Abstract
Initiating new rounds of Escherichia coli chromosome replication requires DnaA-ATP to unwind the replication origin, oriC, and load DNA helicase. In this issue of Genes & Development, Fujimitsu and colleagues (pp. 1221–1233) demonstrate that two chromosomal sites, termed DARS (DnaA-reactivating sequences), recycle inactive DnaA-ADP into DnaA-ATP. Fujimitsu and colleagues propose these sites are necessary to attain the DnaA-ATP threshold during normal growth and are important regulators of initiation timing in bacteria.
Keywords: DnaA reactivation, nucleotide exchange, initiation regulation, functional DNA sequence, protein complex, replication cycle
All cells must coordinate the timing of chromosome duplication with cell division during the cell cycle, to ensure that each daughter cell receives equivalent genomic material. An early step toward accomplishing this task requires assembly of initiator protein complexes that prepare replication origins to begin new rounds of DNA replication (Bell 2002; Leonard and Grimwade 2005; Mott and Berger 2007). Each origin is initially marked by a multisubunit origin recognition complex (ORC) that recruits additional initiator proteins and transitions into a prereplication complex (pre-RC) responsible for loading the replicative helicase (Stillman 2005; Nievera et al. 2006). It is now clear that initiator proteins from all domains of life share structural and functional similarities, including membership in the AAA+ family of proteins whose activity is regulated by binding and hydrolysis of ATP (Erzberger et al. 2002). By switching between ATP- and ADP-bound states, active initiators can be readily inactivated, ensuring their use only once per cell cycle at each origin copy. Despite this convenient mode of regulation, questions remain about the cell's ability to balance the regulatory switches that trigger inactivation of AAA+ initiators with those that produce new initiators for the subsequent cell cycle.
Escherichia coli initiator DnaA: building a bacterial ORC and pre-RC
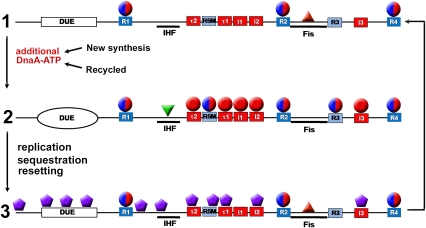
Questions about cycling initiator activity are particularly relevant to DNA replication control in rapidly growing bacteria, since these cells contain multiple origins that must fire synchronously only once per cell division cycle (Skarstad et al. 1986; Boye et al. 2000). Studies of the E. coli AAA+ initiator, DnaA protein, provide insights into the various cellular mechanisms that ensure that the initiator is active and available at the time of initiation and is then prevented from triggering another round of replication until the next cell cycle. DnaA has a high affinity for adenine nucleotides ADP and ATP, but is active only when bound to ATP (Sekimizu et al. 1987). DnaA-ATP assembles into an ordered multimeric pre-RC that unwinds the unique chromosomal origin of replication, oriC (Leonard and Grimwade 2005), and assists in loading replicative DNA helicase (DnaB) on the newly exposed single strands (Fang et al. 1999). Throughout most of E. coli's cell division cycle, DnaA occupies only three high-affinity 9-mer recognition sites (termed R boxes 1, 2, and 4) (Samitt et al. 1989; Cassler et al. 1995), which bind equivalently to active DnaA-ATP and inactive DnaA-ADP (Sekimizu et al. 1987; McGarry et al. 2004). These DNA–protein complexes form a eubacterial version of ORC (Fig. 1, stage 1; Nievera et al. 2006). The transition to a pre-RC requires recruitment of DnaA-ATP to low-affinity 9-mer recognition sites (I sites, τ sites, R5M, and R3) within inter-ORC DNA (Fig. 1, stage 2; Kawakami et al. 2005; Leonard and Grimwade 2005). I and τ sites have a strong preference for DnaA-ATP compared with DnaA-ADP (McGarry et al. 2004; Kawakami et al. 2005), and precisely couple pre-RC assembly with the availability of an active initiator. The behavior of R5M suggests that it is occupied by DnaA-ADP during pre-RC assembly (Grimwade et al. 2007). The conversion from the ORC to a pre-RC in E. coli is also modulated by two DNA-bending proteins, Fis and IHF (Fig. 1), that, respectively, repress and promote DnaA-ATP interactions with low-affinity recognition sites (Leonard and Grimwade 2005). As many as 20 molecules of DnaA-ATP may be required during the ORC-to-pre-RC transition (Crooke et al. 1993), and the number of DnaA molecules required at oriC appears to remain constant at all growth rates (Donachie and Blakely 2003). Recent structural analysis of DnaA reveals that ATP-bound monomers have the ability to form right-handed helical oligomeric filaments, presenting an attractive mechanism to promote ordered pre-RC assembly by extension of filaments as well as to position DNA helicase at the end of an oligomeric structure of fixed length (Erzberger et al. 2006; Mott et al. 2008).
