Abstract
The genetic basis for the evolution of development includes genes that encode proteins expressed on the surfaces of sperm and eggs. Previous studies of the sperm acrosomal protein bindin have helped to characterize the adaptive evolution of gamete compatibility and speciation in sea urchins. The absence of evidence for bindin expression in taxa other than the Echinoidea has limited such studies to sea urchins, and led to the suggestion that bindin might be a sea urchin-specific molecule. Here we characterize the gene that encodes bindin in a broadcast-spawning asterinid sea star (Patiria miniata). We describe the sequence and domain structure of a full-length bindin cDNA and its single intron. In comparison to sea urchins, P. miniata bindin is larger but the two molecules share several general features of their domain structure and some sequence features of two domains. Our results extend the known evolutionary history of bindin from the Mesozoic (among the crown group sea urchins) into the early Paleozoic (and the common ancestor of eleutherozoans), and present new opportunities for understanding the role of bindin molecular evolution in sexual selection, life history evolution, and speciation among sea stars.
INTRODUCTION
Gene products expressed on the surfaces of planktonic gametes mediate reproductive success, sperm competition, sexual selection, reinforcement, and other important demographic and ecological processes in the ocean (Vacquier and Moy 1977; Brandriff et al. 1978; Palumbi 1994; Vacquier et al. 1995; Lessios 2007). Well-studied examples include lysin and bindin expressed in the acrosomal vesicles of mollusk and echinoderm sperm (Swanson and Vacquier 2002). The molecular evolution of bindin (Metz & Palumbi 1996; Biermann 1998; Metz et al. 1998; Zigler et al. 2003; Zigler and Lessios 2003; McCartney and Lessios 2004; Zigler and Lessios 2004) and its role in gamete compatibility and fertilization success (Zigler et al. 2005; Lessios 2007; Zigler 2008) has been carefully analyzed in studies that include bindin sequences from six orders of sea urchins (Echinoidea). Patterns of bindin variation include high rates of amino acid replacement substitutions in coding sequences, highly variable repetitive domain structures, high or very low within-species polymorphism, and strong divergence of alleles within and between some sympatric species (Swanson and Vacquier 1998, 2002; Hellberg et al. 2000; Zigler and Lessios 2003; Moy et al. 2008). These patterns have been interpreted as the products of reinforcement, strong directional selection (and selective sweeps), sperm competition, and sexual conflict (Palumbi 1994, 1999; Swanson and Vacquier 2002; Lessios 2007).
A fuller understanding of the coevolution between bindin and life history traits such as sperm competition has been limited to members of the crown group of sea urchins with a most recent common ancestor about 250 million years ago (Smith 1988; Smith et al. 1995; Ziegler & Lessios 2003). Previous studies (Moy & Vacquier 1979; Minor et al. 1991) using sea urchin bindin sequences as molecular probes have failed to find a bindin homolog among other members of the Eleutherozoa including the sea stars (Asteroidea). Echinoids and asteroids last shared a common ancestor in the early Paleozoic (Sumrall and Sprinkle 1998; Janies 2001; Blair and Hedges 2005).
Discovery of a sea star bindin homolog would greatly extend both the known evolutionary history of this molecule (by more than 200 million years) and the opportunities for studying the coevolution of bindin and life histories. A promising group for such a study is the valvatid sea star family Asterinidae (cushion stars), which includes highly diverse mating systems (gonochoric, hermaphrodite) and modes of fertilization (planktonic, benthic, internal) that might be expected to coevolve with gamete recognition molecules (Swanson and Vacquier 2002; Byrne 2006; Zigler 2008). Here we identify and characterize bindin from the northeast Pacific asterinid Patiria miniata, and compare the structure of the predicted gene product to sea urchin bindin.
MATERIALS AND METHODS
We analyzed bindin from male Patiria miniata collected from an intertidal habitat near the Bamfield Marine Sciences Centre (48° 49′ N, 125° 9′ W). We used the same approach previously described for discovery and characterization of abalone gamete recognition genes (Aagaard et al. 2006) to prepare total RNA from P. miniata testes by guanidinium/cesium chloride centrifugation, isolate mRNA, directionally clone cDNA fragments, sequence randomly selected clones (~1000), process these single-strand sequences, and assemble expressed sequence tags (ESTs) into contigs. BlastX comparison of the ESTs against the nonredundant protein database at the National Center for Biotechnology Information (www.ncbi.nih.gov) was used to identify significant sequence similarities to sea urchin bindin. We used those provisionally identified bindin ESTs as the basis for design of primers (Table 1) for rapid amplification of cDNA ends (RACE; see Aagaard et al. 2006) to extend the partial bindin sequence from the EST library into a full-length coding sequence (GenBank accession FJ439659). We supplemented the RACE results with targeted amplifications of coding sequences from double-stranded cDNA. We used some of the same primers for PCR amplification from genomic DNA: amplicons that were larger than the size predicted from the coding sequence were cloned and sequenced from both directions in order to characterize the associated intron (GenBank accession FJ517556).
