Abstract
Persistent DNA double-strand breaks and telomeres represent genomic hazards, as they can instigate inappropriate repair reactions. Two recent papers by Oza and colleagues (pp. 912–917) and Schober and colleagues (pp. 928–938) show that both types of DNA ends are sequestered from bulk DNA by Mps3, a SUN domain protein that spans the inner nuclear membrane. Anchorage maintains telomere integrity and steers double-strand breaks toward specialized repair pathways. This work defines the nuclear periphery as a subcompartment where dangerous DNA elements can be handled with care.
Keywords: Chromatin, DNA double-strand break, DNA repair, Mps3p, Rad52, 3C
A DNA double-strand break (DSB) is a terribly dangerous thing to acquire. In the best-case scenario, homologous sequences serve as templates for homologous recombination (HR). Budding yeast are particularly adept at this form of DNA repair. Broken DNA ends can also be rejoined by DNA ligase via the nonhomologous end-joining pathway (NHEJ). Humans are particularly proficient at this alternative. The danger lies in the possibility that neither HR nor NHEJ succeed quickly. In this case, alternative attempts at repair can lead to a host of possibilities, like chromosomal rearrangements, only some of which allow the cell to survive. In principle, the telomeric ends of linear chromosomes present the same problem. These too would be prone to initiate repair, including end-to-end chromosomal fusions, if not for mechanisms that prevent telomeres from being recognized as DSBs (Sabourin and Zakian 2008).
New work from research teams led by Craig Peterson (Oza et al. 2009) and Susan Gasser (Schober et al. 2009) uncovers an important yet unappreciated factor in how budding yeast deal with DNA ends. In papers published in the April 15, 2009, issue of Genes & Development (Oza et al. 2009; Schober et al. 2009), the laboratories find that components of the telomere replication machinery link both telomeres and persistent DSBs to Mps3, a SUN domain protein that spans the inner nuclear membrane. The interactions account for much of the well-documented perinuclear targeting of telomeres (e.g., Hediger et al. 2002), and the more recently described perinuclear localization of unrepairable DSBs (Nagai et al. 2008). Critically, Oza et al. (2009) and Schober et al. (2009) show that anchoring by Mps3 influences the ways that telomeres and DSBs are processed. Taken together, the new findings reveal that the nuclear periphery is a specialized subnuclear compartment, analogous to the backroom of a political convention, where crucial decisions are made about the fate of DNA ends.
Persistent DSBs and telomeres are sequestered at the nuclear periphery
While the Peterson study (Oza et al. 2009) focuses on DSBs and the Gasser study (Schober et al. 2009) focuses on telomeres, both laboratories have taken advantage of the ever-popular HO endonuclease. To achieve a mating-type switch, budding yeast express HO to generate a single DSB at the MAT locus, which initiates a gene conversion event with homologous donor sequences at either end of the same chromosome. By commandeering the HO system, much has been learned about the intricacies of DSB repair (Pâques and Haber 1999). Recent microscopy by the Gasser laboratory (Nagai et al. 2008; Bystricky et al. 2009) now indicates that recombination occurs randomly within the nuclear interior, as neither the recipient nor the donor sequences become enriched at the nuclear periphery during the repair interval. This and other observations described below suggest that facile repair by the HR pathway proceeds without a specialized subnuclear compartment.
Without donor sequences, generation of a DSB at MAT leads to the eventual death of most cells. Under these constraints, both laboratories (Oza et al. 2009; Schober et al. 2009) observed a similar striking behavior. The DSB, monitored by GFP proteins tethered to DNA near the break, transported to the peripheral edge of the nucleus where it remained for hours. To complement the cytology, Oza et al. (2009) employed 3C (Chromosome Conformation Capture), a cross-linking approach developed by Job Dekker, an investigator on the Peterson study, that measures the relative frequencies of collision between nonadjacent DNA sequences (Dekker et al. 2002). This revealed that perinuclear localization imposes restrictions on the ability of the DSB to interact with other DNAs. Taken together, the findings indicate that persistent DSBs are not only localized at the nuclear periphery, but also sequestered from the rest of the genome.
Mps3 anchors DSBs and telomeres to the nuclear envelope
In principle, peripheral enrichment could arise by exclusion from the nuclear interior. This is not the case. Oza et al. (2009) discovered that DNA sequestration requires Mps3, a protein that spans the inner nuclear membrane. Mps3 can be cross-linked near break sites, but only when the breaks persist, strengthening the supposition that the nuclear periphery is a destination for damage not easily repaired by HR. Mps3 and its homologs in a variety of other organisms anchor meiotic telomeres. Sue Jaspersen, another investigator on the Peterson study (Oza et al. 2009), found previously that the protein also anchors telomeres in vegetative budding yeast (Bupp et al. 2007), a result now extended by Schober et al. (2009). Collectively, the results indicate that Mps3 is a common docking site for both persistent DSBs and telomeres on the inner surface of the nuclear envelope.