Figure 1.
Nucleoprotein complex assembly at oriC during the cell cycle. (Stage 1) DnaA occupies the three higher-affinity R boxes—R1, R2, and R4—throughout the cell cycle, with Fis bound to its cognate site. This complex forms the bacterial version of the ORC. (Stage 2) As additional DnaA-ATP accumulates in the cell, either by new synthesis or by recycling of DnaA-ADP, lower-affinity sites τ1, τ2, I1, I2, I3, with preference for DnaA-ATP, become occupied, as well as nondiscriminatory sites R5M and R3. Fis is displaced, and IHF binds between R1 and R5M. This results in formation of the pre-RC, with localized DNA strand separation in the duplex unwinding region (DUE). (Stage 3) After new replication forks duplicate oriC, the resulting hemimethylated DNA binds SeqA, allowing resetting of the ORC, but preventing reformation of the pre-RC (sequestration). Sequestration lasts for one third of the cell cycle, and the origin returns to the ORC. Red spheres are DnaA-ATP, red-blue spheres are either nucleotide form of DnaA, purple hexagons are SeqA, and red and green triangles are Fis and IHF, respectively.
Inactivation of DnaA is coupled to movement of the replication fork
Intracellular DnaA levels are high (∼1000 copies per cell) and the protein is stable (Sekimizu et al. 1988; Hansen et al. 1991). Despite these features, DnaA fluctuates between active and inactive forms during the cell cycle, with DnaA-ATP rising to 80% of the total prior to initiation of new rounds of DNA synthesis and dropping to 20% after new rounds are initiated (Kurokawa et al. 1999). Self-hydrolysis of DnaA-ATP is normally too slow to be an effective regulator of initiator activity, but interaction with the moving replication fork stimulates hydrolysis of DNA-associated DnaA-ATP by a mechanism termed RIDA (regulatory inactivation of DnaA) (Katayama and Sekimizu 1999). RIDA is catalyzed by a protein factor, Hda, a DnaA paralog associated with the β-clamp subunit of DNA polymerase III (Kato and Katayama 2001). Although the details of the stimulatory mechanism are not yet clear, RIDA appears to be the dominant means used by E. coli to inactivate DnaA-ATP bound to oriC as well as to ∼300 other chromosomal sites, including titration sites (Hansen et al. 1991; Kitagawa et al. 1996) and binding sites within transcriptional promoters (Messer and Weigel 1997). One particular region of the chromosome, termed datA, has an unusually high capacity for DnaA (>50 molecules) (Kitagawa et al. 1996). It is possible that the heavy occupation of this or similar sites would result in a mass inactivation of DnaA-ATP at a fixed time after each initiation of new DNA synthesis, when the replication fork moved through the titration site. However, any significant increases in the rate of RIDA during the cell cycle have yet to be revealed.