Table 1.
Primer sequences used for bindin amplification
| Name | 5’-Sequence-3’ | Codons | Application1 | Target |
|---|---|---|---|---|
| AGCf | GACACTACATGTCCTGCAGGATGTCG | 25-33 | RACE, cDNA | preprobindin |
| AGCf2 | CATGTCCTGCAGGATGTCG | 27-33 | gDNA | preprobindin |
| TALf | GGTCATTGTATCTACCAACAGCGCTGG | 188-197 | RACE | preprobindin, 1st repeat cluster |
| AAEf | GGCACGACGTGCCGCAGAG | 227-233 | RACE, gDNA | 1st repeat |
| AAEr | CTCTGCGGCACGTCGTGCC | 233-227 | gDNA | preprobindin |
| VEGKf | GTTAAACCAGTGGAAGGGAAGC | 407-414 | gDNA | 2nd repeat cluster |
| VEGKr | GCTTCCCTTCCACTGGTTTAAC | 414-407 | gDNA | 1st repeat |
| QPAf | GGAATCGGAGTCACAACCAGCGG | 925-933 | RACE, gDNA | ‘hot spot’, intron |
| QPAr | CCGCTGGTTGTGACTCCGATTCC | 933-925 | gDNA | non-repetitive sequence, 2nd repeat |
| DLPf | GGAATCTACGAAGATTGACCTTCCTGG | 967-976 | RACE | core region, 3’ UTR |
| VLSr | CGTCGTTTTCATTCATGGCGCTGAGG | 1019-1010 | RACE, cDNA | core region |
| VLSr2 | GTTTTCATTCATGGCGCTGAGG | 1017-1010 | gDNA | core region, intron |
Primers were used for rapid amplification of cDNA ends (RACE), or in forward (f) and reverse (r) pairs for targeted amplification of the coding sequence only (cDNA) or the genomic sequence (gDNA).
We repeated the comparison of the full-length coding sequence to the nonredundant protein database; searched for repetitive domains in RADAR (www.ebi.ac.uk/Radar); searched for other domains in SMART (smart.emblheidelberg.de); searched for peptide cleavage sites in ProP (www.cbs.dtu.dk/services/ProP/); and characterized several features of the predicted bindin peptide in ProtParam (ca.expasy.org/cgi-bin/protparam) and ProtScale (ca.expasy.org/tools/protscale.html). The RADAR results were combined with visual alignment of sequences to generate a nucleotide alignment of codons from repetitive domains. This alignment was used to calculate the ratio of nonsynonymous to synonymous nucleotide substitution rates (ω) between repeats in SNAP (www.hiv.lanl.gov/content/sequence/SNAP).
We confirmed the expression of the predicted bindin protein in P. miniata sperm by the same tandem mass spectrometry (MS/MS) method previously used to successfully identify predicted proteins from complex protein mixtures (e.g., Aagaard et al. 2006). Total sperm proteins from four individual males were precipitated with 25% trichloroacetic acid, washed twice with 100% acetone, and dried in a speedvac. Protein pellets were solubilized, reduced and alkylated, and then digested with trypsin as described previously (Aagaard et al. 2006). The resulting peptides were separated by HPLC and electrosprayed directly into an LTQ linear ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA). The acquired tandem mass spectra were searched against a database containing the 6-reading-frame translations of P. miniata testis ESTs, the full length P. miniata bindin protein, proteins of common contaminants (e.g. trypsin, keratin), and a shuffled decoy database using a parallelized implementation of SEQUEST (MacCoss et al. 2002; Sadygov et al. 2002). The program DTASelect (Tabb et al. 2002) was used to filter the peptide identifications and assemble the peptides and proteins, with DTASelect filters adjusted to produce protein identifications with a false discovery rate of ≤ 5%. Finally, peptides identified from all four sperm protein samples were pooled and mapped to the full bindin protein sequence.
