Abstract
Heterogeneity of cells within exponentially growing populations was addressed in a bacterium, the facultative methylotroph Methylobacterium extorquens AM1. A transcriptional fusion between a well-characterized methanol-inducible promoter (PmxaF) and gfpuv was used with flow cytometry to analyse the distribution of gene expression in populations grown on either succinate or methanol, correlated with forward scatter as a measure of cell size. These cell populations were found to consist of three major subpopulations defined by cells that were actively growing and dividing, newly divided, and non-dividing. Through the use of flow cytometry, it was demonstrated that a significant percentage of the total population did not respond to carbon shift. In addition, these experiments demonstrated that a small subset of the total population was significantly brighter than the rest of the population and dominated fluorimetry data. These results were corroborated with a continuous flow-through system and laser scanning microscopy, confirming that subpopulations, not discernible in the population average, dominate population response. These results demonstrate that the combination of flow cytometry and microscopic single-cell analysis can be effectively used to determine the dynamics of subpopulations in population response. In addition, they support the concept that physiological diversity in isogenic populations can poise some proportion of the population to respond appropriately to changing conditions.
INTRODUCTION
Historically, most prokaryotic cell biology studies have focused on data from bulk culture experiments. While this approach has been powerful in the elucidation of population dynamics and response to perturbation, the caveat of such data is that they represent averages and do not address cell-to-cell variability or the presence of physiologically distinct subpopulations. It is becoming clear that significant cell-to-cell variation in gene expression occurs in bacteria, which can result in bimodal, multimodal or Gaussian distributions within populations (Bongaerts et al., 2002; Elowitz et al., 2002; Siegele & Hu, 1997; Strovas et al., 2007), and it has been suggested that such variability might result in phenotypic differences between cells in isogenic populations (Elowitz et al., 2002; Siegele & Hu, 1997; Balaban et al., 2004; Kuang et al., 2004; Pedraza & van Oudenaarden, 2005; Rosenfeld et al., 2005). Therefore, in order to understand the response of populations, it is important to determine heterogeneity of key parameters within the populations and to identify significant physiologically distinct subpopulations. Such determinations require analysis at the single-cell level for comparison with population dynamics.
The drawback of single-cell studies is the difficulty of obtaining data on large numbers of cells, for large-scale population analysis. In this case, flow cytometry provides an option. However, it is also important to confirm the population-level analyses with single-cell analyses on smaller numbers of cells. Therefore, the combination of flow cytometry and high-resolution microscopy methods provides an attractive approach to characterize heterogeneity of key parameters within cell populations and assess the response of subpopulations to environmental perturbation.
The facultative methylotroph Methylobacterium extorquens AM1 has two strongly contrasting modes of metabolism: growth on multicarbon compounds, which is energy-limited, and growth on one-carbon compounds, which is limited by reducing power (Anthony, 1982; Van Dien & Lidstrom, 2002). Therefore, it is an interesting model system for studying the shift between modes of metabolism. We have previously shown by analysis of individual actively growing M. extorquens cells in a flow-through system that significant cell-to-cell heterogeneity exists in both gene expression and growth rate, in exponentially growing populations (Strovas et al., 2007). In this study, we examined much larger cell populations via flow cytometry and obtained evidence for major subpopulations that respond differently to substrate change.
METHODS
Bacterial strains and growth conditions
M. extorquens.
M. extorquens AM1 strains were grown at 28 °C in 30 ml minimal salts media (Hypho) supplemented with either 0.3 % (v/v) methanol or 0.4 % (w/v) succinate as a growth substrate (Attwood & Harder, 1972; Van Dien et al., 2003). Table 1 lists the strains and plasmids used for this study. Antibiotics were added as appropriate: 50 μg rifamycin ml−1 and 10 μg tetracycline ml−1. The optical density (OD600) of cultures was measured on a Beckman DU 640B spectrophotometer (1 cm path length).
Table 1.
