Abstract
Gene transcription in the cell nucleus is a complex and highly regulated process. Transcription in eukaryotes requires three distinct RNA polymerases, each of which employs its own mechanisms for initiation, elongation, and termination. Termination mechanisms vary considerably, ranging from relatively simple to exceptionally complex. In this review, we describe the present state of knowledge on how each of the three RNA polymerases terminates and how mechanisms are conserved, or vary, from yeast to human.
Keywords: Transcription termination, RNA polymerase I, RNA polymerase II, RNA polymerase III, yeast, mammals
Transcription in eukaryotes is performed by three RNA polymerases, which are functionally and structurally related (Cramer et al. 2008). RNA polymerase II (RNAPII) is responsible for transcription of protein-coding genes and many noncoding RNAs, including spliceosomal small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), microRNA (miRNA) precursors, and cryptic unstable transcripts (CUTs). RNA polymerase I (RNAPI) transcribes the abundant ribosomal RNAs (rRNAs), and RNA polymerase III (RNAPIII) transcribes noncoding RNAs such as transfer RNAs (tRNAs), 5S rRNA, and U6 spliceosomal snRNA. Transcription consists of three main steps: initiation, elongation, and termination, all of which are highly controlled and involve a large number of specific factors. While initiation and elongation have been well studied (Sims et al. 2004), termination was until recently an obscure mechanism, especially for RNAPII transcription. Termination is crucial for the release of RNAP from its template, and is important not only to avoid interference with transcription of downstream genes (Greger et al. 2000), but also to ensure that a pool of RNAPs is available for reinitiation or new transcription. Termination can also prevent formation of antisense RNAs that can interfere with normal pre-RNA production, thereby preventing aberrant gene expression.
Termination by RNAPII does not occur at a conserved site or constant distance from the 3′-end of mature RNAs. In mammals, termination can occur anywhere from a few base pairs to several kilobases downstream from the 3′-end of the mature RNA (Proudfoot 1989). RNAPII transcription termination is coupled to 3′-end processing of the pre-mRNA (Birse et al. 1998; Hirose and Manley 2000; Yonaha and Proudfoot 2000; Proudfoot 2004; Buratowski 2005), and an intact polyadenylation signal has long been known to be necessary for transcription termination of protein-coding genes in human and yeast cells (Whitelaw and Proudfoot 1986; Logan et al. 1987; Connelly and Manley 1988).
RNAPIII and RNAPI termination appear simpler than RNAPII termination. RNAPIII terminates transcription at T-rich sequences located a short distance from the mature RNA 3′-end and seems to involve at most a limited number of auxiliary factors (Cozzarelli et al. 1983). RNAPI terminates at a major terminator located downstream from the rRNA precursor sequence and requires terminator recognition by specific protein factors (Kuhn and Grummt 1989; Lang and Reeder 1995). Recent studies, though, suggest that additional complexities are involved (El Hage et al. 2008; Kawauchi et al. 2008).
The three transcription machineries use different strategies for transcription termination, although they share common features (Cramer et al. 2008; Gilmour and Fan 2008). This review describes advances in our understanding of transcription termination of all three RNAPs, particularly in human and in yeast, where transcription termination mechanisms are somewhat better characterized. We refer the reader to several excellent recent reviews that have dealt with aspects of transcription termination (Rosonina et al. 2006; Lykke-Andersen and Jensen 2007; Gilmour and Fan 2008; Rondon et al. 2008). We begin with the most complex, RNAPII termination.
Termination by RNAPII
3′-End processing machinery
Given the importance of mRNA 3′-end formation in transcription termination, we begin with a brief description of the 3′-end processing reaction and the factors involved. 3′-End processing is an essential step in the maturation of all mRNAs and is coupled to transcription and splicing, as well as termination (Hirose and Manley 2000; Proudfoot et al. 2002; Buratowski 2005). The mammalian pre-mRNA 3′-end processing machinery contains more than 14 proteins giving rise to a 1-MDa complex (for review, see Colgan and Manley 1997; Zhao et al. 1999; Mandel et al. 2008; see also Shi et al. 2009).
