and maintaining surface expression of amino acids transporters (Gottesdiener et al. 1988). Amino acids can directly influence the rate of protein synthesis through stimulation of mTOR. This is mediated by the Rag family of small GTPases, which interact with TORC1 and promote its activation in response to amino acid signals (Kim et al. 2008; Sancak et al. 2008).
Many prominent oncogenes including c-Myc, H-ras, src, and Akt stimulate anabolic metabolism in transformed cells (Dang and Semenza 1999; Elstrom et al. 2004). In particular, the c-Myc proto-oncogene is a potent regulator of multiple metabolic pathways essential for cancer growth (Dang et al. 2008). Myc is a transcription factor activated by growth factor stimulation and is essential for the regulation of proliferation and cell cycle entry in nontransformed cells. Several lines of evidence suggest that oncogenic amplification of c-Myc directly alters glucose metabolism and also regulates specialized biosynthetic activities required for successful cell division. Oncogenic c-Myc promotes increased aerobic glycolysis through the constitutive elevation of LDH-A (Shim et al. 1997; Osthus et al. 2000), as well as expression of enzymes involved in nucleotide and amino acid metabolism (Gordan et al. 2007). Interestingly, Myc transformed cells are exquisitely sensitive to glutamine levels and rapidly deplete their TCA cycle intermediates when glutamine availability is reduced (Yuneva et al. 2007). Recently, a novel role for c-Myc in the up-regulation of glutaminolysis associated with cancer has been proposed (Wise et al. 2008). The generation of potentially toxic levels of ammonia during c-Myc-dependent glutaminolysis may explain why oncogenic Myc expression often induces senescence or is proapoptotic (Wise et al. 2008). On the other hand, c-Myc-induced glutaminolysis provides cells with an abundant supply of NADPH to support anabolic synthesis. The recent identification of mutations in the NADP+-dependent enzyme isocitrate dehydrogenase (IDH1) also suggests that cancer cells select for enzymatic mutations that affect cytoplasmic NADPH production during transformation (Parsons et al. 2008).
Metabolic adaptation by the AMPK
Increased metabolic potential, while supportive of unchecked growth, poses a distinct metabolic challenge for tumor cells. Once a growing tumor outstrips its nutrient supply, it must enact strategies to maintain cellular bioenergetics (Fig. 2). Thus, how tumors engage strategies of metabolic adaptation to survive stress may contribute to cancer progression and outcome (i.e., metastasis). Intracellular metabolic sensors represent key intersection points between the environment and physiology. The AMP-activated protein kinase (AMPK) is a highly conserved heterotrimeric protein kinase complex that plays a central role in the regulation of cellular energy homeostasis, tethering the bioenergetic state of a cell to a number of critical biological functions aimed at preserving cellular energy and viability (Hardie 2007). AMPK was originally identified as a protein kinase allosterically regulated by AMP (Yeh et al. 1980). AMPK is responsive to changes in the cellular AMP:ATP ratio and is activated by stresses that stimulate ATP consumption or inhibit ATP production (Hardie 2003). As a cellular sensor of AMP levels, AMPK is activated under conditions of energetic stress, such as that triggered by nutrient deprivation or hypoxia. AMP binding to the γ subunit promotes both increased kinase activity of the α subunit and phosphorylation at Thr-172 by upstream kinases (Suter et al. 2006). Three upstream AMPK kinases have been identified to date; i.e., the tumor suppressor LKB1 (Hawley et al. 2003), calmodulin-dependent protein kinase kinase β (CamKKβ) (Hawley et al. 2005; Hurley et al. 2005; Woods et al. 2005), and transforming growth factor-β (TGFβ)-activated kinase-1 (TAK1) (Momcilovic et al. 2006; Xie et al. 2006). LKB1 is required for AMPK activation under conditions of bioenergetic stress including glucose withdrawal (Hawley et al. 2003; Woods et al. 2003; Shaw et al. 2004b).
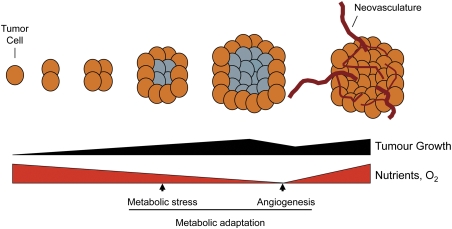
Figure 2.
Metabolic stress during tumor development. A solid tumor can outstrip its nutrient and oxygen supply as it grows, resulting in metabolic stress (tumors experiencing metabolic stress are depicted in gray). As a consequence, tumor cells must undergo a period of metabolic adaptation to survive this metabolic stress or undergo apoptosis. Angiogenesis and neovascularization of the tumor is one strategy of metabolic adaptation used by tumors to relieve this stress.