Mps3 is essential, as it also participates in duplication of the spindle pole body (SPB), the yeast equivalent of the centrosome. The protein is a member of a family of proteins that possess the SUN domain, a conserved peptide motif that resides in the lumen of the nuclear envelope. Most SUN domain proteins also span the inner nuclear envelope. Their associations with KASH domain proteins that span the outer nuclear membrane create mechanical links between the nucleus and cytoplasm. During meiosis in budding yeast, for example, Mps3 transduces the dynamics of cytoplasmic actin cables into DNA movements that drive meiotic chromosome pairing (Conrad et al. 2008; Koszul et al. 2008 and references therein). In fission yeast, forces on these linkages can be so great that if chromosome anchorage does not counterbalance the pull of cytoplasmic microtubules, the nucleus distorts and the SPB disassembles (Tomita and Cooper 2007; King et al. 2008). Whether a nuclear/cytoplasmic connection mediated by Mps3 helps cells contend with persistent DSBs is not known.
Paradoxically, the Gasser laboratory (Schober et al. 2009) recently found persistent DSBs comingling with nuclear pore complexes (NPCs), which have no known connection with Mps3 (Nagai et al. 2008). NPCs normally decorate the surface of the entire nuclear envelope, but in this instance, they were genetically forced into an aggregate that made colocalization with damaged DNA abundantly clear. How can anchoring at both Mps3 and NPCs be reconciled? A simple explanation might be that docking occurs first with Mps3 before the DSB is shuttled to NPCs to attempt certain forms of repair (see below).
Telomeric replication machinery links DSBs and telomeres to Mps3
Telomere anchoring, it turns out, is not a simple affair. There are multiple pathways, the relative contributions of which vary from one telomere to the next, and vary yet again during progression through the cell cycle. Two redundant mechanisms have been described in detail (Hediger et al. 2002; Taddei et al. 2004), and there is room for others. Sir4, a constituent of yeast heterochromatin domains that form near telomeres, defines one of the predominant pathways. Mps3 interacts with Sir4 and mediates the telomeric heterochromatin anchor (Bupp et al. 2007), along with another perinuclear protein named Esc1 (Andrulis et al. 2002). A second predominant anchoring pathway is defined by the Ku70 and Ku80 proteins, which form the Ku telomere-binding (and DSB-binding) complex. During the G1 stage of the cell cycle, Ku recruits telomerase via the enzyme's template RNA (Stellwagen et al. 2003; Fisher et al. 2004). Schober et al. (2009) now show that telomerase is an integral part of the Ku-based anchor. The work was propelled by an assay that measures the intrinsic localization of a nuclear protein. In the assay, a GFP-tagged DNA segment is tethered to a query protein, which drags the GFP reporter to the protein's nuclear address (Taddei et al. 2004). Using this assay, Schober et al. (2009) show that telomerase and Ku localize to the nuclear periphery but at locations distinct from those occupied by NPCs. These data fit nicely with the observation that most telomeres (there is at least one exception) do not colocalize with NPCs (Hediger et al. 2002; Therizols et al. 2006). Schober et al. (2009) go on to show that depleting telomerase components or disrupting interactions between them blocks perinuclear enrichment of Ku. Coupled with the knowledge that Mps3 interacts with the Est1 subunit of telomerase (Antoniacci et al. 2007), the Gasser laboratory (Schober et al. 2009) traced out an entire chain of molecules that begins at telomeric DNA and terminates in the nuclear envelope.
Recruitment of telomerase to telomeres is not left solely to Ku. During S phase, a second recruitment strategy relies on Cdc13, the DNA-binding protein that associates preferentially with the single-stranded TG repeat sequences of telomeres. Oza et al. (2009) find, like the Shore laboratory before (Negrini et al. 2007), that Cdc13 binds the persistent DSBs, even in the absence of recognizable TG-like DNA. More remarkable, Oza et al. (2009) find that persistent DSBs also recruit the telomerase catalytic subunit, and that the recruitment pathway (defined by Cdc13) ultimately leads to contact of the DSBs with Mps3. Some of the details are still murky. It is not known, for example, whether telomerase subunits mediate perinuclear docking or whether telomerase binding merely accompanies it. Nevertheless, the existing data do support the idea that DSBs acquire features of telomeres, which link the broken ends to Mps3 at the nuclear periphery.