M. extorquens AM1 strains and plasmids used in this study
| Strain | Description | Source or reference |
|---|---|---|
| AM1 | RifR derivative | Lab collection |
| CM174 | katA : : (loxP-trrnB-PmxaF-gfpuv-tT7) | Marx & Lidstrom (2004) |
| TSG.10 | katA : : (loxP-trrnB-Plac-gfpuv-tT7) | This study |
| TSG.11-2 | katA : : (loxP-trrnB-PphaA-gfpuv-tT7) | This study |
| TSG.12-2 | katA : : (loxP-trrnB-PsgaA-gfpuv-tT7) | This study |
| TSX | Non-motile mutant (ΔmotA-fliN-fliM-fliG, ΔmotC-motB, ΔpomA-motB) | Strovas et al. (2007) |
| TSXCM174 | Non-motile mutant with katA : : (loxP-trrnB-PmxaF-gfpuv-t17) | Strovas et al. (2007) |
| Plasmid | ||
| pGFPuv | Commercial vector | BD Biosciences (ClonTech) |
| pCR2.1 | PCR cloning vector | Invitrogen |
| pCM139 | Broad-host-range shuttle vector containing Pfae-fae | Vorholt et al. (2000) |
| pCM157 | Broad-host-range cre expression vector (TetR) | Marx & Lidstrom (2002) |
| pCM168 | Insertional expression vector (ttrrnB-MCS-tT7) | Marx & Lidstrom (2004) |
| pCM174 | pCM168 with PmxaF-gfpuv | Marx & Lidstrom (2004) |
| pRK2073 | Helper plasmid expressing IncP tra functions | Lab collection |
| pTS29 | pCR2.1 with PphaA | This study |
| pTS35r | pCR2.1 with PsgaA | This study |
| pTSGex | pCM139 with Plac-gfpuv | This study |
| pTSG.10 | pCM168 with Plac-gfpuv | This study |
| pTSG.11-2 | pCM168 with PphaA-gfpuv | This study |
| pTSG.12-2 | pCM168 with PsgaA-gfpuv | This study |
Carbon shift experiments.
Cultures were grown to mid-exponential phase and centrifuged at 4000 r.p.m. for 10 min at 15 °C. Cells were resuspended in minimal medium with methanol or succinate at an OD600 of approximately 0.2 or 0.5. At 1 h time points over a period of 26 h, 0.5 ml aliquots were taken and analysed by either flow cytometry or fluorimetry. All samples were kept at room temperature prior to analysis. For the carbon shift, cultures were centrifuged as previously described, washed with minimal medium, and recentrifuged. The cultures were then resuspended in medium such that methanol-grown cultures were shifted to succinate or vice versa.
Construction of promoter-GFPuv transcriptional fusion insertional vectors.
The Plac-gfpuv insertional vector, pTSG.10, was made from pCM168 and pTSGex (Marx & Lidstrom, 2004). pTSGex was constructed by inserting GFPuv via HindIII-EcoRI into pCM139, which results in the excision of Pfae-fae (Vorholt et al., 2000). pTSG.10 was made by inserting the PciI–EcoRI Plac-gfpuv into pCM168 via NcoI-EcoRI. The phaA and sgaA promoters were PCR amplified and inserted into pCR2.1 to make pTS29 and pTS35r respectively. The PphaA-gfpuv and PsgaA-gfpuv insertional vectors were made by inserting the 625 bp BamHI–NsiI PphaA fragment and the ∼700 bp BamHI–NsiI PsgaA fragment into pTSG.10 via BglII-SbfI, excising Plac to make pTSG.11-2 and pTSG.12-2 respectively.
Construction of promoter-GFPuv transcriptional fusion strains.
Promoter-GFPuv transcriptional fusion strains containing the fusions inserted into the M. extorquens AM1 katA chromosomal insertion site were constructed via triparental matings as previously described and screened for tetracycline resistance and kanamycin sensitivity (Marx & Lidstrom, 2004). Antibiotic markers were removed from mutants by introduction of pCM158 by triparental matings (Marx & Lidstrom, 2002). All mutants were confirmed by PCR and fluorimetry analysis.
Fluorimetry analysis.
Fluorescence measurements were carried out with a Shimadzu RF-5301PC fluorimeter. GFPuv excitation was conducted at 405 nm and emissions were monitored at 509 nm. Emissions from the cells of CM174 and TSXCM174 were measured with slit widths of 5 : 5 and emissions from all other strains carrying promoter fusions were measured with slit widths of 10 : 10. Lower slit widths were used to measure PmxaF activity in order to prevent the fluorescence signal from saturating the detector. Promoter activities were calculated as previously described by plotting fluorescence versus OD600 (Leveau & Lindow, 2001). PmxaF activity was normalized to compensate for different slit-width settings based on calibration curves generated with rhodamine-123 (data not shown).
Flow cytometry analysis.
Flow cytometry experiments (n≥6) were conducted on a BD Biosciences LSR cytometer controlled by BD CellQuest Pro. Excitation was conducted with a 488 nm argon laser and fluorescence emission detected with a 530/28 nm bandpass filter in the FL1 channel. Data analysis was conducted with WinMDI 2.8 [Joseph Trotter (http://facs.scripps.edu/)], MatLab 6.5 (The MathWorks Inc.) and Excel (Microsoft).
Microscopy.
Microscopy experiments were conducted on a Zeiss LSM 510 META using a 100×1.45 N.A. objective and an FCS2 flow-through cell (Bioptechs) as described previously (Strovas et al., 2007).