RESULTS
One contig from the EST library (EX452594, 1484 bp) was similar to a large number of sea urchin bindin genomic and cDNA sequences (expectation values as low as E = 0.0003). The major source of this similarity was a sequence corresponding to the so-called invariant core region of sea urchin bindin (Vacquier et al. 1995; Zigler and Lessios 2005). The 3’ RACE sequences included the stop codon, a 703 bp 3’ untranslated region, and the poly-A tail. Initial 5’ RACE results showed several different repetitive regions, plus sequences that matched an EST (EX452859, 810 bp) from near the 5’ end of the gene for which we found no significant BlastX or BlastN similarity to other sequences in the databases. Because the repetitive region between the invariant core and the start codon was large and included some long repeats (~100 bp), we were not able to use RACE amplicons alone to sequence through the entire repetitive region and obtain a confidently aligned complete coding sequence. For this reason, we supplemented the EST and RACE results with sequences from PCR products amplified from cDNA using additional primers designed from the preliminary RACE results (see Fig. 1, Table 1).
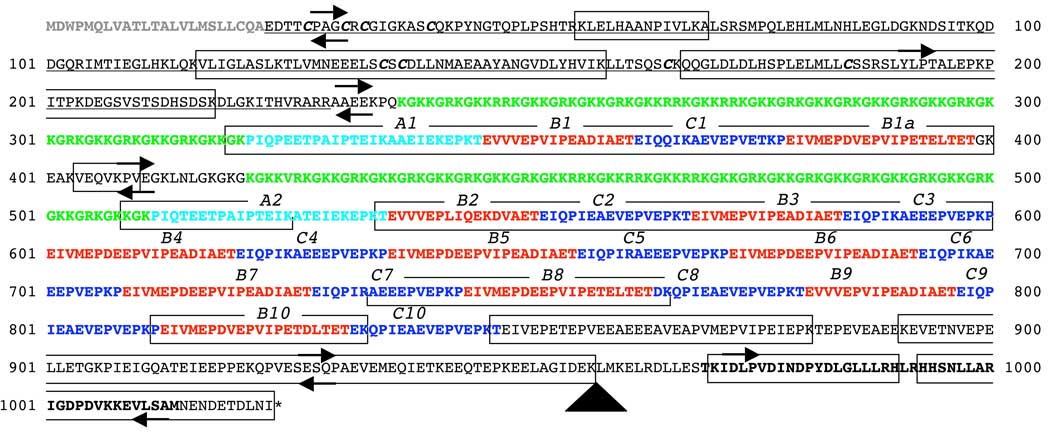
Fig. 1.
Inferred amino acid sequence for the complete Patiria miniata bindin cDNA. Domains are distinguished by type face: signal sequence (gray), collagenlike (green), A (cyan), B (red), C (blue), core (bold). Preprobindin sequence (ending in the probable cleavage motif RARR) is underlined. Highly conserved cysteine residues in preprobindin are shown in bold italics. Repeats are numbered above each sequence. Arrows show location and direction of PCR primers (Table 1) used in RACE experiments or in amplification from cDNA or genomic DNA. Large arrowhead shows the location of a single 1005 bp intron in codon 959 (leucine). Boxes show peptide fractions identified in sperm by tandem mass spectrometry.
The complete bindin coding sequence was 3072 bp (1024 codons; Fig. 1). The most common amino acids of the predicted protein were glutamic acid (180 of 1024), lysine (145), and proline (103); phenylalanine (0), tryptophan (1), and tyrosine (5) were absent or rare. The predicted molecular weight was 114 kD.