Roles for perinuclear anchoring in DSB processing and telomere stability
Undoubtedly, the most important finding from the studies by Oza et al. (2009) and Schober et al. (2009) is that perinuclear anchoring matters. The fate of a persistent DSB or integrity of telomeric DNA depends on Mps3. Both laboratories (Oza et al. 2009; Schober et al. 2009) took advantage of mps3 mutants that either trimmed the portion of the protein that protrudes into the nucleus or challenged that domain in a dominant-negative fashion. To evaluate roles for Mps3 in telomere maintenance, Schober et al. (2009) sensitized cells to telomeric insults by eliminating Tel1, the budding yeast ATM checkpoint kinase, which resulted in slightly shortened telomeric TG repeat sequences. They found that disruption of Mps3 in this background imposed a senescence-like phenotype normally associated with telomerase loss. Cultures became enriched in large-budded cells that appeared to be trapped at the G2/M transition. Surprisingly, the TG repeats added by telomerase remained largely intact. Instead, the repetitive blocks of dsDNA next to yeast telomeres known as Y′ elements increased in copy number. It is unlikely that these amplifications cause the senescence phenotype. Rather, the amplifications may be symptomatic of a larger defect in telomere structure, like recombination intermediates, which would account for the telomere cross-bridges that Schober et al. (2009) mention. Telomeric Y′ sequences have been known to amplify to extraordinary lengths to survive loss of telomerase (Lundblad and Blackburn 1993). It will be interesting to see how the situation differs in these arresting cells. Even without a full accounting of the molecular defect, it is clear that Mps3 is important for telomere integrity.
To evaluate roles for Mps3 in processing persistent DSBs, both Oza et al. (2009) and Schober et al. (2009) assayed for gross chromosomal rearrangements (GCRs). In yeast, these usually take the form of de novo telomere additions, but nonreciprocal translocations, deletions, and isoduplications are also seen. The assay relies on spontaneous DSBs thought to arise from replication fork mishaps. A variety of mechanisms suppress GCRs. In particular, GCRs due to de novo telomere formation are blocked by Pif1, a helicase that antagonizes engaged telomerase (Schulz and Zakian 1994; Pennaneach et al. 2006). Oza et al. (2009) now find that GCR levels recede when Mps3 is disrupted in a pif1 mutant. The result implies that Mps3 anchorage is critical for rescue of persistent DSBs by de novo telomere addition.
GCR formation is also suppressed by Slx5 and Slx8, proteins that combine to form a SUMO-dependent ubiquitin-conjugating enzyme. In recent work, Gasser and colleagues (Nagai et al. 2008) showed that Slx5/Slx8 associates with the Nup84 nuclear pore subcomplex, and that both together bind persistent DSBs at NPCs. In this locale, SUMO-dependent ubiquitylation of proteins may activate an alternative repair pathway that suppresses GCRs. Oza et al. (2009) now find that disruption of Mps3 eliminates GCRs caused by Slx5 loss. Since Mps3 does not suppress GCRs caused by other mutations, Oza et al. (2009) conclude that Mps3 and Slx5 operate in the same pathway. As described above, a model consistent with these data is that Mps3 serves as the preliminary docking site before persistent DSBs transfer to NPCs for attempts at repair.
Finally, Oza et al. (2009) show that recruitment of a DSB to Mps3 at the periphery can also influence HR recombination. In one of two examples, they used a system in which gene conversion of a DSB at MAT requires donor sequences on a separate chromosome. The reaction proceeds slower than a normal mating-type switch because it takes longer for the two sites to locate one another. Oza et al. (2009) found that this DNA break was recognized as a persistent DSB and transported to the nuclear periphery. Remarkably, the donor sequence eventually moved to the periphery, too. Repair of the break by HR initiated, albeit at a slower rate due to anchorage by Mps3. These results indicate that the perinuclear sequestration can intercede if HR is delayed. However, targeting to the periphery does not represent an irreversible path that blocks HR should the opportunity arise.
The nuclear periphery as a depot for genomic hazards
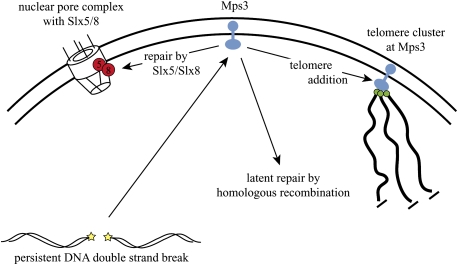
The work by Oza et al. (2009) strengthens a growing perception that evolution selected the nuclear periphery as a subcompartment to handle hazardous nuclear events. Sequestration of persistent DSBs and telomeres away from bulk DNA decreases the probability that they will find inappropriate partners. Moreover, docking at the nuclear envelope ensures that DNA ends are processed in an orderly way. A persistent DSB that reaches Msp3 sits at the nexus of at least three choices (Fig.1). If the break exposes a telomeric “seed,” telomerase could heal the DSB by de novo telomere synthesis. The newly protected DNA end would remain with other native telomeres at Mps3, where irreconcilable recombination is suppressed. If telomerase cannot initiate synthesis, the DSB could instead be shuttled to nuclear pores for an attempt at Slx5/Slx8-mediated repair. Finally, if repair can be attained by latent HR—say, by the delayed arrival of a donor—repair intermediates could be released back into the nucleoplasm.