In mammals, cleavage and polyadenylation occur ∼10–30 nucleotides (nt) downstream from a conserved hexanucleotide, AAUAAA, and ∼30 nt upstream of a less conserved U- or GU-rich region. The AAUAAA signal is recognized by the cleavage and polyadenylation specificity factor (CPSF), which contains five subunits: CPSF-30, CPSF-73, CPSF-100, CPSF-160, and Fip1. Cleavage stimulation factor (CstF) contains three subunits—CstF-50, CstF-77, and CstF-64—the latter of which directly recognizes the U/GU-rich element. The endonucleolytic cleavage that precedes polyadenylation is carried out by CPSF-73 (Ryan et al. 2004; Mandel et al. 2006). CPSF-73 contains a metallo-β-lactamase domain and is a member of the β-CASP protein superfamily (Callebaut et al. 2002). CPSF-73 is also likely responsible for cleavage of nonpolyadenylated histone pre-mRNAs (Marzluff et al. 2008; Yang et al. 2009). CPSF-100 also belongs to the β-CASP superfamily (Callebaut et al. 2002) but lacks certain key catalytic residues and plays an unknown role in 3′-end cleavage. However, mutations affecting the metallo-β-lactamase domain of both CPSF-73 and CPSF-100 impair the endonucleolytic cleavage of histone mRNA precursors (Kolev et al. 2008). CPSF-160, which directly recognizes AAUAAA, binds CPSF-100, CstF-77, poly(A) polymerase (PAP), and Fip1 (Murthy and Manley 1995; Kaufmann et al. 2004). In addition, the cleavage complex contains CFI (cleavage factor I), a dimeric protein implicated in the regulation of poly(A) site selection (Ruegsegger et al. 1998; Brown and Gilmartin 2003; Venkataraman et al. 2005), and CFII, containing Pcf11 and Clp1. Clp1, which interacts with the CFI and CPSF complexes (de Vries et al. 2000), also possesses an RNA kinase activity (Weitzer and Martinez 2007), although the significance of this to 3′-end formation is unknown. Another protein, symplekin, is also part of the 3′-end processing complex and appears to function as a scaffold (Takagaki and Manley 2000). PABP [poly(A)-binding protein] protects the pre-mRNA from exonuclease degradation by binding the poly(A) tail and is required for correct and efficient poly(A) tail synthesis (Minvielle-Sebastia et al. 1997).
Several factors help link 3′-end processing to transcription. For example, the C-terminal domain (CTD) of the RNAPII largest subunit also plays an important role in 3′-end processing, likely by mediating interactions with 3′-end processing factors (McCracken et al. 1997; Hirose and Manley 1998; Barilla et al. 2001; Fong and Bentley 2001). The Paf1 complex (Paf1C), which was first identified in yeast as an elongation factor and plays a role in transcription-associated chromatin modification (Krogan et al. 2002; Squazzo et al. 2002; Simic et al. 2003; Mueller et al. 2004; Rozenblatt-Rosen et al. 2005), has been shown to be involved in mRNA 3′-end formation (Penheiter et al. 2005; Rosonina and Manley 2005) by interacting with CPSF, CstF, and symplekin (Nordick et al. 2008; Rozenblatt-Rosen et al. 2009). It is notable that one Paf1C subunit, Cdc73 or parafibromin, has been identified as a tumor suppressor protein, and the Paf1 protein is overexpressed in many cancers (Chaudhary et al. 2007).
In yeast, noncanonical nuclear PAPs, Trf4/5 (Haracska et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006), function in a nuclear RNA surveillance pathway as part of the TRAMP (Trf–Air–Mtr4 polyadenylation) complex, which activates degradation by the exosome of a variety of aberrant nuclear precursors of many types of RNAs after addition of a poly(A) tail (LaCava et al. 2005). Although not well studied, this RNA surveillance function is likely conserved in humans since homologs of the TRAMP and exosome complexes are present in mammals.
CTD of the RNAPII large subunit
At least in mammals, the CTD plays an important role in 3′-end processing as first determined by experiments examining the effects on processing of CTD deletions in transfected cells (McCracken et al. 1997) and the requirement of the CTD for efficient 3′ cleavage in vitro (Hirose and Manley 1998). The CTD of RNAPII is flexibly linked to the core enzyme and consists of heptapeptide repeats (25 or 26 repeats in yeast, 52 in mammals) with the consensus sequence Tyr1–Ser2–Pro3–Thr4–Ser5–Pro6–Ser7 (YSPTSPS). The CTD displays different phosphorylation patterns during the transcription cycle (Phatnani and Greenleaf 2006; Egloff and Murphy 2008). RNAPII is recruited to the promoter in an unphosphorylated form (RNAPIIA) that becomes extensively phosphorylated during transcription (RNAPIIO). Phosphorylation of Ser5 by the cyclin-dependant kinase CDK7, a subunit of the general transcription factor TFIIH, occurs at the initiation step of transcription to facilitate promoter release and recruitment of capping factors (Komarnitsky et al. 2000; Schroeder et al. 2000) and the methyltransferase Set1 that is responsible for trimethylation of Lys4 on histone 3 (H3-K4) (Hampsey and Reinberg 2003). Phosphorylation of Ser2 by the positive transcription elongation factor P-TEFb, comprising the kinase CDK9 (Ctk1 in yeast) and cyclin T, occurs after initiation. Ser2 phosphorylated CTD helps to recruit and/or stabilize polyadenylation factors, thereby facilitating coupling of transcription and mRNA 3′-end formation. For instance, in yeast, Ser2 phosphorylation of the CTD potentiates interaction with the essential polyadenylation factor Pcf11 (de Vries et al. 2000; Barilla et al. 2001; Licatalosi et al. 2002; Ahn et al. 2004). The H3-K36 methyltransferase Set2 binds elongating RNAPII, recognizing the doubly phosphorylated CTD (Ser5/Ser2) (Hampsey and Reinberg 2003; Kizer et al. 2005). Interestingly, a genomic analysis employing tilling arrays in mammalian cells showed that H3-K36 methylation often decreases near the poly(A) site during or prior to RNAPII release, leading to the intriguing possibility that H3-K36 methylation plays a role in transcription termination (Lian et al. 2008).