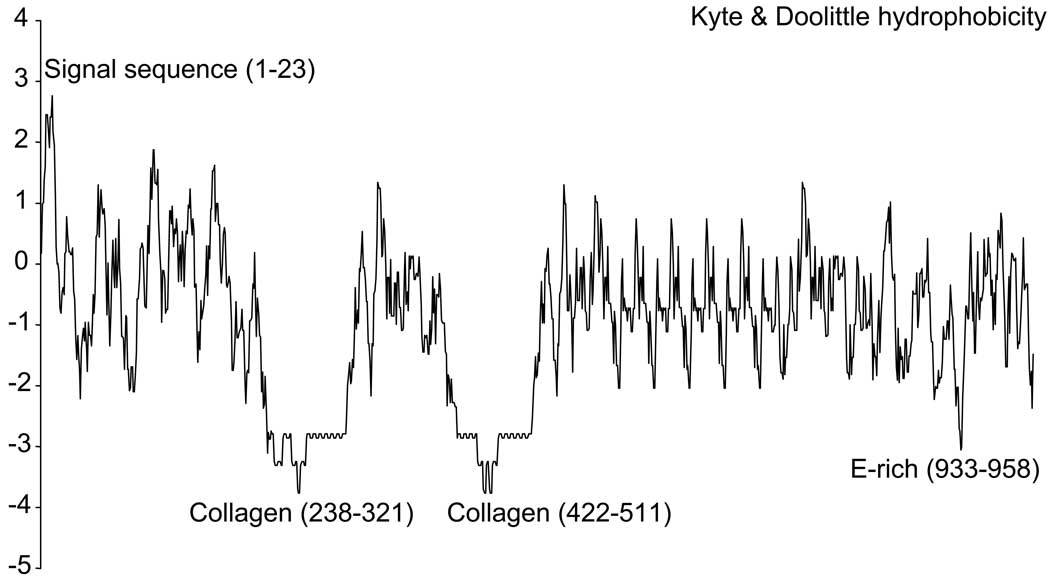
The signal peptide (codons 1–23) was the only region of the predicted amino acid sequence with hydrophobicity scores > 2 (Fig. 2). A 642 bp non-repetitive domain (codons 24–237) lacked substantial sequence similarity (E ≥ 0.03) to nucleotide or protein sequences in the databases but had several structural and functional features similar to the preprobindin domain of sea urchins: (1) a short amino acid motif (CSCD, codons 136–139) identical to part of the preprobindin of sea urchins (Gao et al. 1986; Minor et al. 1991); (2) six additional cysteine codons in a conserved arrangement relative to the CSCD motif; and (3) a highly basic RARR motif (codons 227–230) that ProP identified as a furin-type peptide cleavage site similar to the trypsin-like cleavage site in sea urchin preprobindins. These functional similarities suggest that the N-terminal domain is cleaved from the mature bindin, which is predicted to start at codon 231 (alanine) and have a molecular weight of 88 kD.
Two domains (codons 238–321; 422–511) each included 13 copies of a KGKK(G/R)R repeat; the glycine at most of the second and fifth codons in these repeats made the sequences highly similar (BlastP E ≤ 1×10−6) to sea urchin alpha collagens. Both domains ended with a unique KGKKGK motif; they differed from each other by two G↔R amino acid substitutions and by an additional KGKKVR repeat at the 5’ end of the second domain. In contrast to the signal sequence, the two collagen-like domains were highly hydrophilic (Fig. 2).
Fig. 2.
Hydrophobicity of Patiria miniata bindin using the Kyte and Doolittle (1982) method.
The collagen-like domains were each followed by a complex of three larger tandemly repeated domains (A, B, C) that were not similar to protein or nucleotide sequences in the databases. Repeat domains A (starting at codons 328, 512) were PIQ(P/T)EETPAIPTEIKA(A/T)EIEKEPKT motifs. The two A repeats differed at just two (non-synonymous) nucleotide sites. These two domains were each followed by tandem repeats of domains B (16 or 20 codons) and C (16 codons; Fig. 1). The repeat structure following the first collagen-like domain was ABCB; the second longer repeat structure was A(BC)9. The overall structure of this large repetitive region of the gene was consistent with duplication of an ancestral collagen-A-B-C repeat, followed by addition of a B repeat (B1a) to the first cluster and tandem duplication of BC pairs in the second cluster.
Among 11 B repeats no copies differed by more than 14 of 60 aligned sites; the average relative rate of nonsynonymous substitutions was ω = 0.036. Four of the B repeats shared a four-codon insertion-deletion (DEEP, Fig. 1); one of these (B2) included unique V↔L, P↔Q, and A↔K substitutions (codons 543, 545, 547) and may be under selection for a different function from five other B repeats (B3–B7; ω= 4.9). Among 10 C repeats no copies differed by more than 9 of 48 sites in total (average pairwise ω = 0.36); many ω values were undefined for pairs of repeats with 1–4 nonsynonymous differences but no synonymous differences. The C10 repeat was followed by a 363 bp sequence in which RADAR identified two other domains (codons 864–882; 915–950) with amino acid motifs similar to parts of the B and C repeats but with more problematic alignments. This region included a lysine- and glutamate-rich region (codons 933–958) that was the only other part of the gene predicted to be as hydrophilic as the collagen domains (Fig. 2).
The last distinctive bindin domain (codons 970–1014) was strongly (10−17 < E < 10−7) similar to the central part of the invariant core domain of sea urchin bindin. Of those 45 codons, 40 encoded amino acids identical to the sea urchin core region. The central portion of this core domain (the LGLLLRHLRHHSNLLARI motif) showed just one amino acid difference from the so-called ‘B18’ region of sea urchin bindin that is invariant among sea urchin orders and is thought to mediate sperm-egg fusion (Zigler 2008). Ten codons separated the 3’ end of the core domain from the stop codon.