Figure 1.
A model for the routing of persistent DSBs and telomeres at the nuclear periphery. A persistent DSB (yellow) first docks with Mps3 (blue) at the inner nuclear envelope. If a telomeric sequences can be added, the DNA end remains tethered to Mps3 within a telomere cluster (green). If a telomere cannot be added, Slx5/Slx8 (red) attempts repair at nuclear pore complexes. If donor sequences become available, HR initiates at a location within the nuclear periphery (not yet specified) before the DNA segment returns to the nucleoplasm.
The precise specificity of repair by HR, usually between adjacent sister chromatids, is so rapid and accurate that it appears to proceed unencumbered throughout the nuclear interior. However, not all HR is safe. In yeast, the ribosomal RNAs are encoded by a tandem array of 100–200 identical rDNA genes. Unequal sister chromatid exchange between the repeats can produce arrays that are either too long or too short for optimal growth. To compensate, pathways have evolved to prevent recombination between the repeats. Recently, Moazed and coworkers (Mekhail et al. 2008) showed that one suppression mechanism involves tethering the array to the nuclear envelope, in this case by a pair of different inner membrane-spanning proteins that are also evolutionarily conserved. rDNA repeats are highly dynamic, departing their nucleolar compartment at the edge of the nucleus on a regular basis to permit repair by HR when necessary (Torres-Rosell et al. 2007). Indeed, if nucleolar departures are blocked by genetic manipulation, recombination between repeats elevates to inordinately high levels. Thus, while there may be differences in the details particular to stabilizing the rDNA or to stabilizing persistent DSBs at other loci, the underlying concept is still the same: All go to the periphery to avoid reactions between unintended partners.
Sequestration of DNA at the nuclear periphery bears a similarity to another compartmentalized nuclear event: the heterochromatic repression of genes. Heterochromatin proteins are targeted to specific chromosomal locations, where they assemble cooperatively into structures that span large domains, irrespective of the underlying sequence. Perinuclear sequestration is among the host of mechanisms that impart specificity to these generalized repressors. When anchorage is disrupted, heterochromatin domains and proteins disperse throughout the nucleus (Gartenberg et al. 2004), causing untimely and promiscuous gene repression (Taddei et al. 2009). Thus, sequestration of heterochromatin at the nuclear periphery, like the sequestration of DNA ends, offers a secure destination for events on DNA that must remain nuclear but that would otherwise be problematic for the rest of the genome.
Acknowledgments
I thank Jim Haber for valuable comments on the manuscript. This work was funded by grants from the NIH and March of Dimes.
Note added in proof
A new publication by Jentsch and colleagues (Kalocsay et al. 2009) also reports that persistant DSBs contact Mps3 at the nuclear periphery, and that this event requires sumolyation of H2A.Z, a histone variant that binds early yet transiently to break sites.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1805309.
References
- Andrulis E.D., Zappulla D.C., Ansari A., Perrod S., Laiosia C.V., Gartenberg M.R., Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. MCB. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci L.M., Kenna M.A., Skibbens R.V. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6:75–79. doi: 10.4161/cc.6.1.3647. [DOI] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1–UNC-84 domain protein Mps3. J. Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Van Attikum H., Montiel M.D., Dion V., Gehlen L., Gasser S.M. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol. Cell. Biol. 2009;29:835–848. doi: 10.1128/MCB.01009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Chao G., Shinohara M., Kosaka H., Shinohara A., Conchello J.A., Dresser M.E. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Fisher T.S., Taggart A.K., Zakian V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Gartenberg M.R., Neumann F.N., Laroche T., Blaszczyk M., Gasser S.M. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 2004;119:955–967. doi: 10.1016/j.cell.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N.J., Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- King M.C., Drivas T.G., Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E. An alternative pathway for yeast telomere maitenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Mekhail K., Seebacher J., Gygi S.P., Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Ribaud V., Bianchi A., Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: Implications for telomere capping and telomerase regulation. Genes & Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes & Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V., Putnam C.D., Kolodner R.D. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol. Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Sabourin M., Zakian V.A. ATM-like kinases and regulation of telomerase: Lessons from yeast and mammals. Trends Cell Biol. 2008;18:337–346. doi: 10.1016/j.tcb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M. Yeast telomerase and SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes & Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V.P., Zakian V.A. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Stellwagen A.E., Haimberger Z.W., Veatch J.R., Gottschling D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes & Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Bauer C., Gasser S.M. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Nagai S., Erb I., van Nimwegen E., Gasser S.M. The functional importance of telomere clustering: Global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Cooper J.P. The telomere bouquet controls the meiotic spindle. Cell. 2007;130:113–126. doi: 10.1016/j.cell.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]