In order for RIDA to be effective, synthesis of new DnaA (rapidly converted to DnaA-ATP) must also be repressed during the post-initiation phase of the cell cycle. E. coli uses another replication fork-dependent mechanism, termed sequestration, to accomplish this task (Campbell and Kleckner 1990). E. coli chromosomes carry many copies of the palindrome GATC target for DNA adenine methyltransferase (DAM methylase). Semiconservative replication converts fully methylated GATC to the hemimethylated state, which is recognized by the sequestration protein SeqA (Lu et al. 1994). Both the dnaA promoter region and low-affinity DnaA recognition sites in oriC contain an unusually high number of GATC copies, and remethylation of SeqA-bound dnaAp and oriC is delayed for about one-third of the cell cycle (Campbell and Kleckner 1990). During this period, SeqA blocks dnaA transcription (Campbell and Kleckner 1990) as well as DnaA-ATP rebinding to the low-affinity sites in oriC (Fig. 1, stage 3; Nievera et al. 2006). GATC is not found at R1, R2, and R4 in oriC, allowing immediate reset of ORC during the sequestration period (Fig. 1, stage 3; Nievera et al. 2006). Following remethylation, synthesis of DnaA-ATP resumes and oriC once again becomes accessible for pre-RC assembly (Campbell and Kleckner 1990). Hemimethylated oriC DNA also interacts with E. coli membranes, and these interactions may be important for some aspect of oriC function during the sequestration period (Ogden et al. 1988).
Keeping up: does E. coli synthesize enough DnaA-ATP?
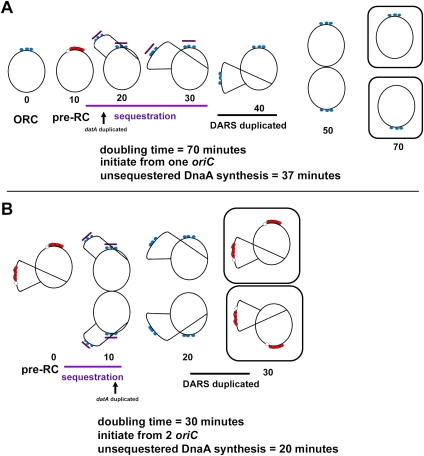
Although RIDA and sequestration ensure that DnaA-ADP levels increase during the post-initiation phase of the cell cycle, new synthesis of DnaA-ATP must ultimately outpace the rate of inactivation in order to rebuild pre-RC during subsequent cycles. New DnaA-ATP synthesis is required for each new round (protein synthesis inhibitors effectively block DNA replication at the initiation step) (Helmstetter 1996), but the data presented by Fujimitsu et al. (2009) in this issue of Genes & Development suggest that new synthesis might not be sufficient to produce enough DnaA-ATP under all growth conditions. Outpacing RIDA may be particularly problematic during rapid growth in E. coli when ongoing chromosomal DNA replication overlaps the initiation of new rounds (Fig. 2B). E. coli normally requires ∼40 min to duplicate its genome (and an additional 20 min to prepare for cell division) (Helmstetter 1996). For this reason, cells growing with doubling times faster than 40 min must initiate new rounds of DNA synthesis prior to the completion of previous rounds (Fig. 2B), leading to an increased copy number of oriC. The multiple copies of oriC are initiated synchronously (Skarstad et al. 1986), even though RIDA, associated with the previous round, is ongoing at the time of initiation. Although increased oriC copy number is accompanied by a comparable increase of nearby dnaA copies, RIDA might still overwhelm new synthesis. The dnaA promoter is moderately strong, but the translation efficiency of DnaA mRNA is very low (only about one to four molecules per transcript) (Hansen et al. 1991; Macian et al. 1994; Kaguni 2006). In addition, ongoing replication causes an increase in the number of DnaA titration sites on the chromosome, and these sites have a higher affinity for DnaA than the low-affinity sites in oriC (Kitagawa et al. 1998).
Figure 2.
Duplication of the DARS sites relative to time of unsequestered DnaA-ATP synthesis and time of initiation varies as a function of growth rate. (A) At slow growth rates, only one origin is initiated before the DARS sites are duplicated. (B) In rapidly growing cells, pre-RCs form at multiple copies of oriC, but there is less time for synthesis of new DnaA-ATP. The DARS sites in these cells are replicated just prior to formation of the pre-RC, and may contribute to the DnaA-ATP needed for this step. Blue circles represent DnaA-ATP or DnaA-ADP bound to high-affinity oriC sites, red circles represent DnaA-ATP bound to oriC in the pre-RC, the open circle indicates localized oriC DNA strand separation, and the purple bar indicates a sequestered oriC.