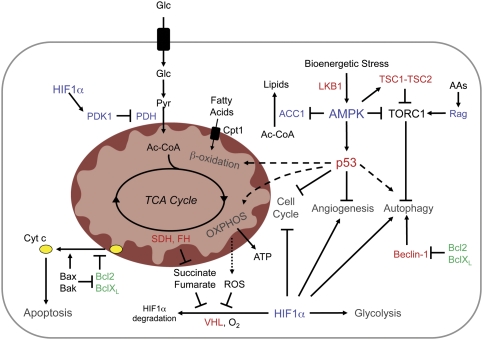
AMPK appears to function across evolution as a low energy checkpoint, triggering multiple biological adaptations designed to preserve energy and adapt to metabolic stress before the cell reaches a state of bioenergetic catastrophe that triggers cell death. In Saccharomyces cerevisiae, the yeast AMPK homolog Snf1 is essential for the metabolic shift required to grow on alternate or nonfermentable carbon sources (Carlson et al. 1981). In Drosophila, disruption of AMPK signaling results in developmental defects due to impaired activation of energy checkpoints (Mandal et al. 2005; J.H. Lee et al. 2007). AMPK regulates energy homeostasis in mammalian cells through multiple pathways. These include inhibition of lipid synthesis by Acetyl-CoA carboxylase-1 (ACC1) phosphorylation (Davies et al. 1990), inhibition of mTOR-dependent protein translation through the regulation of the TSC1–TSC2 complex or the mTOR-binding protein Raptor (Inoki et al. 2003; Corradetti et al. 2004; Shaw et al. 2004a; Gwinn et al. 2008), or through the regulation of macroautophagy (Buzzai et al. 2007; J. Liang et al. 2007; Lippai et al. 2008). Several of the downstream effects of AMPK on metabolic adaptation can be attributed to the AMPK-dependent activation of the p53 tumor suppressor (Fig. 3). AMPK phosphorylates p53 at Ser-15, which stimulates both its transcriptional activity and initiates a metabolic cell cycle checkpoint (Jones et al. 2005). This metabolic stress response is reinforced by the p53 target genes sestrin1 and sestrin2, which activate AMPK as part of a feedback loop (Budanov and Karin 2008). Other downstream targets likely involved in AMPK-dependent metabolic control include Foxo3 and PGC1α (Greer et al. 2007; Jager et al. 2007). The interaction of AMPK with several transcription factors indicates that transcriptional regulation may be central to AMPK-dependent stress responses.
Figure 3.
Strategies of metabolic adaptation in cancer. Intracellular sensors of energy, nutrients, and oxygen promote metabolic adaptation to stress during tumorigenesis. Major physiological strategies of metabolic adaptation include cell cycle inhibition, inhibition of biosynthetic pathways (lipid, protein synthesis), increases in bioenergetic pathways (β-oxidation, glycolysis, and OXPHOS), and induction of autophagy. Bioenergetic stress activates AMPK through an LKB-dependent manner. AMPK mediates metabolic adaptation through several pathways, including down-regulation of fatty acid synthesis through inhibition of ACC1, translation inhibition by blocking TORC1 activity, and activation of p53. Rag GTPases regulate TORC1 activity and autophagy in response to amino acid deprivation. Low HIF1α protein levels are maintained in cells under normoxia through the action of VHL-mediated proteosomal degradation. Low O2, ROS, or accumulation of the TCA substrates succinate or fumarate promotes HIF1α protein stabilization and transcriptional activity. HIF1α blocks pyruvate entry into the mitochondrion by up-regulating the PDH antagonist PDK1. Beclin-1 is required for autophagy induction and is antagonized by the anti-apoptotic proteins Bcl2/BclXL. Failure to manage or correct bioenergetic imbalance leads to cytochrome c release from mitochondria and induction of apoptosis. Oncogenes are displayed in green and tumor suppressors in red.
The emerging view across several fields is that AMPK, as an evolutionarily conserved energy sensor, is a key player in stress resistance at both cellular and organismal levels (Hardie et al. 1998; Baena-Gonzalez and Sheen 2008). The position of AMPK both downstream from and upstream of defined tumor suppressors—LKB1 and p53, respectively—suggests that AMPK may function as a tumor suppressor; however, the exact contribution of AMPK to tumor progression is unclear. LKB1 is inactivated in patients with Peutz-Jegher's syndrome, a condition that predisposes patients to gastrointestinal hamartomas and malignant tumors in a variety of tissues (Giardiello et al. 1987; Hearle et al. 2006). Mice with heterozygous disruption of LKB1 develop intestinal neoplasia that mimics human disease in Peutz-Jegher's patients (Bardeesy et al. 2002), and complete loss of LKB1 results in resistance to senescence and the development of aggressive cancers (Ji et al. 2007; Contreras et al. 2008; Gurumurthy et al. 2008). The contribution of AMPK to these phenotypes, however, remains unclear as AMPKα1 and α2 are just two of 13 enzymes of the AMPK-related family of kinases activated by LKB1 (Lizcano et al. 2004). AMPK activation may be positive or detrimental to tumor growth depending on p53 context. When p53 is present, AMPK induces a metabolic cell cycle checkpoint that restricts cell proliferation (Jones et al. 2005). However, in the absence of p53, treatment with the blood glucose-lowering drug metformin, an inhibitor of mitochondrial respiration, delays tumor progression, suggesting that the AMPK–p53 signaling axis is essential for tumors to adapt to metabolic stress in vivo (Buzzai et al. 2007; Huang et al. 2008).