Differential CTD phosphorylation patterns throughout genes appear to coordinate the different steps of transcription, and dephosphorylation also contributes to this. For example, Fcp1 dephosphorylates Ser2 preferentially while Ssu72 (SCP1 in humans), which is a component of the yeast 3′-end machinery, dephosphorylates Ser5 (Meinhart et al. 2005). Dephosphorylation of Ser2 is also critical for RNAPII recycling (Cho et al. 1999). Both unphosphorylated and Ser2 phosphorylated CTD accumulate near transcription termination sites, suggesting that CTD dephosphorylation occurs during or prior to RNAPII release (Lian et al. 2008). Chromatin immunoprecipitation (ChIP) experiments localized Ssu72 predominantly to the 3′-ends of genes (Nedea et al. 2003), but it was also detected at promoter regions (Ansari and Hampsey 2005). Ssu72 might then be involved in coupling transcription termination and reinitiation via gene looping (see below).
For many years, Ser2 was considered the major site of phosphorylation on the CTD at the 3′-ends of genes (Komarnitsky et al. 2000; Cho et al. 2001). However, it was recently demonstrated that mammalian RNAPII can also be phosphorylated at Ser7 of the CTD (Chapman et al. 2007; Egloff et al. 2007). High levels of Ser7 phosphorylation were detected at promoter regions of protein-coding genes, with increasing levels toward the 3′-region (Chapman et al. 2007), as well as at spliceosomal snRNAs genes (Egloff et al. 2007). The role played by CTD phosphorylated at Ser7 in transcription, and whether this modification occurs in yeast, remains to be elucidated; however, association with the 3′-end of protein-coding genes (Chapman et al. 2007) may suggest a role in 3′-end processing and/or termination.
CTD phosphorylation seems to modify CTD function by orchestrating the recruitment of specific factors. It is an intriguing possibility that changes in CTD phosphorylation contribute to termination.
Transcription termination of protein-coding genes
Allosteric/anti-terminator or torpedo: a unified model for transcription termination
Two models were proposed more than 20 years ago to explain the role of 3′-end processing in RNAPII transcription termination (Logan et al. 1987; Connelly and Manley 1988; for review, see Proudfoot 2004; Buratowski 2005; Rosonina et al. 2006). The first model, known as the allosteric or anti-terminator model, proposes that transcription through the poly(A) site leads to conformational changes of the elongation complex (EC) by dissociation of elongation factors and/or association of termination factors (Logan et al. 1987). The second model, the torpedo model, is based on the observed rapid degradation of the 3′ RNA after cleavage at the poly(A) site. Cleavage creates an entry site for a 5′–3′ exonuclease at the uncapped 5′-monophosphate, which degrades the RNA and, according to this model, in some way promotes RNAPII release upon “catching up” with RNAPII (Connelly and Manley 1988). However, as we shall see an emerging view is that the termination mechanism more likely reflects a combination of both models (Fig. 1; Luo et al. 2006; Kaneko et al. 2007; West et al. 2008).
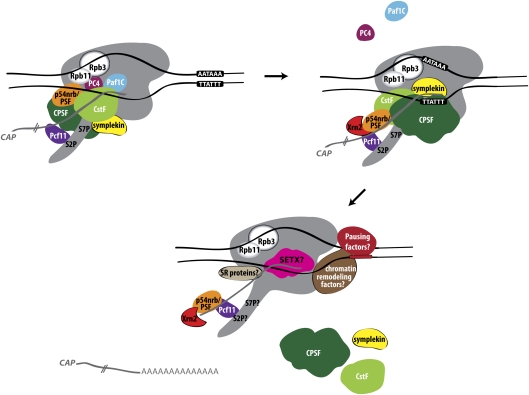
Figure 1.