RESULTS
Reporter validation and fluorimetry-based gene-expression analysis in bulk cultures
GFPuv is a convenient reporter of gene expression from promoters in aerobic bacteria. In order to validate GFPuv as a dynamic reporter in M. extorquens AM1, three previously characterized methylotrophy promoters were utilized and compared to results obtained with other reporters (Bongaerts et al., 2002; Albano et al., 1998; Kalyuzhnaya & Lidstrom, 2003; Lissemore et al., 2000; Marx & Lidstrom, 2001; Zhang & Lidstrom, 2003). Three new transcriptional fusion strains were constructed as chromosomal insertions in the wild-type background, in which GFPuv was transcribed from a standard Plac promoter and from two native M. extorquens AM1 promoters, PphaA (driving genes for poly-β-hydroxybutyrate biosynthesis; Korotkova et al., 2002) and PsgaA (driving genes for the serine cycle for formaldehyde assimilation; Kalyuzhnaya & Lidstrom, 2003). These strains, along with the previously constructed PmxaF-gfpuv strain CM174 (Marx & Lidstrom, 2004), were used to determine the relationship between fluorescence intensity and culture optical density in cells grown on methanol and, in the case of the mxaF promoter, also cells grown on succinate. A linear relationship between relative fluorescence and cell density (OD600) was observed for all transcriptional fusion strains, as exemplified for CM174 in Fig. 1. Promoter activities for all mutant strains were calculated as described by Leveau & Lindow (2001) and are shown in Table 2. The relative activities correlated well with those obtained previously using a xylE reporter, such that Plac : xylE=32±12 nmol min−1 (mg protein)−1 (Marx & Lidstrom, 2001), PphaA : xylE=135 mU (milliunits, Korotkova et al., 2002), PsgaA : xylE=281±33 mU (Kalyuzhnaya & Lidstrom, 2003), PmxaF : xylE activity was 1580±210 nmol min−1 (mg protein)−1 on methanol and 1170±10 nmol min−1 (mg protein)−1 on succinate (Marx & Lidstrom, 2001).
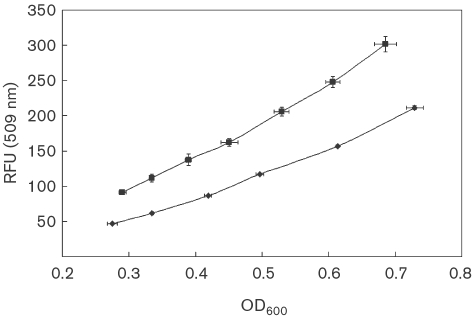
Fig. 1.
Representative RFU versus OD600 plots for CM174 (containing the mxaF-gfp promoter fusion) grown on succinate (▪) and methanol (⧫). Means±sd are plotted (n=3).
Table 2.
Calculated promoter activities from transcriptional fusions with GFPuv in M. extorquens AM1
Activities (P) were calculated from P=fss×μ×(1+μ/m), where μ=growth rate, m=maturation half-life of GFPuv, and fss=ΔRFU/ΔOD600 (Leveau & Lindow, 2001). The data represent means±sd.
| Promoter | Promoter activity (RFU OD600−1 h−1) | Growth condition | n |
|---|---|---|---|
| Plac | 5.6±0.9 | Methanol | 13 |
| PphaA | 7.1±1.7 | Methanol | 4 |
| PsgaA | 16.0±2.5 | Methanol | 6 |
| PmxaF | 349.1±82.6 | Methanol | 9 |
| 264.3±10.3 | Succinate | 6 | |
| Carbon shift activities for PmxaF | |||
| 232.3±17.1 | Succinate→Methanol (pre-shift) | 3 | |
| 402.4±28.7 | Succinate→Methanol (post-shift) | 3 | |
| 290.2±11.2 | Methanol→Succinate (pre-shift) | 3 | |
| 364.5±6.0 | Methanol→Succinate (post-shift) | 3 |
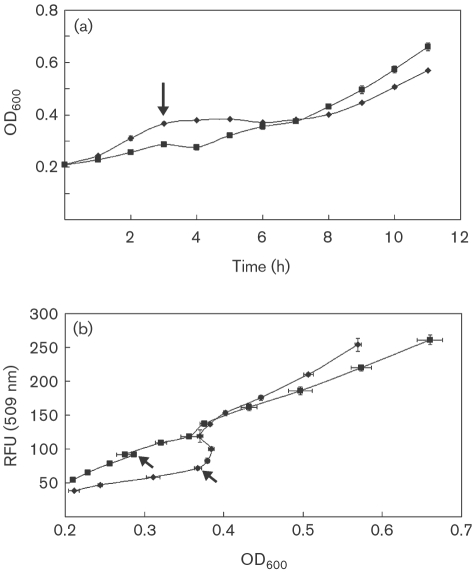