p53 and metabolic control
p53 is a transcription factor that functions in a complex signaling network to mediate cellular adaptation to stress (Levine 1997). In response to diverse cellular insults including DNA damage, hypoxia, and oxidative stress, p53 becomes post-translationally modified, which promotes both its stabilization and activation, and activates the expression of genes that induce cell cycle arrest, senescence, and apoptosis (Vogelstein et al. 2000). Only recently has p53 been implicated in metabolic control, coordinating stress responses with changes in cellular metabolism and angiogenic potential (Fig. 4). Activation of p53 by metabolic stress is driven by AMPK-dependent phosphorylation and influenced by mTOR (Feng et al. 2005; C.H. Lee et al. 2007), although other pathways likely contribute. Moreover, p53 activation can reinforce AMPK-dependent responses through a series of p53-dependent feedback loops that in turn activate AMPK (Feng et al. 2005; Budanov and Karin 2008). Activation of the AMPK-p53 pathway by metabolic stress triggers multiple effects on catabolic metabolism, including enhanced β-oxidation of fatty acids and macroautophagy (Buzzai et al. 2007). Metabolic stress stimulates β-oxidation through the action of the carnitine palmitoyltransferase (Cpt1) enzymes (Deberardinis et al. 2006), which regulate mitochondrial fatty acid import. Genotoxic stress also stimulates p53-dependent up-regulation of the damage-regulated autophagy modulator (DRAM), a p53 target gene that encodes a lysosomal protein that induces macroautophagy (Crighton et al. 2006). However, while p53 can trigger autophagy in response to multiple stresses, its influence is likely cell- and context-specific (Tasdemir et al. 2008b). Thus, in addition to having well-documented roles in cell cycle regulation and apoptosis, p53 regulates stress-induced transcriptional programs that function to regulate pathways of energy homeostasis. Determining both the stimuli that trigger the metabolic activation of p53 and the specific p53 target genes that function in p53-dependent responses to metabolic stress remain to be determined.
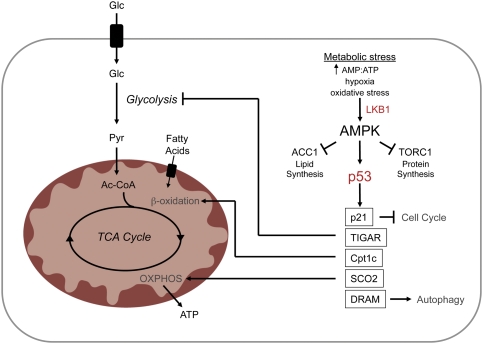
Figure 4.
The AMPK–p53 pathway in metabolic adaptation. Activation of the tumor suppressor p53 by AMPK promotes physiological adaptation to metabolic stress by multiple mechanisms. AMPK-dependent p53 activation negatively regulates cell cycle progression via the cyclin-dependent kinase (cdk) inhibitor p21. Impairment of glycolytic flux by TIGAR, combined with increased activation of β-oxidation and electron transport through expression of carnitine palmitoyltransferase Ic (Cpt1c) and SCO2, respectively, results in a p53-dependent metabolic shift toward OXPHOS. Transcriptional regulation of DRAM by p53 induces autophagy, the products of which can also fuel OXPHOS.
p53 can also influence the metabolic balance in cells between glycolysis and OXPHOS. p53 activity favors the production of ATP by OXPHOS through the transcriptional regulation of the fructose-2,6-bisphosphatase TP53-induced glycolysis regulator (TIGAR) and the synthesis of cytochrome c oxidase (SCO2) subunit of complex IV of the electron transport chain (Bensaad et al. 2006; Matoba et al. 2006). TIGAR negatively regulates glycolysis by degrading fructose-2,6-bisphosphate, an allosteric activator of the glycolytic regulatory enzyme PFK1. SCO2 is required for assembly of the cytochrome c oxidase (COX) complex. Loss of p53-dependent regulation of SCO2 impairs the mitochondrial respiratory chain and promotes a switch in ATP production from OXPHOS to glycolysis. In addition, p53 activation has been reported to lead to the induction of potent anti-angiogenic factors that limit tumor growth (Teodoro et al. 2006). Thus, in terms of metabolism, loss of p53 may provide a significant growth advantage to cancer cells. p53-deficient tumors engaged in aerobic glycolysis are able to more readily access energetic and biosynthetic pathways that support anabolic synthesis. By disengaging anti-angiogenic programs, p53 loss may rapidly promote greater access to nutrients to fuel these pathways. However, this growth advantage comes at a cost as loss of p53 renders cancer cells more sensitive to metabolic stress.