DnaA-reactivating sequence (DARS): a novel mechanism to recycle DnaA-ADP
One possible mechanism to provide additional DnaA-ATP beyond the synthetic capabilities of the cell is the recycling of inactive DnaA-ADP produced during the previous cell cycle by RIDA. Reactivation of DnaA-ADP requires nucleotide exchange in the presence of ATP. Although acidic phospholipids in fluid bilayers were shown previously to be capable of catalyzing this exchange in vitro (Crooke 2001; see below), it remains to be determined whether significant amounts of membrane-dependent DnaA-ADP recharging takes place in vivo. In this issue of Genes & Development, Fujimitsu et al. (2009) provide strong evidence that E. coli requires recycling mechanisms that reactivate DnaA-ADP in order to properly time the initiation of DNA synthesis. Remarkably, the major components of this recycling mechanism are two sets of nucleotide sequence motifs located on the chromosome.
The current report provides an extensive follow-up of an observation made by the Katayama laboratory (Fujimitsu and Katayama 2004) that a 70-base-pair (bp) fragment containing three DnaA box-like sequences obtained from ColE1-derived plasmids was capable of catalyzing DnaA-ADP nucleotide exchange in vitro. Although the presence of these plasmid sequences had little affect on E. coli growth in vivo, a search for similar sequence motifs on the chromosome successfully led to the identification of two DnaA-reactivating sequences termed DARS1 and DARS2. DARS1 is located within intergenic regions between bioD-uvrB (17.49 min), and DARS2 is located between ygpD and mutH (63.96 min). Both DARS1 and DARS2 catalyze nucleotide exchange and reactivation of DnaA-ADP to DnaA-ATP in vitro, and the initiation of chromosome replication was pushed to earlier times during the cell cycle by raising the copy number of these sites in vivo. Ongoing protein synthesis is not required for DARS activity, but DARS2 is stimulated by a cell extract, suggesting it is regulated by a protein factor. DARS deletion strains remain viable, but loss of DARS activity enhances the thermosensitivity of underinitiating dnaAts strains. DARS deletion also rescues mutant strains whose growth is severely perturbed by overinitiation due to the loss of RIDA (hda deletion strains). These results support a significant contribution for DARS in the normal regulation of the levels of intracellular DnaA-ATP, reducing the need for new synthesis.
The arrangement of DnaA recognition sites within DARS appears to be important for nucleotide exchange, suggesting that specific oligomeric assemblies are involved in DARS activity. At least three DnaA recognition sites are mapped within DARS regions, but an unusual arrangement of two closely spaced recognition sites in opposite orientations to one another is an important conserved feature. It may be that by arranging available DnaA-ADP in specific configurations, DARS can alter a particular structural feature of DnaA that is required for high-affinity nucleotide binding. In support of this idea, Fujumitsu et al. (2009) report that both Domain III (containing Walker and Sensor motifs) and Domain IV (containing the DNA-binding domain) are required for DARS activity.
Membrane-dependent DnaA activity
As mentioned previously, DARS is not the sole mechanism for recycling DnaA-ADP into the active form in E. coli. Acidic phospholipids in a fluid bilayer are also capable of performing the nucleotide exchange reaction (Crooke 2001; Boeneman and Crooke 2005); in the presence of oriC DNA, DnaA-ADP can be rejuvenated to the ATP-bound form (Crooke et al. 1992). Although the contribution of membranes to DnaA recharging in vivo remains unclear, there can be little doubt that membranes are an important part of the DnaA life cycle. In the absence of ongoing acidic phospholipid biosynthesis, cells ultimately suffer growth arrest due to the inability to initiate new rounds of DNA synthesis (Xia and Dowhan 1995). This arrest is suppressed by expression of versions of DnaA (for example, DnaA L366K) harboring mutations within the membrane interaction domain located between Domains III and IV (Zheng et al. 2001).