Mammalian RNAPII termination at protein-coding genes. Poly(A) site recognition leads to changes in the EC. Some factors associated with RNAPII through elongation that may function as anti-terminators are released (Paf1C, PC4) upon passage through the poly(A) site. At the same time, other factors, such as Xrn2, are recruited to the EC. After cleavage, Xrn2, most likely recruited by p54nrb/PSF, degrades the downstream RNA, “catches up” with RNAPII, and, perhaps with the aid of the helicase SETX, terminates transcription by releasing RNAPII from the template DNA. Involvement of chromatin remodeling factors and pausing sequences and factors are depicted. The RNAPII subunits Rpb3 and Rpb11 are also shown to play a role in termination by perhaps transducing a “termination signal.” The possible involvement of SR proteins in termination is also indicated.
Rat1/Xrn2: the transcription termination torpedo
The torpedo model is strongly supported by studies that demonstrated a role for the yeast 5′–3′ exonuclease Rat1 and its human homolog, Xrn2, in RNAPII termination (Kim et al. 2004b; West et al. 2004). In both cases, inactivation of the exonuclease resulted in defects in termination, as measured by ChIP and/or nuclear run-on experiments, both methods measuring RNAPII density throughout transcribed genes. Rat1 and its cofactor Rai1 (see Xiang et al. 2009) are strongly associated with the 3′-end of genes, although in a manner independent of Ser2 CTD phosphorylation since a deletion of CTK1 does not affect their association (Kim et al. 2004b). Recruitment of Rat1 is more likely indirect, through 3′-end processing factors. Rtt103 is a Ser2 phosphorylated CTD-binding protein that cross-links to the 3′-ends of genes in ChIP experiments, and copurifies with Rat1 and other subunits of the cleavage/polyadenylation complex (Kim et al. 2004b). The Caenorhabditis elegans homolog of Rtt103, CIDS-1, was recently identified in a genome-wide RNAi screen for factors involved in termination, and deletion of cids-1 resulted in reduced 3′-end cleavage, supporting the connection between cleavage and termination (Cui et al. 2008). Additionally, Rat1 recruitment to the 3′-end of a protein-coding gene was reduced by Pcf11 inactivation. Similarly Rat1 is required for the recruitment of Pcf11 and Rna15, the yeast CstF-64 equivalent, to the 3′-end of genes (Luo et al. 2006). These data suggest that Rat1 is important for the efficient recruitment of 3′-end processing factors and/or vice versa. However, depletion of Rat1 does not affect cleavage at the poly(A) site (Kim et al. 2004b).
Rat1 activity is likely not sufficient to bring about RNA and RNAPII release from the template. Depletion of Rat1 inhibits but does not abolish termination (Kim et al. 2004b), and digestion of RNAPII-associated RNA is not sufficient to disassemble the EC in vitro (Gu et al. 1996; Kireeva et al. 2000). Consistent with this, the abundant and mostly cytoplasmic exonuclease Xrn1, which is similar to Rat1, is able to complement the exonuclease activity of a Rat1-deficient strain when targeted to the nucleus, but unable to complement the termination defect (Luo et al. 2006). Moreover, a defect in termination in a RAT1 mutant strain (Kim et al. 2006) is not associated with stabilization of the 3′ downstream product, providing evidence that degradation of the nascent 3′ product is not sufficient to promote termination (Luo et al. 2006).
The above results suggest that another activity, perhaps associated with Rat1, is required to efficiently dismantle the EC. Indeed, Kawauchi et al. (2008) provided evidence that Sen1, an RNA/DNA-dependant ATPase and a member of the superfamily I of RNA/DNA helicases (Kim et al. 1999), cooperates with Rat1 to promote efficient RNAPII termination at two protein-coding genes. They suggested that Sen1 might expose the downstream RNA product generated after cleavage to facilitate its degradation by Rat1. Another possibility is that it could function at the site of termination to help disrupt the ternary complex. Furthermore, even though Sen1 was previously detected at snoRNAs genes (Kim et al. 2006), read-through at noncoding and some coding genes in a SEN1 mutant strain supports Sen1 involvement in termination of both class of RNAs (Steinmetz et al. 2006b). Sen1 is a large, low-abundance nuclear protein that interacts with the CTD (Ursic et al. 2004) and is implicated in processing of a variety of noncoding RNAs (Winey and Culbertson 1988; Ursic et al. 1995).
Senataxin (SETX) is the putative human homolog of Sen1 and possesses a conserved helicase domain (Moreira et al. 2004). Interestingly, mutations in SETX lead to two neurological disorders, an autosomal dominant form, ALS4 (amyotrophic lateral sclerosis type 4) (Blair et al. 2000), and an autosomal recessive form, AOA2 (ataxia-oculomotor apraxia type 2) (Moreira et al. 2004; Chen et al. 2006). Although the only characterized role of SETX in humans is its involvement in DNA damage repair (Suraweera et al. 2007), it will be interesting to determine whether SETX also functions in transcription termination, particularly in neurons.