Adaptation through oxygen sensing
Like nutrient depletion, reduced oxygen availability—hypoxia—stimulates metabolic adaptation in cells to promote energy production and reduce ATP consumption. Two major phenotypic consequences of hypoxia are increased glucose consumption and lactate production caused by a switch to anaerobic glycolysis. This metabolic shift is synchronized by hypoxia-inducible factor 1 (HIF1), a transcription factor complex activated by hypoxic stress (Gordan and Simon 2007; Semenza 2007). Under normal oxygen tension, HIF1α accumulation is suppressed through modification by prolyl hydroxylation, which results in its ubiquitination by the von Hippel-Lindau (VHL) tumor suppressor and subsequent proteosomal degradation (Fig. 3). Under low oxygen, prolyl hydroxylation of HIF1α is reduced, resulting in HIF1α protein stabilization, association with the HIF1β subunit (ARNT), and increased HIF1α transcription. HIF1α accumulation can also result from reactive oxygen species (ROS) generated from mitochondrial OXPHOS, which inhibit prolyl hydroxylase activity to promote HIF1α stabilization (Brunelle et al. 2005; Guzy et al. 2005; Mansfield et al. 2005). HIF1 activity triggers the up-regulation of a number of genes involved in aerobic glycolysis, including glucose transporters, glycolytic enzymes, and LDH-A (Semenza et al. 1994). In addition, HIF1 promotes the shunting of pyruvate away from mitochondria by up-regulating pyruvate dehydrogenase kinase 1 (PDK1) (Kim et al. 2006; Papandreou et al. 2006), a negative regulator of pyruvate dehydrogenase (PDH), the rate-limiting enzyme for pyruvate to acetyl-CoA conversion. This serves three functions: It effectively diverts pyruvate away from mitochondrial biosynthetic pathways, reduces electron flux through OXPHOS, and, as a result, reduces oxidative stress derived from mitochondrial metabolism. By directing pyruvate away from mitochondria in favor of lactate and NAD+ production, HIF1α-dependent regulation of PDK1 may contribute to the Warburg effect in tumor cells.
Recent findings suggest a broader role for HIF1 in stress adaptation beyond oxygen sensing. In addition to hypoxic stress, HIF1 activity is stimulated by oxidative, energetic, inflammatory, and oncogenic stresses. Moreover, elevated concentrations of the TCA cycle intermediates succinate and fumarate induce constitutive HIF1α stabilization by interfering with prolyl hydroxylase function (Koivunen et al. 2007). Loss-of-function mutations in either succinate dehydrogenase (SDH) subunits or fumarate hydratase (FH) promote HIF1α stabilization under normoxic conditions due to the buildup of these metabolites (Isaacs et al. 2005; Selak et al. 2005). The emergence of SDH and FH as tumor suppressors highlights the importance of HIF1α regulation for proper homeostatic regulation. Other proteins involved in metabolic adaptation, including AMPK and p53, can also be activated by hypoxia (Graeber et al. 1994; Laderoute et al. 2006), suggesting the potential for communication across pathways. Interestingly, while primarily regulated by ubiquitin-dependent protein degradation, HIF1α is transcriptionally regulated by growth factors through PI3K signaling (Jiang et al. 2001). Thus, in tumors harboring activating PI3K mutations, the combined effect of growth factor signaling and stress-dependent HIF1α stabilization may act to enforce HIF1-dependent adaptation strategies for growth and survival.
Another important physiological response to hypoxia is the rapid down-regulation of protein translation, which is both reversible and HIF1-independent. Regulation of mRNA translation is thought to be important for energy conservation in oxygen-poor environments subjected to chronic hypoxia, as well as for selectively reducing the translation of mRNAs associated with a “growth” phenotype that would be detrimental during periods of hypoxic stress. Multiple signaling pathways converge on hypoxic translation inhibition. Hypoxia promotes the acute activation of the endoplasmic reticulum (ER)-resident kinase PERK, a central component of the unfolded protein response, leading to inhibition of mRNA translation through phosphorylation of the translation initiation factor eIF2α (Harding et al. 1999; Koumenis et al. 2002). Second, chronic hypoxia promotes disruption of the cap-binding eIF4F complex, effectively blocking translation by decreasing the association of mRNA with ribosomes (Koritzinsky et al. 2006). AMPK, which is also acutely regulated by hypoxia, leads to translation inhibition through two additional methods: TSC2-dependent inhibition of mTOR (Liu et al. 2006), and inhibition of peptide elongation through the activation of eukaryotic elongation factor 2 (eEF2) kinase (Browne et al. 2004). These tumor suppressor pathways do not likely function in isolation and rather work together to regulate HIF1-independent mechanisms of energy conservation to promote survival under hypoxic stress.