Based on our current knowledge, it appears that there are two mechanisms for recycling DnaA-ADP in E. coli cells: One is located in the membrane and is mediated by acidic phospholipids, and the other is located on the chromosome. Why should there be this redundancy? The key to answering this question may depend on the location of the DnaA-ADP that interacts with the separate mechanisms. One possibility is that DARS provides a general recharging mechanism for free DnaA-ADP in the cytoplasm, while membranes provide a specific recharging mechanism for DnaA–DNA complexes that interact with the membrane, thus recharging these complexes in situ. Such complexes might include newly replicated oriC whose high-affinity ORC sites have become occupied by DnaA-ADP. Since it is hard to imagine how oriC might prohibit binding by DnaA-ADP at these sites, reactivating it to DnaA-ATP by membranes during sequestration might be required to build the next round of pre-RC.
Questions to ponder about DnaA recycling
The novel DARS mechanism described by Fujimitsu et al. (2009) in this issue of Genes & Development forces a new look at the dogma that the rate of accumulation of newly synthesized DnaA-ATP alone determines the timing of chromosome duplication during the E. coli cell cycle. The contribution of DARS will obviously lower the amount of new DNA synthesis that is required, making initiation earlier in the cell cycle. However the existence of only two regions carrying DARS on the E. coli chromosome leads to the interesting speculation that their location may also be important for cell cycle regulation. Each replicore contains one site, located at a similar distance from oriC. This placement may be a feature related to the progression of replication forks during the cell cycle. It will be interesting to see if it matters where DARS are located on the genome.
If DARS duplication plays a role in determining the time of initiation, then its function may be more critical at certain growth rates (Fig. 2). Although the time required to replicate the genome remains constant over a range of growth rates, the time of initiation, the number of origins, and the configurations of replication forks during the cell cycle vary at different doubling times (Fig. 2; Helmstetter 1996). This means that the time of DARS duplication, relative to the duration of dnaAp sequestration, the end of RIDA, and the time of initiation, will also vary at different growth rates. During slow growth, chromosome replication and RIDA are completed before cell division, and cells have a G1-like period before the next initiation of DNA synthesis at a single copy of oriC (Fig. 2A). Under these conditions, new synthesis may contribute more of the DnaA-ATP needed for initiation than during rapid growth. In fast-growing cells, RIDA is continuous, pre-RCs must be formed at multiple oriC copies, and there is less time for new DnaA-ATP synthesis (Fig. 2B). In this case, duplication of DARS sites before initiation might play a greater role in supplying the required level of DnaA-ATP. Additional factors may also fine-tune the regulation of DARS to accommodate all growth rates.
On a related theme, questions are raised about the contribution of RIDA to DARS activity. Although it is expected that RIDA would be the sole source of inactive DnaA-ADP for recycling by DARS, this may not be the case. For example, in Figure 5B of Fujimitsu et al. (2009), the investigators show that a ΔoriC strain lacking Hda, DARS1, and DARS2 contained nearly 70% DnaA-ADP. (In this strain, chromosome replication was controlled by a plasmid, since the hda deletion would result in lethal overinitiation in the wild-type background.) Since RIDA should not take place in these cells, where did the DnaA-ADP come from? The data suggest that there may be an additional, but unidentified, source for DnaA-ADP.
Finally, important and fundamental questions remain about how the arrangement of DnaA-binding sites in a DNA region affects assembly of oligomeric DnaA complexes with specific functions. Fujimitsu et al. (2009) provide evidence that relative positions and orientations of DnaA may be sufficient to promote assembly of an oligomeric complex that disrupts nucleotide binding. Different configurations of binding sites might assemble complexes that lead to different outcomes, including pre-RC assembly or DnaA titration. Perhaps rules for these dynamic spatial relationships will be one of the products from future studies of DARS.
No matter what the future holds, this study reveals that even though multiple laboratories have spent the last 40 years dissecting the mechanisms that regulate the initiation of DNA replication, there are still a few surprises. Although the passage of time will no doubt reveal an increasingly complex set of features, those who work on this fascinating topic are eager to welcome a recharging from time to time.
Acknowledgments
We thank our many colleagues for their insightful discussions with us over the years. A.C.L. and J.E.G. are supported by NIH GM54042.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1809909.