To further validate batch culture results, the PmxaF-gfpuv fusion construct CM174 was tested in carbon shift experiments, assessing kinetics of response before and after shifts to and from the inducing substrate (methanol). Cultures growing exponentially on methanol or succinate were shifted to the alternative carbon source, as described in Methods, and the intensity of GFPuv fluorescence was measured. Switching from succinate to methanol resulted in a growth lag of approximately 5 h (Fig. 2a, diamonds), consistent with previous results (Strovas et al., 2007; Marx et al., 2005). For this condition, an abrupt increase in fluorescence was observed (Fig. 2b), after which a linear correlation between fluorescence and cell density was restored, corresponding to a higher relative promoter activity (Table 2). Switching from methanol to succinate resulted in a growth lag of approximately 1 h (Fig. 2a, squares). In this case, a smaller increase in fluorescence occurred immediately after the shift, but the promoter activity remained fairly constant, with a small decrease in rate. This may be a combined result of continued high rates of transcription during the transition between methanol- and succinate-growth modes, consistent with the results from flux analysis (Pedraza & van Oudenaarden, 2005), and of negligible degradation of GFPuv over time due to an in vivo half-life that is >24 h (Leveau & Lindow, 2001; Andersen et al., 1998; Zhao et al., 1999).
Fig. 2.
Dynamics of growth and gene expression for CM174 during carbon shift experiments. (a) Growth curves; (b) RFU versus OD600. Arrows indicate carbon shift at 3 h (▪, methanol→succinate; ⧫, succinate→methanol). Means±sd are plotted (n=3).
Flow cytometry-based analysis of population distribution in bulk cultures
Flow-cytometric analysis was employed to investigate the distribution of cells in bulk cultures in terms of relative size (estimated by forward scatter analysis) and fluorescence intensity. Forward scatter data were collected for wild-type M. extorquens AM1 and for the GFPuv-expressing strain CM174 at multiple time points during growth-curve experiments in batch culture. For the single-cell validation studies a non-motile mutant is necessary, to facilitate long-term observations of individual cells (Strovas et al., 2007). Growth-curve and gene-expression studies were carried out with this non-motile strain (TSXCM174) and compared to CM174, and the results for growth rate, dynamics of the growth curve and gene expression could not be distinguished between the two strains (data not shown). Therefore, further work was carried out only with the non-motile strain TSXCM174, to allow direct comparisons with previously published single-cell results (Strovas et al., 2007). During the exponential phase of growth on either succinate or methanol, cells exhibited lognormal distributions for both forward scatter and fluorescence emission. During the transition to stationary phase, cells from succinate-grown cultures distributed lognormally for forward scatter and fluorescence, whereas cells from methanol-grown cultures exhibited Gaussian distributions for both parameters upon the transition to stationary phase, which may reflect the specific regulation pattern for PmxaF. Shifts from lognormal to Gaussian distribution were observed previously by Banerjee et al. (2004) using the Plac-gfp system in Escherichia coli.