Autophagy as a tumor suppressor mechanism
As mentioned previously, autophagy is a central metabolic stress response conserved throughout evolution from yeast to man and represents a key pathway in metabolic stress adaptation. Macroautophagy involves the sequestration of internal components and organelles into double-membrane structures known as autophagic vesicles, and subsequent degradation by the lysosome. Autophagy performs an important physiological role as a waste disposal system for the removal of aged or damaged organelles, particularly mitochondria (Lum et al. 2005). However, when faced with metabolic stress such as nutrient depletion (amino acids, glucose) or hypoxia, cells activate the autophagic pathway. The degradation products derived from macroautophagy can be used to fuel catabolic metabolic processes for energy generation (Levine and Klionsky 2004; Lum et al. 2005; Buzzai et al. 2007), thus promoting cell viability. mTOR is a central regulator of this process. Inhibition of TORC1 activity, either through AMPK activation, decreased amino acid availability, or rapamycin, stimulates autophagy.
As with metabolic transformation, oncogene and tumor suppressor pathways display opposing effects on autophagy. Oncogene pathways including Akt, mTOR, and Bcl2 appear to function as inhibitors of autophagy, while tumor suppressors including PTEN, TSC2, LKB1, and HIF1α promote autophagy (Maiuri et al. 2009). The role of p53 in autophagy remains controversial. p53 activation has been shown to promote autophagy in response to stress through transcriptional control of autophagy regulators such as DRAM (Crighton et al. 2006), while cytoplasmic p53 appears to function as a negative regulator of autophagy (Tasdemir et al. 2008a,b). Thus, the role of p53 in autophagy induction may depend on its cellular localization, transcriptional activity, and mechanism of activation. Studies involving Beclin-1, which is required for autophagy induction in mammalian cells, suggest that autophagy functions as an important tumor suppressor mechanism. Beclin-1 haploinsufficiency promotes tumorigenesis in mouse models and is associated with breast and ovarian cancers in humans (Aita et al. 1999; Liang et al. 1999; Qu et al. 2003). Other modifiers of Beclin-1 activity such as UVRAG and Bif display tumor suppressor properties (C. Liang et al. 2007; Takahashi et al. 2007), while anti-apoptotic Bcl-2 inhibits Beclin-1 function (Pattingre et al. 2005). These data suggest that, even at a basal or background state, autophagy can act as a tumor suppressor mechanism, and that suppression of autophagy is an important gateway to tumorigenesis. The importance of autophagy in tumor suppression may actually stem from its role in the degradation of long-lived proteins and organelles, rather than its role in stress responses. However, any subsequent inability to induce autophagy under conditions of nutrient or oxygen deprivation could render tumors more susceptible to apoptosis by metabolic stress.
Conclusions
Oncogenic transformation or loss of tumor suppressor function drives unchecked growth and proliferation in cancer cells, which often coincide with both an increase in bioenergetic potential and the ability to take up nutrients in a cell-autonomous fashion. Reprogramming of core metabolism in tumors confers a selective growth advantage (i.e., enhanced proliferation, resistance to apoptosis) that promotes tumor progression. In retrospect, it seems obvious that cellular metabolism is of great physiological importance to tumor growth, and that genes involved in tumor progression would be intricately linked with its regulation. Oncogenic pathways that promote proliferation and survival in normal cells confer “metabolic permission” in transformed cells by stimulating bioenergetic pathways and anabolic growth. Tumor suppressors such as p53, LKB1, and TSC2 function as a brake on cell growth by antagonizing many of the same biosynthetic pathways stimulated by oncogenes under conditions of limiting intracellular glucose or amino acid supplies. It is still debated whether these metabolic changes are a consequence or a cause of tumorigenesis. However, the fact that metabolic enzymes such as SDH and FH function as tumor suppressors in humans suggests that metabolic deregulation can be an initiating event in cancer. The recent discovery of activating mutations in isocitrate dehydrogenase (IDH1) in human glioblastoma demonstrates that an activating mutation in a metabolic enzyme can be and are selected during carcinogenesis (Parsons et al. 2008). The emergence of metabolic enzymes (i.e., PK-M2, LDHA, SDH, FH, PDK1, and IDH1) as important regulators of cancer cell growth suggests that metabolic control is a key element of tumor progression.