Tandem mass spectrometry confirmed the expression of the predicted protein in sperm. We identified peptides corresponding to most of the predicted mature bindin protein (Fig. 1) plus some regions of the prepropeptide. Total coverage was 488/1024 residues (47.7%). The expressed peptides in the mature bindin sequence included both of the A repeats, most of the B and C repeats, most of the nonrepetitive region downstream of the C10 repeat, and all but two residues of the core region. Notably absent from all of the identified peptides were the two collagen-like repeats. We found that an unquantified but large majority of the total sperm proteins could not be solubilized prior to HPLC, and we were therefore not able to quantify the expression of bindin in sperm relative to other commonly expressed proteins.
All but one PCR products from genomic DNA using the RACE and cDNA PCR primers (Fig. 1) were of the size predicted from the coding sequence. The exception was a large amplicon that spanned the coding region between the QPAf primer (downstream of the last C repeat; Table 1) and the VLSr2 primer (in the core domain) and included a 1005 bp intron. The splice site for the intron was inferred to be between the first (T) and second (T) nucleotides in codon 959 (a TTG leucine): the cDNA nucleotide sequence of the three codons 958–960 spanning the intron site was AAATTGATG, and the 5’ and 3’ ends of the corresponding genomic intron sequence were, respectively, AAATgtaag… and …gacagTGATG.
DISCUSSION
Patiria miniata bindin shares some of the domain and genomic features of sea urchin bindin but is considerably larger. Sea urchins express a ~50–80 kD bindin molecule that contains a signal sequence and a cysteine-rich prepropeptide sequence that are separated from several repetitive domains and the invariant core region by a trypsin-like RKKR or RQRR enzymatic cleavage site. The prepropeptide is cleaved from the C-terminal mature protein (~25-40 kD) in sperm. Bindin of P. miniata has a similar overall structure: a cysteine-rich prepropeptide, an RARR cleavage site (for furin rather than trypsin, similar to the RVRR cleavage site of the sand dollar Encope stokesi; Zigler and Lessios 2003), several different repetitive elements, and a domain similar to the sea urchin core region. These similarities even extend to the location of the single intron near the 5’ end of the core domain, although the sea urchin bindin intron occurs in a conserved valine codon (rather than in a leucine codon in the sea star). The mature peptide is predicted to be about twice as large (88 kD) as the largest known sea urchin bindin (Zigler and Lessios 2003), and similar in size to the major acrosomal vesicle protein (presumably bindin) isolated from sperm of a forcipulate sea star, Pisaster ochraceus (95 kD; Christen 1985).
The conservation of the preprobindin amino acid sequence in the mature P. miniata protein suggests a conserved function in localization to the acrosomal vesicle. Others (Minor et al. 1991; Vacquier et al. 1995) have noted the conservation of eight cysteine residues (at codons 32, 39, 41, 48; 102, 104; 128, 147; Minor et al. 1991) in sea urchin preprobindin, the absence of cysteine (and disulphide bridges) and tryptophan in mature sea urchin bindin (Zigler and Lessios 2003), and the probability of limited stable secondary structure in the functional protein (Biermann 1998). Patiria miniata bindin also lacks tryptophan, and shows remarkable conservation of the relative positions of the same eight cysteine residues in the N-terminal portion (at codons 28, 32, 34, 41; 136, 138; 166, 185) of the protein, but not of the phenylalanine, tryptophan, and tyrosine residues with aromatic side chains that are conserved among sea urchin preprobindins (Minor et al. 1991).
Specific amino acid sequence similarities to sea urchins were limited to a very small number of codons: the CSCD motif in preprobindin; six additional cysteine codons; the RARR peptide cleavage motif; and 40 codons of the core domain (54 total, 5.3%). Apart from these specific amino acid sequence similarities (in preprobindin and the core domain), P. miniata bindin would not be identifiable as a sea urchin bindin homologue. Comparable sequence divergence outside the core domain of mature bindin sequences occurs between sea urchin orders as well, and has been interpreted as evidence for the importance of bindin repeat insertion-deletions (in addition to amino acid substitutions) in the evolution of bindin function and the specificity of fertilization.