The Rat1 homolog in humans is Xrn2, and its function in termination appears to be conserved. West et al. (2004), using siRNA-mediated knockdown, provided evidence that Xrn2 degrades the downstream 3′ RNA of the β-globin transcript and promotes termination. However, the effect of Xrn2 on β-globin pre-mRNA termination is specific to a cotranscriptional cleavage (CoTC) activity and not to the poly(A) site. Indeed, the β-globin gene contains a specific CoTC element, ∼1 kb downstream from the poly(A) site, that is required for termination (Dye and Proudfoot 1999, 2001). The CoTC sequence undergoes a rapid cleavage event that exposes a free 5′ monophosphorylated downstream RNA that serves as a substrate for Xrn2 (Teixeira et al. 2004; West et al. 2004). It is important to note that CoTC appears to be a specific termination signal for some globin genes (it is also found in the ε-globin 3′ flanking region) (Dye and Proudfoot 2001) that has not yet been identified in other genes. In most genes, which lack CoTC, cleavage at the poly(A) site generates the entry site for Xrn2.
Like Rat1, Xrn2 interacts with processing factors that might facilitate its recruitment to the 3′-ends of genes. Kaneko et al. (2007) showed that Xrn2 interacts with CstF-64 and p54nrb/PSF (protein-associated splicing factor). p54nrb and PSF are related multifunctional proteins that are involved in transcription, splicing, and polyadenylation (Rosonina et al. 2005; Liang and Lutz 2006). ChIP experiments revealed that, while p54nrb/PSF, cross-linked along the length of the β-actin gene, Xrn2 cross-linked primarily downstream from the poly(A) site, a result consistent with Rat1 localization along yeast genes (Kim et al. 2004b; Luo et al. 2006). In vitro cleavage assays demonstrated that while Xrn2 and p54nrb/PSF are required for degradation of the 3′ cleavage product, they are not necessary for cleavage itself (Kaneko et al. 2007). Coupling of Xrn2 to 3′ processing, however, was shown to stimulate exonuclease activity in the nuclear extract. These experiments, together with the observation that knockdown of p54nrb interferes with Xrn2 recruitment and termination, suggest that Xrn2 is recruited to the cleavage/polyadenylation machinery via p54nrb/PSF. The involvement of p54nrb/PSF in termination was recently confirmed in a genome-wide RNAi screen in C. elegans (Cui et al. 2008).
Evidence supporting the hybrid model of termination came from recent experiments that separated pre-mRNA associated with the DNA template from the released RNA. West et al. (2008) provided evidence that poly(A) site cleavage can occur while RNAPII is still bound to the template, and that Xrn2-mediated degradation of the RNAPII-associated downstream RNA precedes RNAPII release from the template. Alternatively, cleavage at the CoTC site in the terminator region of the β-globin gene allows Xrn2 entry and leads to RNAPII release before cleavage at the poly(A) site. However, in both cases, poly(A) site recognition, which occurs rapidly, is essential for termination. These data reinforce the idea that transcription termination is a combination of both the allosteric and torpedo models (Fig. 1).
Pausing the polymerase: another factor in RNAPII termination
ChIP analysis of human genes frequently shows a high density of RNAPII at promoter regions that decreases throughout the gene body and increases downstream from the poly(A) site (Glover-Cutter et al. 2008). This increased RNAPII density at the 3′-end may reflect pausing of RNAPII that precedes termination. Indeed, transcription pause sites located downstream from poly(A) sites appear to play a role in RNAPII 3′-end processing and termination, at least in mammalian genes, such as α-globin and γ-globin genes (Enriquez-Harris et al. 1991; Plant et al. 2005). A G-rich sequence (MAZ element: G5AG5) was identified downstream from the poly(A) site of the liver-specific C2 complement gene (Ashfield et al. 1994; Yonaha and Proudfoot 1999, 2000). This sequence is a potential binding site for the transcription factor MAZ and promotes poly(A)-dependant termination. Indeed, siRNA-mediated depletion of Xrn2 decreased MAZ-dependent termination efficiency (Gromak et al. 2006). Additionally, Gromak et al. (2006) described another G-rich region downstream from the β-actin poly(A) site that acts as a pause site-mediated terminator. Interestingly, the strength of the poly(A) site correlates with efficient pausing-dependent termination (Plant et al. 2005; Gromak et al. 2006).