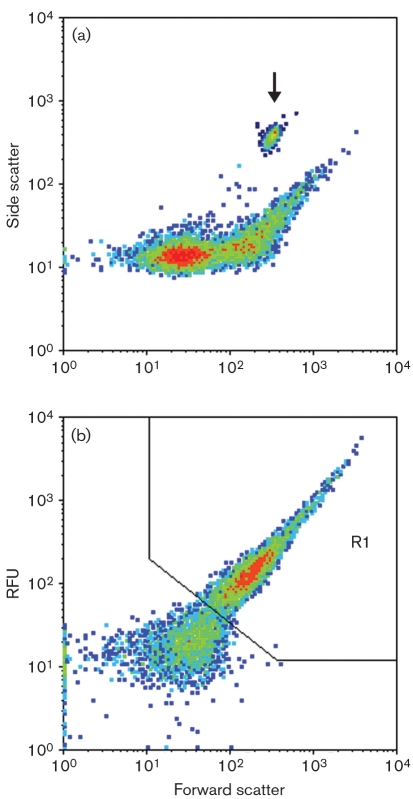
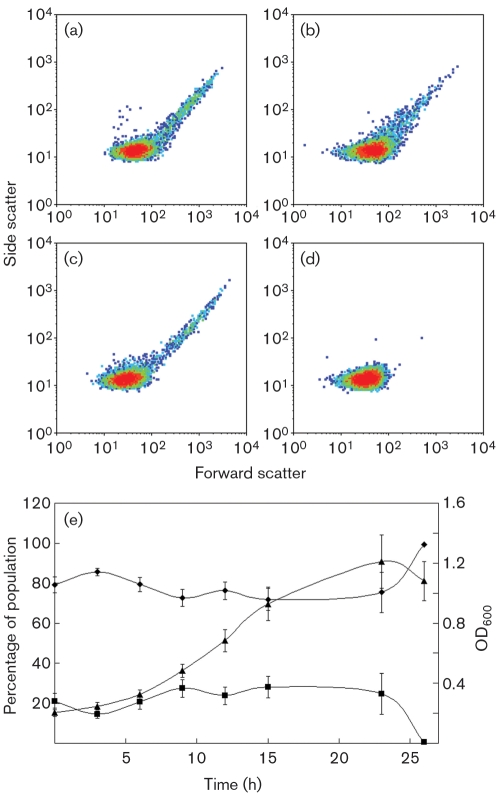
By plotting fluorescence or side scatter versus forward scatter, two subpopulations could be discerned for cultures grown on succinate or methanol (Fig. 3a, b). The subpopulation within the gated region (R1) consisted of 14–28 % of the events detected and exhibited a lognormal distribution during the exponential phase of growth (data not shown). The majority of the events detected (72–86 %) were below this gated region, and these exhibited a Gaussian distribution (data not shown). Fluorescence data summed from cells that were in the 103–104 RFU (relative fluorescence units) range were found to be 3–45 times greater than cells that were in the 100–103 RFU range. When methanol cultures transitioned into the stationary phase, only cells below the gated region could be detected (Fig. 4d, e), further defining the two subpopulations and suggesting that the cells below the gated region included those that were not actively growing and dividing. In succinate-grown cultures, the disappearance of cells within the gated region in stationary phase was not observed (Fig. 4b).
Fig. 3.
Representative distributions during exponential growth, which exhibit two subpopulations within bulk cultures, from methanol-grown cultures. (a) Side scatter versus forward scatter, which contains an overlay of calibration data obtained from 3 μm beads (indicated by arrow). (b) Fluorescence from the FL1 channel versus forward scatter. The R1 region in the fluorescence versus forward scatter plots was used to gate data for further analysis to gauge PmxaF response.
Fig. 4.
Side scatter versus forward scatter distributions for succinate-grown (a and b) and methanol-grown (c and d) cells in exponential phase (a and c) and stationary phase (b and d). (e) Distributions throughout a growth curve for methanol-grown cells; ⧫, non-gated region; ▪, gated region; ▴, OD600. Means±sd are plotted (n=12).
These data are consistent with the smaller lognormally distributed subpopulation within the region R1 representing actively growing and dividing cells, and the Gaussian-distributed subpopulation outside of region R1 representing a mixture of newly divided and non-dividing cells (cells that are not actively dividing at the time the sample was taken; Fig. 4d).
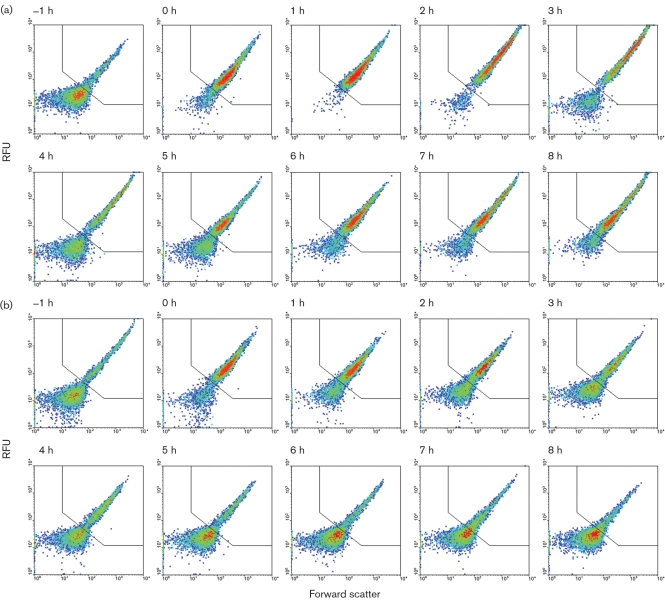
During carbon shift experiments, the switch from succinate to methanol resulted in a rapid increase in GFPuv expression, with most of the population (∼ 85 %) shifting into the gated region R1 after the first hour (Fig. 5a). The remaining ∼15 % of the total population did not respond to the carbon shift to methanol. Time points 0–3 h showed continued increase in GFPuv expression. Four hours after the shift to methanol as a carbon source, the resumption of growth was observed by optical density measurements and the reappearance of cells outside of region R1, presumably as a result of cell division.
Fig. 5.