While much attention has been paid to the prevalence of aerobic glycolytic “Warburg” metabolism in cancer, the metabolic pathways that support cell growth during tumorigenesis are likely far more complex. Tumors often display genetic and phenotypic heterogeneity within subpopulations of tumor cells. Thus, it is possible that that individual cells within a tumor could display distinct metabolic signatures. The metabolic response of an individual tumor cell is dependent on both the combination of oncogenic and/or tumor suppressor mutations and the environmental conditions (nutrient depletion, hypoxia) encountered within the tumor microenvironment. By way of example, ∼30% of cancers cannot be detected by FDG-PET, suggesting that these tumors either display a glycolytic rate below the limit of detection or utilize alternative (nonglucose) metabolic pathways for energy production. Thus, defining the mechanisms by which cancer cells coordinate their metabolic activities to support tumor growth and adapt to varying environmental conditions will aid in our understanding of cancer development and progression.
Cancer therapy is increasingly shifting toward individualized therapeutic approaches based on the genetic abnormalities exhibited by transformed cells. In cancer cells, a given transformation event (i.e., oncogene activation) may play a qualitatively different or essential role in a given pathway compared with its role in normal cells, such that cancer cells become dependent on the activity of a specific oncogene for function and survival, a concept known as oncogene addiction. From this logic, oncogene-driven signal transduction may promote greater dependence on specific metabolic pathways such as glycolysis to maintain tumor growth. When combined with the loss of tumor suppressors that promote metabolic adaptation, cancer cells risk developing greater sensitivity to bioenergetic stress. The LKB1-AMPK-p53 pathway best illustrates exposure of such a metabolic “Achilles' heel” and its impact on tumor survival. LKB or p53 deficiency, while permissive for unchecked cell growth, renders cells more susceptible to death by metabolic stress. Moreover, the ability of tumor cells to survive metabolic stress in vivo requires AMPK-dependent activation of p53 (Buzzai et al. 2007), which may explain why metformin, an anti-diabetic therapy, can be anti-tumorigenic (Evans et al. 2005; Huang et al. 2008). Understanding the pathways that regulate cancer cell metabolism may lead to greater understanding of cancer development and progression, and has the potential to open a new vista of metabolic therapy for cancer treatment.
Acknowledgments
We thank Ben Fuerth, Jessica Joh, Erin Podewils, and Ralph DeBerardinis for help in completing this manuscript. This work was supported by the McGill University Faculty of Medicine (to R.G.J.), the Canadian Institutes of Health Research (to R.G.J.), and the National Institutes of Health (RO1 CA105463-01 to C.B.T.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1756509.
References
- Aita V.M., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E., Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N., Sinha M., Hezel A.F., Signoretti S., Hathaway N.A., Sharpless N.E., Loda M., Carrasco D.R., DePinho R.A. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- Bauer D.E., Harris M.H., Plas D.R., Lum J.J., Hammerman P.S., Rathmell J.C., Riley J.L., Thompson C.B. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 2004;18:1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D.E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Browne G.J., Finn S.G., Proud C.G. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J. Biol. Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- Brunelle J.K., Bell E.L., Quesada N.M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R.C., Chandel N.S. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Budanov A.V., Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M., Jones R.G., Amaravadi R.K., Lum J.J., DeBerardinis R.J., Zhao F., Viollet B., Thompson C.B. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- Cairns P., Okami K., Halachmi S., Halachmi N., Esteller M., Herman J.G., Jen J., Isaacs W.B., Bova G.S., Sidransky D. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- Carlson M., Osmond B.C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., Cantley L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008a;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk H.R., Vander Heiden M.G., Wu N., Asara J.M., Cantley L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008b;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Contreras C.M., Gurumurthy S., Haynie J.M., Shirley L.J., Akbay E.A., Wingo S.N., Schorge J.O., Broaddus R.R., Wong K.K., Bardeesy N., et al. Loss of Lkb1 provokes highly invasive endometrial adenocarcinomas. Cancer Res. 2008;68:759–766. doi: 10.1158/0008-5472.CAN-07-5014. [DOI] [PubMed] [Google Scholar]
- Corradetti M.N., Inoki K., Bardeesy N., DePinho R.A., Guan K.L. Regulation of the TSC pathway by LKB1: Evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes & Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P.R., Gasco M., Garrone O., Crook T., Ryan K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Dang C.V., Semenza G.L. Oncogenic alterations of metabolism. Trends Biochem. Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- Dang C.V., Kim J.W., Gao P., Yustein J. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- Davies S.P., Sim A.T., Hardie D.G. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur. J. Biochem. 1990;187:183–190. doi: 10.1111/j.1432-1033.1990.tb15293.x. [DOI] [PubMed] [Google Scholar]