Other cis-acting protein-binding sites have been shown to function in termination. It has been suggested that the poly(A) signal itself can direct RNAPII pausing. The AAUAAA-dependent pausing is mediated by CPSF but does not require the CTD and any other downstream pausing elements (Orozco et al. 2002; Park et al. 2004; Nag et al. 2006, 2007). Another sequence, a CCAAT-box in the adenovirus late promoter is necessary for termination of the upstream gene (Connelly and Manley 1989). West et al. (2006b) showed that multiple termination sequence elements in the mouse serum albumin (MSA) 3′-flanking region promote termination. Many MSA transcripts appear to be cleaved cotranscriptionally and, interestingly, seem to be adenylated and processed by the TRAMP/exosome complex (West et al. 2006a,b). Whether these sequence elements are implicated in RNAPII pausing remains to be elucidated. Pausing also plays a role in RNAPI and RNAPIII termination (see below), making it a general feature of the termination process. However, whether or not it is required at every gene remains unclear.
Trans-acting factor dynamics at the poly(A) site affect transcription termination
Transcription termination of protein-coding genes requires recognition of the polyadenylation signal. This provides a signal for binding many factors involved in 3′-end formation, which in turn renders RNAPII competent for termination (Whitelaw and Proudfoot 1986; Logan et al. 1987; Connelly and Manley 1988; Birse et al. 1997; West et al. 2008). In yeast, mutations and conditional depletion of factors involved in pre-mRNA cleavage and polyadenylation, including homologs of CstF-64 (Rna15), CstF-77 (Rna14), Pcf11, CPSF160 (Yhh1), CPSF-73 (Ysh1), and Ssu72, result in read-through at the 3′-end of protein-coding genes (Birse et al. 1998; Dichtl et al. 2002; Steinmetz and Brow 2003; Garas et al. 2008). Accordingly, ChIP assays suggest that 3′-end processing factors, including Pcf11, Rna14, and Rna15, become associated with the RNAPII EC at the poly(A) site, in a Ser2 phosphorylated CTD-dependant manner (Ahn et al. 2004; Kim et al. 2004a; Luo et al. 2006). Other components of the yeast polyadenylation machinery, including homologs of CPSF-100, CPSF-30, and symplekin, are also associated with the 3′-ends of protein-coding genes (Kim et al. 2004a). ChIP analysis in human cells also revealed the association of components of the 3′-end processing complex, including CPSF-73 and CstF-64 at the 3′-end of genes (Swinburne et al. 2006; Glover-Cutter et al. 2008). Interestingly, mutations in RNAPII subunits Rbp3 and Rpb11 induce read-through at an mRNA and snoRNA terminator, leading Steinmetz et al. (2006a) to propose that a “termination signal” is transduced through the Rpb3/Rpb11 heterodimer. An intriguing possibility is that the “termination signal” involves interactions with 3′-end processing factors (Fig. 1).
While some 3′-end processing factors and RNAPII itself behave as termination factors, other proteins function as anti-terminators. In ChIP experiments, factors such as the Paf1C and the TREX/THO complex, which functions in mRNP biogenesis and maintenance of genome integrity (Garcia-Rubio et al. 2008), cross-link throughout genes, but their levels are reduced on passage through the poly(A) site, suggesting they are released from the EC (Ahn et al. 2004; Kim et al. 2004a). Dissociation of a CstF64-interacting protein, the transcriptional coactivator PC4 (human homolog of Sub1), at the poly(A) site renders RNAPII competent for termination, and genetic evidence in yeast suggests that Sub1/PC4 has an evolutionarily conserved anti-terminator activity (Calvo and Manley 2001). The yeast mRNA-binding protein Npl3 is thought to compete for mRNA binding with cleavage and polyadenylation factors, thereby preventing termination until transcription of the poly(A) site (Bucheli and Buratowski 2005; Bucheli et al. 2007). It is intriguing to note that Npl3, like p54nrb/PSF, plays multiple functions in pre-mRNA metabolism (Dermody et al. 2008; Kress et al. 2008), and all three contain related RNP-type RNA-binding domains.
Pcf11 activity may be particularly important for transcription termination. Pcf11 interacts with nascent RNA, contains a conserved CTD-interacting domain (CID), and mutations or depletion of Pcf11 cause defects in cleavage and termination (Licatalosi et al. 2002; Sadowski et al. 2003; Meinhart and Cramer 2004; Zhang et al. 2005). Using purified yeast RNAPII ECs, Zhang et al. (2005) showed that the CID of Pcf11 bridges the CTD to pre-mRNA, and can dismantle the EC in vitro, although this activity is independent of its activity in cleavage, and its physiological significance is unclear (Sadowski et al. 2003; Zhang et al. 2005). In Drosophila, depletion of Pcf11 causes transcriptional read-through, and Drosophila Pcf11 can also dismantle the EC in vitro (Zhang and Gilmour 2006). Zhang et al. (2007) also provided evidence that Pcf11 causes premature termination of an HIV provirus by limiting RNAPII processivity. However, in yeast, Pcf11 mutants defective in cleavage also showed transcriptional read-through of the ADH1 and PMA1 genes (Kim et al. 2006). In contrast to the previous experiments, this result argues that Pcf11 function in cleavage is necessary for efficient termination. Furthermore, depletion of Pcf11 in human cells by siRNA reduced the efficiency of RNAPII termination and stabilized the 3′-product of pre-mRNA cleavage, suggesting a role of Pcf11 in degradation of the 3′ RNA (West and Proudfoot 2008). These experiments together suggest that Pcf11 may also facilitate Xrn2-mediated degradation of the downstream RNA and efficient transcription termination, but the mechanism remains unclear.