The repetitive structure of P. miniata bindin also suggests that it might experience selection for adaptive evolution similar to selection acting in some sea urchins. In sea urchins, the self-affinity of bindin (to form a kind of molecular glue between sperm and egg) and its species-specific interaction with the egg receptor (to mediate fertilization) depend in part on the number and length of repetitive elements around the core domain (Vacquier et al. 1995). Zigler (2008) noted that such bindin repeat variation has only been discovered in genera of the order Echinoida and is consistently associated with positive selection for between-species bindin divergence (Minor et al. 1991; Biermann 1998). Analogous repeat differences between congeners are an important feature of variation in sea urchin genes that encode the egg bindin receptor (Kamei and Glabe 2003). The number and amino acid sequences of some repeats may have important functional consequences at the level of interactions between molecules and between gametes (Biermann et al. 2004; McCartney and Lessios 2004; Zigler et al. 2005; Levitan and Ferrell 2006). The nucleotide and amino acid sequences of P. miniata bindin repeat domains bear no resemblance to sea urchin bindin repeats, and are restricted to the region 5’ of the core. Functional studies of this repetitive variation (e.g., Minor et al. 1991) are needed to determine the significance of the four different repetitive elements relative to the fertilization specificity of bindin, and comparative studies are needed to determine whether other repeat domain types are found in other sea star bindins (as in sea urchins).
Specific candidate targets for selection include the two highly hydrophilic collagen-like domains that are likely to be displayed on the bindin surface (Fig. 2). Because collagens self-assemble into fibrils (Kadler et al. 1996), the presence of collagen-like domains in P. miniata bindin could mediate self-affinity among bindin molecules in a way that affects the orientation of other elements (including repeats and the core domain) toward the egg receptor. Self-affinity among collagen-like domains of bindin proteins could also promote the formation of insoluble bindin masses (Vacquier and Moy 1977) on the activated sperm head and in vitro (as we observed in sperm protein extractions). The third highly hydrophilic domain, between the C10 repeat and the core, is also in the same location (relative to repetitive elements, the intron, and the core domain) as the glutamate-rich ‘hot spot’ of high ω values between some closely related sea urchin species (Biermann 1998; Zigler and Lessios 2003). Another likely target of selection is the highly divergent B2 repeat, which may have experienced positive selection for amino acid replacements relative to other B paralogs.
The discovery and characterization of sea star bindin provides the basis for several kinds of comparative studies relative to the evolution of life histories and variation in the strength of sexual selection among sea star species and clades. For example, species in several asterinid genera (Aquilonastra, Asterina, Cryptasterina, Parvulastra) show parallel evolution of benthic self-fertilization by small hermaphrodite adults (Byrne 1996, 2005, 2006). If the rate and pattern of bindin molecular evolution is strongly dependent on the strength of sexual selection among males, then surveys of bindin variation may reveal lower rates of within-species molecular evolution consistent with weaker sexual selection among ‘males’ in these species compared to large-bodied gonochoric asterinids with group spawning, higher potential for strong sperm competition, and sexual selection among males (as in some sea urchins; e.g., Levitan and Ferrell 2006; Levitan 2008). Comparable patterns are known from other gene systems in copulating vertebrates and arthropods, especially male-expressed seminal fluid proteins and their female-expressed receptors that mediate sperm competition, cryptic female choice, and sexual conflict in the female reproductive tract (Clark et al. 2006; Panhuis et al. 2006; Calkins et al. 2007; Haerty et al. 2007; Almeida and DeSalle 2008; Dean et al. 2008; Findlay et al. 2008). In general, the evolution of highly derived developmental patterns among closely related species of asterinids and other sea stars (e.g., Flowers and Foltz 2001; Foltz et al. 2008) provides rich opportunities for understanding the co-evolution of bindin (and other gamete recognition loci; Matsumoto et al. 1999), gamete interactions, mating systems, and other aspects of reproduction and early development.
ACKNOWLEDGMENTS
Thanks to Jenn Sunday for collecting adult sea stars, and to two anonymous reviewers for constructive criticisms. We are supported by grants from the Natural Sciences and Engineering Research Council (MWH), the National Science Foundation, and the National Institutes of Health (WJS), and by a scholarship from the Consejo Nacional de Ciencia y Tecnología (SP).