Fluorescence from the FL1 channel versus forward scatter for carbon shift experiments. Carbon shifts were performed 30 min prior to the 0 h time point and data were plotted on a log scale. (a) Carbon shift from succinate→methanol. (b) Carbon shift from methanol→succinate.
Cultures shifted from methanol to succinate showed a steady decline of cells within the gated region (Fig. 5b). A brief spike in GFPuv expression was observed 0–2 h after the shift to succinate.
Analysis of population heterogeneity at the single-cell level
The flow cytometry data suggested that a significant proportion of the cells in exponentially growing populations might be non-dividing, but the actual proportion cannot be determined from flow cytometry experiments, since it is not possible to follow individual cells through growth and division. Therefore, experiments were carried out to microscopically observe single cells of M. extorquens AM1 growing on methanol or succinate, using a flow-through culture system and a non-motile mutant of M. extorquens AM1 (TSXCM174) (Strovas et al., 2007).
As previously described (Strovas et al., 2007), the cells were attached by their flagella to a poly-l-lysine-coated glass slide in the flow-through system, and were observed during multiple divisions. Daughter cells were swept away by the flow, allowing observation of division in only the originally attached cell. Cell division was observed by laser scanning microscopy, and the proportion of actively dividing cells was calculated (Table 3). In keeping with the flow cytometry results, a significant proportion of the cells did not divide over the course of 12–30 h observation, while during this time period other cells divided multiple times. In addition, during the course of observation, a small proportion that was actively dividing stopped dividing for the duration of the experiment and occasionally these non-dividing cells were observed to lyse as inferred by the loss of GFPuv fluorescence. The proportion of cells that were non-dividing was 52 % and 58 % during growth on succinate and methanol, respectively, in this flow-through microscope-based system, and these cells were observed to be attached at the cell surface rather than by flagella. Lastly, it should be noted that no clumping of cells was observed during all flow-through culture experiments.
Table 3.
Proportions of actively dividing cells, non-dividing cells, and dividing cells that become non-dividing for populations of M. extorquens AM1 grown on either succinate or methanol, as determined from microscopy experiments
| Carbon source | Dividing | Non-dividing | Dividing→Non-dividing | Total cells |
|---|---|---|---|---|
| Succinate | 44.1 % | 52.0 % | 3.9 % | 413 |
| Methanol | 36.9 % | 57.6 % | 5.5 % | 396 |
DISCUSSION
In this study we employed the fluorescent protein GFPuv to address population heterogeneity in exponentially growing cultures of M. extorquens AM1. First, fluorimetry was used to follow gene expression in bulk cultures and validate GFPuv as a reporter system for M. extorquens AM1 gene-expression dynamics. With the reporter system validated, flow cytometry and laser scanning microscopy were then employed to investigate population distribution in terms of cell size and fluorescence intensity.
The flow cytometry experiments revealed the presence of subpopulations of cells rather than a homogeneous or Gaussian distribution of cells. Based on the data collected from cultures grown on succinate and methanol during exponential and stationary phase, populations could be placed into two subpopulations, designated according to gating cytometry data: 14–28 % of the total population consisted of larger cells expressing high levels of GFP, consistent with actively growing cells undergoing division, while the remainder of the population consisted of cells that were relatively smaller and expressing lower levels of GFP, consistent with either newly divided or non-dividing cells. Only ∼85 % of the total population responded to carbon shift, further supporting the suggestion that the population consisted partly of cells that were metabolically active and partly of cells that were much less active. The presence of a subpopulation of non-active cells is consistent with a previous study showing that 15–25 % of the cells from mid-exponential phase in batch culture on methanol did not exhibit detectable staining with the redox dye Redox Sensor Green, indicating low or no metabolic activity (Kalyuzhnaya et al., 2008). Furthermore, staining of M. extorquens AM1 cells with Redox Sensor Green followed by observation over time with a laser scanning microscope, using a growth chamber, showed that cells that did not accumulate the redox-sensing dye exhibited no growth (M. Kalyuzhnaya, personal communication). These results taken together suggest that in the order of 15 % of the cells in the population are non-dividing and have low or no metabolic activity. Direct confirmation of these subpopulations was obtained by observing cells in a flow-through system. In this environment, a large proportion of the cells did not divide over the course of at least three divisions by other cells. However, in the flow-through system, only the tethered cell is observed, not the newly divided daughter cell in each case. In addition, the non-dividing cells were observed to be attached differently than the dividing cells, by the cell surface instead of by the flagellum. It is possible that this tethered flow-through culture system resulted in an artificially high proportion of non-dividing cells becoming attached, which would in turn bias the proportion of non-dividing cells in the population that was observed. However, these results show that some part of the population in an actively growing population is non-dividing. These cells may be non-viable, or they may be dormant. In the microscopy studies, such cells remained intact for up to 30 h, while others were observed to lyse, suggesting that the intact cells may be dormant.