Many factors involved in 3′-end formation and termination also associate with promoter regions (Calvo and Manley 2003). This has been observed in yeast (Bentley 2002; Calvo and Manley 2003; He et al. 2003) and humans (Dantonel et al. 1997; Venkataraman et al. 2005; Swinburne et al. 2006; Zhang et al. 2007; Glover-Cutter et al. 2008; Rozenblatt-Rosen et al. 2009), and suggests a cooperation between the transcription initiation and 3′-end processing/termination machineries. One scenario is that this facilitates recruitment of polyadenylation factors to active genes, and that these factors travel with the EC during transcription (e.g., Dantonel et al. 1997). Alternatively, studies in yeast suggest that gene looping brings promoter and terminator regions together during early stages of transcription, facilitating RNAPII reinitiation (O'Sullivan et al. 2004; Ansari and Hampsey 2005; Singh and Hampsey 2007). Moreover, these studies showed that gene looping is dependent on components of the cleavage/polyadenylation machinery and the general transcription factor TFIIB. In mammals, gene looping has been proposed to regulate transcription of the human BRCA1 gene (Tan-Wong et al. 2008). Additionally, Perkins et al. (2008) showed the existence of gene looping at an integrated HIV-1 provirus, which required a functional poly(A) signal.
Cooperation between the splicing machinery and termination might also take place. An intriguing study in C. elegans showed that an SR protein, SRp20, is implicated in mRNA transcription termination. Cui et al. (2008) propose that SRp20 functions either in degradation of the cleaved 3′-product or release of RNAPII from the template. ChIP assays combined with high-density microarrays showed that the splicing factor polypyrimidine tract-binding protein (PTB) is found at the 3′-ends of many genes and in some cases many kilobases past the mature 3′-end of the mRNA (Swinburne et al. 2006). However, PTB, a splicing repressor, is a multifunctional protein that also functions, for example, in 3′-end formation (e.g., Moreira et al. 1998). Thus, further experiments will be needed to confirm the role of PTB, and SRp20, in termination, and whether this, in fact, reflects a link with splicing. Notably, West and Proudfoot (2009) showed that termination enhances gene expression by promoting more efficient pre-mRNA processing, including post-transcriptional splicing.
Histone mRNA termination
In contrast to plant and yeast histone mRNAs, mammalian histone proteins are encoded by mRNAs that do not possess a poly(A) tail at their 3′-end, but rather a stem–loop structure (Marzluff et al. 2008). The 3′-ends of histone mRNAs are generated by endonucleolytic cleavage 3′ to a dinucleotide CA, positioned downstream from a conserved stem–loop and upstream of a purine-rich histone downstream element (HDE), which is recognized by base-pairing with the 5′-end of the U7 snRNA (Schaufele et al. 1986; Bond et al. 1991; for review, see Dominski and Marzluff 2007). Interestingly, like polyadenylated mRNAs, cleavage is most likely performed by CPSF-73 (Dominski et al. 2005a; Yang et al. 2009). The ends of mature histone mRNAs consist of a stem–loop followed by a short single-strand tail. The stem–loop structure is bound by SLBP (stem–loop-binding protein) (also called HBF [hairpin-binding factor]), which helps recruitment of U7 snRNP. A zinc finger protein (ZFP100) bridges SLPB and Lsm11, a component of the U7-snRNP (Dominski et al. 2002; Pillai et al. 2003). In addition to these, another factor is required for histone pre-mRNA processing. Kolev and Steitz (2005) isolated a Heat-Labile Factor (HLF), which is required for histone pre-mRNA cleavage and remarkably was found to contain polyadenylation factors, including symplekin. As suggested for polyadenylated RNAs (Takagaki and Manley 2000), symplekin appears to be a scaffold protein that brings the seven other subunits of HLF, which include all CPSF subunits, CstF77 and CstF64, to the 3′-end of histone pre-mRNA.