- Deberardinis R.J., Lum J.J., Thompson C.B. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J. Biol. Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- DeBerardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Edinger A.L., Thompson C.B. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom R.L., Bauer D.E., Buzzai M., Karnauskas R., Harris M.H., Plas D.R., Zhuang H., Cinalli R.M., Alavi A., Rudin C.M., et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Evans J.M., Donnelly L.A., Emslie-Smith A.M., Alessi D.R., Morris A.D. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Feng Z., Zhang H., Levine A.J., Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Energy deregulation: Licensing tumors to grow. Science. 2006;312:1158–1159. doi: 10.1126/science.312.5777.1158. [DOI] [PubMed] [Google Scholar]
- Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Giardiello F.M., Welsh S.B., Hamilton S.R., Offerhaus G.J., Gittelsohn A.M., Booker S.V., Krush A.J., Yardley J.H., Luk G.D. Increased risk of cancer in the Peutz-Jeghers syndrome. N. Engl. J. Med. 1987;316:1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- Gordan J.D., Simon M.C. Hypoxia-inducible factors: Central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan J.D., Thompson C.B., Simon M.C. HIF and c-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesdiener K.M., Karpinski B.A., Lindsten T., Strominger J.L., Jones N.H., Thompson C.B., Leiden J.M. Isolation and structural characterization of the human 4F2 heavy-chain gene, an inducible gene involved in T-lymphocyte activation. Mol. Cell. Biol. 1988;8:3809–3819. doi: 10.1128/mcb.8.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber T.G., Peterson J.F., Tsai M., Monica K., Fornace A.J., Jr, Giaccia A.J. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E.L., Oskoui P.R., Banko M.R., Maniar J.M., Gygi M.P., Gygi S.P., Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Griffiths M., Keast D. The effect of glutamine on murine splenic leukocyte responses to T and B cell mitogens. Immunol. Cell Biol. 1990;68:405–408. doi: 10.1038/icb.1990.54. [DOI] [PubMed] [Google Scholar]
- Guppy M., Greiner E., Brand K. The role of the Crabtree effect and an endogenous fuel in the energy metabolism of resting and proliferating thymocytes. Eur. J. Biochem. 1993;212:95–99. doi: 10.1111/j.1432-1033.1993.tb17637.x. [DOI] [PubMed] [Google Scholar]
- Gurumurthy S., Hezel A.F., Sahin E., Berger J.H., Bosenberg M.W., Bardeesy N. LKB1 deficiency sensitizes mice to carcinogen-induced tumorigenesis. Cancer Res. 2008;68:55–63. doi: 10.1158/0008-5472.CAN-07-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy R.D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K.D., Simon M.C., Hammerling U., Schumacker P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S., Turk B.E., Shaw R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. Minireview: The AMP-activated protein kinase cascade: The key sensor of cellular energy status. Endocrinology. 2003;144:5179–5183. doi: 10.1210/en.2003-0982. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hardie D.G., Carling D., Carlson M. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G., Zhao F., Bauer D.E., Andreadis C., Shaw A.N., Dhanak D., Hingorani S.R., Tuveson D.A., Thompson C.B. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hawley S.A., Boudeau J., Reid J.L., Mustard K.J., Udd L., Makela T.P., Alessi D.R., Hardie D.G. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Pan D.A., Mustard K.J., Ross L., Bain J., Edelman A.M., Frenguelli B.G., Hardie D.G. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Hearle N., Schumacher V., Menko F.H., Olschwang S., Boardman L.A., Gille J.J., Keller J.J., Westerman A.M., Scott R.J., Lim W., et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin. Cancer Res. 2006;12:3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- Holley R.W., Kiernan J.A. Control of the initiation of DNA synthesis in 3T3 cells: Low-molecular weight nutrients. Proc. Natl. Acad. Sci. 1974;71:2942–2945. doi: 10.1073/pnas.71.8.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Wullschleger S., Shpiro N., McGuire V.A., Sakamoto K., Woods Y.L., McBurnie W., Fleming S., Alessi D.R. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem. J. 2008;412:211–221. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- Hurley R.L., Anderson K.A., Franzone J.M., Kemp B.E., Means A.R., Witters L.A. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K., Zhu T., Guan K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Isaacs J.S., Jung Y.J., Mole D.R., Lee S., Torres-Cabala C., Chung Y.L., Merino M., Trepel J., Zbar B., Toro J., et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Jager S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Ramsey M.R., Hayes D.N., Fan C., McNamara K., Kozlowski P., Torrice C., Wu M.C., Shimamura T., Perera S.A., et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Jia S., Liu Z., Zhang S., Liu P., Zhang L., Lee S.H., Zhang J., Signoretti S., Loda M., Roberts T.M., et al. Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.H., Jiang G., Zheng J.Z., Lu Z., Hunter T., Vogt P.K. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–369. [PubMed] [Google Scholar]
- Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M.J., Thompson C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kim J.W., Tchernyshyov I., Semenza G.L., Dang C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., Guan K.L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P., Hirsila M., Remes A.M., Hassinen I.E., Kivirikko K.I., Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: Possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M., Magagnin M.G., van den Beucken T., Seigneuric R., Savelkouls K., Dostie J., Pyronnet S., Kaufman R.J., Weppler S.A., Voncken J.W., et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C., Naczki C., Koritzinsky M., Rastani S., Diehl A., Sonenberg N., Koromilas A., Wouters B.G. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Pouyssegur J. Tumor cell metabolism: Cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Laderoute K.R., Amin K., Calaoagan J.M., Knapp M., Le T., Orduna J., Foretz M., Viollet B. 5′-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol. Cell. Biol. 2006;26:5336–5347. doi: 10.1128/MCB.00166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Inoki K., Karbowniczek M., Petroulakis E., Sonenberg N., Henske E.P., Guan K.L. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 2007;26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Koh H., Kim M., Kim Y., Lee S.Y., Karess R.E., Lee S.H., Shong M., Kim J.M., Kim J., et al. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Oh B.H., Jung J.U. UVRAG: A new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- Liang J., Shao S.H., Xu Z.X., Hennessy B., Ding Z., Larrea M., Kondo S., Dumont D.J., Gutterman J.U., Walker C.L., et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Lippai M., Csikos G., Maroy P., Lukacsovich T., Juhasz G., Sass M. SNF4Aγ, the Drosophila AMPK γ subunit is required for regulation of developmental and stress-induced autophagy. Autophagy. 2008;4:476–486. doi: 10.4161/auto.5719. [DOI] [PubMed] [Google Scholar]
- Liu L., Cash T.P., Jones R.G., Keith B., Thompson C.B., Simon M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano J.M., Goransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J.J., DeBerardinis R.J., Thompson C.B. Autophagy in metazoans: Cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Maiuri M.C., Tasdemir E., Criollo A., Morselli E., Vicencio J.M., Carnuccio R., Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;7:3056–3061. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., Rong L., Yoshida K., Ler L.W., Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Mandal S., Guptan P., Owusu-Ansah E., Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Mansfield K.D., Guzy R.D., Pan Y., Young R.M., Cash T.P., Schumacker P.T., Simon M.C. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L., Kenneth N.S., White R.J. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Momcilovic M., Hong S.P., Carlson M. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 2006;281:25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- Osthus R.C., Shim H., Kim S., Li Q., Reddy R., Mukherjee M., Xu Y., Wonsey D., Lee L.A., Dang C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Papandreou I., Cairns R.A., Fontana L., Lim A.L., Denko N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Pardee A.B. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S.M., Riggins G.J., et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Samuels Y., Diaz L.A., Jr, Schmidt-Kittler O., Cummins J.M., Delong L., Cheong I., Rago C., Huso D.L., Lengauer C., Kinzler K.W., et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak M.A., Armour S.M., MacKenzie E.D., Boulahbel H., Watson D.G., Mansfield K.D., Pan Y., Simon M.C., Thompson C.B., Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- Semenza G.L., Roth P.H., Fang H.M., Wang G.L. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- Shaw R.J., Cantley L.C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Shaw R.J., Bardeesy N., Manning B.D., Lopez L., Kosmatka M., DePinho R.A., Cantley L.C. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004a;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Shaw R.J., Kosmatka M., Bardeesy N., Hurley R.L., Witters L.A., DePinho R.A., Cantley L.C. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. 2004b;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Dolde C., Lewis B.C., Wu C.S., Dang G., Jungmann R.A., Dalla-Favera R., Dang C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M., Riek U., Tuerk R., Schlattner U., Wallimann T., Neumann D. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J. Biol. Chem. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H.D., Sun M., Sato Y., Liang C., Jung J.U., Cheng J.Q., Mul J.J., et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E., Maiuri M.C., Morselli E., Criollo A., D'Amelio M., Djavaheri-Mergny M., Cecconi F., Tavernarakis N., Kroemer G. A dual role of p53 in the control of autophagy. Autophagy. 2008a;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- Tasdemir E., Maiuri M.C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008b;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro J.G., Parker A.E., Zhu X., Green M.R. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science. 2006;313:968–971. doi: 10.1126/science.1126391. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K.W. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.Y., Pfeiffer H.K., Nissim I., Daikhin E., Yudkoff M., McMahon S.B., et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Johnstone S.R., Dickerson K., Leiper F.C., Fryer L.G., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Woods A., Dickerson K., Heath R., Hong S.P., Momcilovic M., Johnstone S.R., Carlson M., Carling D. Ca2+/calmodulin-dependent protein kinase kinase-β acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xie M., Zhang D., Dyck J.R., Li Y., Zhang H., Morishima M., Mann D.L., Taffet G.E., Baldini A., Khoury D.S., et al. A pivotal role for endogenous TGF-β-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc. Natl. Acad. Sci. 2006;103:17378–17383. doi: 10.1073/pnas.0604708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh L.A., Lee K.H., Kim K.H. Regulation of rat liver acetyl-CoA carboxylase. Regulation of phosphorylation and inactivation of acetyl-CoA carboxylase by the adenylate energy charge. J. Biol. Chem. 1980;255:2308–2314. [PubMed] [Google Scholar]
- Yuneva M., Zamboni N., Oefner P., Sachidanandam R., Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]