Clearly, in order to maintain a significant population of non-dividing cells in an actively growing population, some of the dividing cells must convert to non-dividing cells. This phenomenon was observed microscopically in the flow-through system, confirming that it can occur, although the frequency was low. The frequency of this transition in batch culture is not known, but the flow cytometry results would suggest it could be up to 15 % per generation. The generation of dormant cells in cultures is a well-known phenomenon and has been studied in E. coli with respect to antibiotic persistence (Balaban et al., 2004; Lewis, 2007). However, in that case the frequency is in the order of 10−5 in the wild-type, increasing to 10−2 in mutants (high-frequency persistence mutants; Moyed & Bertrand, 1983). The high frequency of apparently dormant cells in growing cultures of M. extorquens AM1 suggests that this bacterium may naturally have a high-frequency dormancy phenotype. Alternatively, a high frequency of cells may lose viability under these growth conditions.
The presence of subpopulations in significantly different physiological states suggests that response to perturbations such as carbon shift might differ for these subpopulations. The switch from multi-carbon growth to methylotrophic growth involves major changes in both assimilatory and dissimilatory metabolic pathways (Anthony, 1982), and a shift from energy-limited to reducing power-limited growth. Our results suggest that these subpopulations did respond differently to carbon shift.
As noted above, a portion of the population (∼15 %) does not respond to carbon shift and is potentially the same portion that is small, non-dividing, and characterized by low or no respiration rate by a redox-sensing dye. Most of the population (∼85 %) responds. However, within the subpopulation of larger, higher GFPuv-expressing, and dividing cells, a significant number of cells have extremely high fluorescence (103–104 RFU from cytometry data). It was calculated that the sum of the fluorescence from these cells dominated the total fluorescence from the population (up to ∼3–45-fold brighter), even though they represented between 3 % and 15 % of the total population with no correlation to the optical density of the culture (data not shown). The large subpopulation that is small and dim contributes relatively little to the total fluorescence from the GFPuv fusion. This phenomenon was especially apparent during the carbon shift experiments and suggests that this small very bright subset of the population dominates bulk fluorimeter data, skewing the contributions of fluorescence response from the majority of the population.
The data we present here indicate that, in M. extorquens AM1, observing bulk population response to stimuli, as an average, does not fully describe the underlying contributions of the individuals that make up the population. Data averages in this study were dominated by the response of only a small subset of the population, based on fluorescence data from cytometric experiments, while a large portion of the population is either not responding or contributing little to the total population response. By observing population dynamics with flow cytometry during carbon shift experiments, it was possible to directly observe the significant proportion of the population that was non-dividing and did not respond to the shift.
It has been suggested that physiological heterogeneity in bacterial cultures provides a selective advantage in fluctuating environments (Kussell & Leibler, 2005; Acar et al., 2008) and in response to stress (Booth, 2002; Aertsen & Michiels, 2005). Carbon shift is an environmental perturbation that often creates dramatic metabolic change. In this study, we show that when M. extorquens AM1 is shifted from one metabolic mode to another, the population response is dominated by a relatively minor proportion of cells, supporting the concept that physiological diversity allows a physiologically distinct subset of a bacterial population to respond in a way that sacrifices a subset of the population but benefits the population as a whole.
Acknowledgments
We would like to thank Joseph Chao for assistance with microscopy, Fred Lewis and Michele Black for assistance with cell cytometry, members of the Lidstrom group and of the Microscale Life Sciences Center for helpful advice. This work was supported by a grant from NHGRI (P50 HG02360) for a Center of Excellence in Genomic Sciences.
Abbreviations
RFU, relative fluorescence units
References
- Acar, M., Mettetal, J. T. & van Oudenaarden, A. (2008). Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40, 471–475. [DOI] [PubMed] [Google Scholar]
- Aertsen, A. & Michiels, C. W. (2005). Diversify or die: generation of diversity in response to stress. Crit Rev Microbiol 31, 69–78. [DOI] [PubMed] [Google Scholar]
- Albano, C. R., Randers-Eichhorn, L., Bentley, W. E. & Rao, G. (1998). Green fluorescent protein as a real time quantitative reporter of heterologous protein production. Biotechnol Prog 14, 351–354. [DOI] [PubMed] [Google Scholar]
- Andersen, J. B., Sternberg, C., Poulsen, L. K., Bjorn, S. P., Givskov, M. & Molin, S. (1998). New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64, 2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, C. (1982). The Biochemistry of Methylotrophs. London: Academic Press.