REFERENCES
- Aagaard JE, Yi XH, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc. Natl. Acad. Sci. USA. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida FC, DeSalle R. Evidence of adaptive evolution of accessory gland proteins in closely related species of the Drosophila repleta group. Mol. Biol. Evol. 2008;25:2043–2053. doi: 10.1093/molbev/msn155. [DOI] [PubMed] [Google Scholar]
- Bierman CH. The molecular evolution of sperm bindin in six species of sea urchins (Echinoida: Strongylocentrotidae) Mol. Biol. Evol. 1998;15:1761–1771. doi: 10.1093/oxfordjournals.molbev.a025902. [DOI] [PubMed] [Google Scholar]
- Biermann CH, Marks JA, Vilela-Silva ACES, Castro MO, Castro PAS. Carbohydrate-based species recognition in sea urchin fertilization: another avenue for speciation? Evol. Dev. 2004;6:353–361. doi: 10.1111/j.1525-142X.2004.04043.x. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Brandriff B, Moy GW, Vacquier VD. Isolation of sperm bindin from the oyster (Crassotrea gigas) Gamete Res. 1978;1:89–99. [Google Scholar]
- Byrne M. Viviparity and intragonadal cannibalism in the diminutive asterinid sea stars Patiriella vivipara and P. parvivipara. Mar. Biol. 1996;125:551–567. doi: 10.2307/1543057. [DOI] [PubMed] [Google Scholar]
- Byrne M. Viviparity in the sea star Cryptasterina hystera (Asterinidae)—conserved and modified features in reproduction and development. Biol. Bull. 2005;208:81–90. doi: 10.2307/3593116. [DOI] [PubMed] [Google Scholar]
- Byrne M. Life history diversity and evolution in the Asterinidae. Integr. Comp. Biol. 2006;46:243–254. doi: 10.1093/icb/icj033. [DOI] [PubMed] [Google Scholar]
- Calkins JD, El-Hinn D, Swanson WJ. Adaptive evolution in an avian reproductive protein: ZP3. J. Mol. Evol. 2007;65:555–563. doi: 10.1007/s00239-007-9034-8. [DOI] [PubMed] [Google Scholar]
- Christen R. Isolation of acrosomal vesicles and their surrounding membranes from starfish sperm. Devel. Growth Differ. 1985;27:529–538. doi: 10.1111/j.1440-169X.1985.00529.x. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Dean MD, Good JM, Nachman MW. Adaptive evolution of proteins secreted during sperm maturation: An analysis of the mouse epididymal transcriptome. Mol. Biol. Evol. 2008;25:383–392. doi: 10.1093/molbev/msm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Yi XH, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:1417–1426. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Foltz DW. Reconciling molecular systematics and traditional taxonomy in a species-rich clade of sea stars (Leptasterias subgenus Hexasterias) Mar. Biol. 2001;139:475–483. [Google Scholar]
- Foltz DW, Nguyen AT, Kiger JR, Mah CL. Pleistocene speciation of sister taxa in a North Pacific clade of brooding sea stars (Leptasterias) Mar. Biol. 2008;154:593–602. [Google Scholar]
- Gao B, Klein LE, Britten RJ, Davidson EH. Sequence of messenger RNA coding for bindin, a species-specific sea urchin sperm protein required for fertilization. Proc. Natl. Acad. Sci. USA. 1986;83:8634–8638. doi: 10.1073/pnas.83.22.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, et al. Evolution in the fast lane: Rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg ME, Moy GW, Vacquier VD. Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Mol. Biol. Evol. 2000;17:458–466. doi: 10.1093/oxfordjournals.molbev.a026325. [DOI] [PubMed] [Google Scholar]
- Janies D. Phylogenetic relationships of extant echinoderm classes. Can. J. Zool. 2001;79:1232–1250. [Google Scholar]
- Kadler KE, Holmes DF, Trotter JA, Chapman JA. Collagen fibril formation. Biochem. J. 1996;316:1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei N, Glabe CG. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lessios HA. Reproductive isolation between species of sea urchins. Bull. Mar. Sci. 2007;81:191–208. [Google Scholar]
- Levitan DR. Gamete traits influence the variance in reproductive success, the intensity of sexual selection, and the outcome of sexual conflict among congeneric sea urchins. Evolution. 2008;62:1305–1316. doi: 10.1111/j.1558-5646.2008.00378.x. [DOI] [PubMed] [Google Scholar]
- Levitan DR, Ferrell D. Selection on gamete recognition proteins depends on sex, density, and genotype frequency. Science. 2006;312:267–269. doi: 10.1126/science.1122183. [DOI] [PubMed] [Google Scholar]