References
- Bell SP. The origin recognition complex: From simple origins to complex functions. Genes & Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Boeneman K, Crooke E. Chromosomal replication and the cell membrane. Curr Opin Microbiol. 2005;8:143–148. doi: 10.1016/j.mib.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Boye E, Lobner-Olesen A, Skarstad K. Limiting DNA replication to once and only once. EMBO Rep. 2000;1:479–483. doi: 10.1093/embo-reports/kvd116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- Cassler MR, Grimwade JE, Leonard AC. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995;14:5833–5841. doi: 10.1002/j.1460-2075.1995.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E. Escherichia coli DnaA protein–phospholipid interactions: In vitro and in vivo. Biochimie. 2001;83:19–23. doi: 10.1016/s0300-9084(00)01224-4. [DOI] [PubMed] [Google Scholar]
- Crooke E, Castuma CE, Kornberg A. The chromosome origin of Escherichia coli stabilizes DnaA protein during rejuvenation by phospholipids. J Biol Chem. 1992;267:16779–16782. [PubMed] [Google Scholar]
- Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J Mol Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- Donachie WD, Blakely GW. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol. 2003;6:146–150. doi: 10.1016/s1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- Fujimitsu K, Katayama T. Reactivation of DnaA by DNA sequence-specific nucleotide exchange in vitro. Biochem Biophys Res Commun. 2004;322:411–419. doi: 10.1016/j.bbrc.2004.07.141. [DOI] [PubMed] [Google Scholar]
- Fujimitsu K, Senriuchi T, Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes & Dev. 2009 doi: 10.1101/gad.1775809. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade JE, Torgue JJ, McGarry KC, Rozgaja T, Enloe ST, Leonard AC. Mutational analysis reveals Escherichia coli oriC interacts with both DnaA-ATP and DnaA-ADP during pre-RC assembly. Mol Microbiol. 2007;66:428–439. doi: 10.1111/j.1365-2958.2007.05930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen FG, Christensen BB, Atlung T. The initiator titration model: Computer simulation of chromosome and minichromosome control. Res Microbiol. 1991;142:161–167. doi: 10.1016/0923-2508(91)90025-6. [DOI] [PubMed] [Google Scholar]
- Helmstetter CE. Timing of synthetic activities in the cell cycle. Escherichia coli and Salmonella: Cellular and molecular biology. In: Neidhardt FC, et al., editors. ASM Press; Washington, DC: 1996. pp. 1627–1639. [Google Scholar]
- Kaguni JM. DnaA: Controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- Katayama T, Sekimizu K. Inactivation of Escherichia coli DnaA protein by DNA polymerase III and negative regulations for initiation of chromosomal replication. Biochimie. 1999;81:835–840. doi: 10.1016/s0300-9084(99)00213-8. [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Mitsuki H, Okazaki T, Ogawa T. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol Microbiol. 1996;19:1137–1147. doi: 10.1046/j.1365-2958.1996.453983.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes & Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Building a bacterial orisome: Emergence of new regulatory features for replication origin unwinding. Mol Microbiol. 2005;55:978–985. doi: 10.1111/j.1365-2958.2004.04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N. SeqA: A negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Macian F, Perez-Roger I, Armengod ME. An improved vector system for constructing transcriptional lacZ fusions: Analysis of regulation of the dnaA, dnaN, recF and gyrB genes of Escherichia coli. Gene. 1994;145:17–24. doi: 10.1016/0378-1119(94)90317-4. [DOI] [PubMed] [Google Scholar]
- McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W, Weigel C. DnaA initiator—also a transcription factor. Mol Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: Mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievera C, Torgue JJ, Grimwade JE, Leonard AC. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol Cell. 2006;24:581–592. doi: 10.1016/j.molcel.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden GB, Pratt MJ, Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Samitt CE, Hansen FG, Miller JF, Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989;8:989–993. doi: 10.1002/j.1460-2075.1989.tb03462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- Sekimizu K, Yung BY, Kornberg A. The dnaA protein of Escherichia coli. Abundance, improved purification, and membrane binding. J Biol Chem. 1988;263:7136–7140. [PubMed] [Google Scholar]
- Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Xia W, Dowhan W. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc Natl Acad Sci. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Li Z, Skarstad K, Crooke E. Mutations in DnaA protein suppress the growth arrest of acidic phospholipid-deficient Escherichia coli cells. EMBO J. 2001;20:1164–1172. doi: 10.1093/emboj/20.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]