- Attwood, M. M. & Harder, W. (1972). A rapid and specific enrichment procedure for Hyphomicrobium spp. Antonie Van Leeuwenhoek 38, 369–377. [DOI] [PubMed] [Google Scholar]
- Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. [DOI] [PubMed] [Google Scholar]
- Banerjee, B., Balasubramanian, S., Ananthakrishna, G., Ramakrishnan, T. V. & Shivashankar, G. V. (2004). Tracking operator state fluctuations in gene expression in single cells. Biophys J 86, 3052–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts, R. J., Hautefort, I., Sidebotham, J. M. & Hinton, J. C. (2002). Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol 358, 43–66. [DOI] [PubMed] [Google Scholar]
- Booth, I. R. (2002). Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int J Food Microbiol 78, 19–30. [DOI] [PubMed] [Google Scholar]
- Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183–1186. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya, M. G. & Lidstrom, M. E. (2003). QscR, a LysR-type transcriptional regulator and CbbR homolog, is involved in regulation of the serine cycle genes in Methylobacterium extorquens AM1. J Bacteriol 185, 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya, M. G., Lidstrom, M. E. & Chistoserdova, L. (2008). Real-time detection of actively metabolizing microbes via redox sensing as applied to methylotroph populations in Lake Washington. ISME J 2, 696–706. [DOI] [PubMed] [Google Scholar]
- Korotkova, N., Chistoserdova, L. & Lidstrom, M. E. (2002). Poly-β-hydroxybutyrate biosynthesis in the facultative methylotroph Methylobacterium extorquens AM1: identification and mutation of gap11, gap20, and phaR. J Bacteriol 184, 6174–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, Y., Biran, I. & Walt, D. R. (2004). Simultaneously monitoring gene expression kinetics and genetic noise in single cells by optical well arrays. Anal Chem 76, 6282–6286. [DOI] [PubMed] [Google Scholar]
- Kussell, E. & Leibler, S. (2005). Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078. [DOI] [PubMed] [Google Scholar]
- Leveau, J. H. & Lindow, S. E. (2001). Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J Bacteriol 183, 6752–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5, 48–56. [DOI] [PubMed] [Google Scholar]
- Lissemore, J. L., Jankowski, J. T., Thomas, C. B., Mascotti, D. P. & deHaseth, P. L. (2000). Green fluorescent protein as a quantitative reporter of relative promoter activity in E. coli. Biotechniques 28, 82–89. [DOI] [PubMed] [Google Scholar]
- Marx, C. J. & Lidstrom, M. E. (2001). Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147, 2065–2075. [DOI] [PubMed] [Google Scholar]
- Marx, C. J. & Lidstrom, M. E. (2002). Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Marx, C. J. & Lidstrom, M. E. (2004). Development of an insertional expression vector system for Methylobacterium extorquens AM1 and generation of null mutants lacking mtdA and/or fch. Microbiology 150, 9–19. [DOI] [PubMed] [Google Scholar]
- Marx, C. J., Van Dien, S. J. & Lidstrom, M. E. (2005). Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol 3, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed, H. S. & Bertrand, K. P. (1983). hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155, 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza, J. M. & van Oudenaarden, A. (2005). Noise propagation in gene networks. Science 307, 1965–1969. [DOI] [PubMed] [Google Scholar]
- Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S. & Elowitz, M. B. (2005). Gene regulation at the single-cell level. Science 307, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Siegele, D. A. & Hu, J. C. (1997). Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A 94, 8168–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strovas, T. J., Sauter, L. M., Guo, X. & Lidstrom, M. E. (2007). Cell-to-cell heterogeneity in growth and gene expression in Methylobacterium extorquens AM1. J Bacteriol 189, 7127–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dien, S. J. & Lidstrom, M. E. (2002). Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C3 and C4 metabolism. Biotechnol Bioeng 78, 296–312. [DOI] [PubMed] [Google Scholar]
- Van Dien, S. J., Okubo, Y., Hough, M. T., Korotkova, N., Taitano, T. & Lidstrom, M. E. (2003). Reconstruction of C3 and C4 metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology 149, 601–609. [DOI] [PubMed] [Google Scholar]
- Vorholt, J. A., Marx, C. J., Lidstrom, M. E. & Thauer, R. K. (2000). Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol 182, 6645–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. & Lidstrom, M. E. (2003). Promoters and transcripts for genes involved in methanol oxidation in Methylobacterium extorquens AM1. Microbiology 149, 1033–1040. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Duong, T., Huang, C. C., Kain, S. R. & Li, X. (1999). Comparison of enhanced green fluorescent protein and its destabilized form as transcription reporters. Methods Enzymol 302, 32–38. [DOI] [PubMed] [Google Scholar]