- MacCoss MJ, Wu CC, Yates JR., 3rd Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Briones A, Nishigaki T, Hoshi M. Sequence analysis of cDNAs encoding precursors of starfish asterosaps. Dev. Gen. 1999;25:130–136. doi: 10.1002/(SICI)1520-6408(1999)25:2<130::AID-DVG7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- McCartney MA, Lessios HA. Adaptive evolution of sperm bindin tracks egg incompatibility in neotropical sea urchins of the genus Echinometra. Mol. Biol. Evol. 2004;21:732–745. doi: 10.1093/molbev/msh071. [DOI] [PubMed] [Google Scholar]
- Metz EC, Gómez-Gutiérrez G, Vacquier VD. Mitochondrial DNA and bindin gene sequence evolution among allopatric species of the sea urchin genus Arbacia. Mol. Biol. Evol. 1998;15:185–195. doi: 10.1093/oxfordjournals.molbev.a025914. [DOI] [PubMed] [Google Scholar]
- Metz EC, Palumbi SR. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol. Biol. Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- Minor JE, Fromson DR, Britten RJ, Davidson EH. Comparison of the bindin proteins of Strongylocentrotus franciscanus, Strongylocentrotus purpuratus, and Lytechinus variegatus – sequences involved in the species specificity of fertilization. Mol. Biol. Evol. 1991;8:781–795. doi: 10.1093/oxfordjournals.molbev.a040690. [DOI] [PubMed] [Google Scholar]
- Moy GW, Springer SA, Adams SL, Swanson WJ, Vacquier VD. Extraordinary intraspecific diversity in oyster sperm bindin. Proc. Natl. Acad. Sci. USA. 2008;15:1993–1998. doi: 10.1073/pnas.0711862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy GW, Vacquier VD. Immunoperoxidase localization of bindin during the adhesion of sperm to sea urchin eggs. Curr. Top. Dev. Biol. 1979;13:31–44. doi: 10.1016/s0070-2153(08)60688-2. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Genetic divergence, reproductive isolation, and marine speciation. Ann. Rev. Ecol. Syst. 1994;25:547–572. [Google Scholar]
- Palumbi SR. All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc. Natl. Acad. Sci. USA. 1999;96:12632–12637. doi: 10.1073/pnas.96.22.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis TM, Clark NL, Swanson WJ. Rapid evolution of reproductive proteins in abalone and Drosophila. Phil. Trans.R. Soc. B. 2006;361:261–268. doi: 10.1098/rstb.2005.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR. Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- Smith AB. Phylogenetic relationship, divergence times, and rates of molecular evolution for camarodont sea urchins. Mol. Biol. Evol. 1988;5:345–365. [Google Scholar]
- Smith AG, Littlewood DTJ, Wray GA. Comparing patterns of evolution: larval and adult life history stages and ribosomal RNA of post-Paleozoic echinoids. Phil. Trans. R. Soc. Lond. B. 1995;349:11–18. [Google Scholar]
- Sumrall CD, Sprinkle J. Phylogenetic analysis of living Echinodermata based on primitive fossil taxa. In: Mooi R, Telford M, editors. Echinoderms: San Francisco. Rotterdam: A. A. Balkema; 1998. pp. 81–87. [Google Scholar]
- Swanson WJ, Vacquier VD. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science. 1998;28:710–712. doi: 10.1126/science.281.5377.710. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat. Rev. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD, Moy GW. Isolation of bindin: the protein responsible for adhesion of sperm to sea urchin eggs. Proc. Natl. Acad. Sci. USA. 1977;74:2456–2460. doi: 10.1073/pnas.74.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD, Swanson WJ, Hellberg ME. What have we learned about sea urchin sperm bindin? Devel. Growth Differ. 1995;37:1–10. doi: 10.1046/j.1440-169X.1995.00001.x. [DOI] [PubMed] [Google Scholar]
- Zigler KS. The evolution of sea urchin bindin. Int. J. Dev. Biol. 2008;52:791–796. doi: 10.1387/ijdb.072521kz. [DOI] [PubMed] [Google Scholar]
- Zigler KS, Lessios HA. 250 million years of bindin evolution. Biol. Bull. 2003;205:8–15. doi: 10.2307/1543440. [DOI] [PubMed] [Google Scholar]
- Zigler KS, Lessios HA. Speciation on the coasts of the new world: phylogeography and the evolution of bindin in the sea urchin genus Lytechinus. Evolution. 2004;58:1225–1241. doi: 10.1111/j.0014-3820.2004.tb01702.x. [DOI] [PubMed] [Google Scholar]
- Zigler KS, McCartney MA, Levitan DR, Lessios HA. Sea urchin bindin divergence predicts gamete compatibility. Evolution. 2005;59:2399–2404. [PubMed] [Google Scholar]
- Zigler KS, Raff EC, Popodi E, Raff RA, Lessios HA. Adaptive evolution of bindin in the genus Heliocidaris is correlated with the shift to direct development. Evolution. 2003;57:2293–2302. doi: 10.1111/j.0014-3820.2003.tb00241.x. [DOI] [PubMed] [Google Scholar]