Histone transcription continues for at least a few hundred nucleotides past the 3′-end of the mRNA (Chodchoy et al. 1987, 1991). The terminator of one mouse histone gene consists of a GC-rich region located ∼600 base pairs (bp) downstream from the mature 3′-end RNA (Chodchoy et al. 1991). Termination of histone gene transcription is, like other protein-coding genes, dependent on 3′-end processing. Indeed, deletion of the stem–loop or HDE results in transcription termination read-through (Chodchoy et al. 1991; Gu and Marzluff 1996). However, in vitro and in vivo experiments suggested that cleavage of histone mRNA precursors is not necessary for termination (Chodchoy et al. 1991; Gu and Marzluff 1996).
It is also likely that histone mRNA termination requires a 5′–3′ exonuclease. UV irradiation of a histone mRNA precursor containing a unique radiolabeled nucleotide at the cleavage site incubated in HeLa cell extract, revealed a cross-link with CPSF-73 in an HDE- and U7 snRNA-dependent manner. CPSF-73 also cross-linked to the 5′-end of the downstream cleavage product (DCP) (Dominski et al. 2005a). Recently, Yang et al. (2009) provided evidence that the DCP generated during 3′-end processing of histone pre-mRNAs is degraded 5′ to 3′ by a U7-dependent exonuclease activity. They proposed that CPSF-73, rather than Xrn2, is the 5′–3′ exonuclease that degrades the DCP, possibly implicating CPSF-73 in termination of histone pre-mRNAs. However, some data show that the degradation of the DCP can be uncoupled from cleavage (Walther et al. 1998; Dominski et al. 2005a), and no direct evidence that CPSF-73 possesses exonuclease activity has yet been presented.
Termination of mRNA transcription is a remarkably complex process involving not only many 3′-end processing factors but also elongation factors, possibly splicing factors, an exonuclease, and perhaps a helicase. Cis-acting sequences, including the AAUAAA sequence, pause sites, and histone 3′ processing signals are also crucial elements of transcription termination. Even if the mechanism of termination of protein-coding genes is better understood now than it was a few years ago, many aspects remain to be elucidated. Why do some genes terminate closer to the poly(A) site than the others? How often do promoter/enhancer elements of downstream genes contribute to termination? Are the downstream genes important elements in the choice of a termination site? Rat1/Xrn2 function is important for termination but apparently not essential. Indeed, some data suggest that endonucleolytic cleavage at the poly(A) site is not a prerequisite for transcription termination (Osheim et al. 1999, 2002; Zhang et al. 2005). Is it then possible that the mechanism of transcription termination can vary among genes? This is perhaps not surprising, given precedents from prokaryotes, such as rho-independent and rho-dependent mechanisms of termination (Nudler and Gottesman 2002). Although many elements and protein factors important for termination have been discovered recently, it remains difficult to draw a detailed common picture of RNAPII termination.
Termination of short RNAPII transcripts
Spliceosomal snRNAs
Mammalian RNAPII-transcribed spliceosomal snRNAs (U1, U2, U4, and U5) are intronless and nonpolyadenylated noncoding RNAs. Formation of their 3′-ends depends on specific snRNA promoters but does not require a poly(A) site or HDE, like histone mRNAs, but rather a 3′-box element located 9–19 nt downstream from the mature 3′-end of snRNAs (de Vegvar et al. 1986; Hernandez and Weiner 1986). The 3′-box is important for proper transcription of snRNAs and was shown to be required for 3′-end processing of U1 and U2 snRNAs (Cuello et al. 1999). After endonucleolytic cleavage downstream from the 3′-box, snRNAs carry an extended 3′-end that is trimmed after their export to the cytoplasm (for review, see Egloff et al. 2008).
Like protein-coding genes, the RNAPII CTD is required for transcription and 3′-processing of U1 and U2 snRNAs (Uguen and Murphy 2003). CTD truncation and CTD kinase inhibitors affect 3′-box recognition of U2 but not transcription termination (Medlin et al. 2003; Jacobs et al. 2004). Medlin et al. (2005) showed that P-TEFb and phosphorylation of both Ser2 and Ser5 are required for cotranscriptional recognition of the 3′-box. Interestingly, phosporylation of Ser7 of the CTD is required for the recruitment of the Integrator complex and, therefore, for snRNAs transcription and processing (Egloff et al. 2007). Integrator is a large complex that contains homologs of some CPSF subunits and has been shown to play a role in pre-snRNA 3′-end formation (Baillat et al. 2005). The CPSF-73 homolog, Int11/RC-68, interacts with the CTD phosphorylated at Ser7. Mutation of Ser7 to alanine specifically affects 3′ processing of snRNAs but not coding mRNAs (Chapman et al. 2007; Egloff et al. 2007).
The 3′-ends of snRNAs are generated by endonucleolytic cleavage, by an endonuclease containing a catalytic β-lactamase domain, and one of the polypeptides of the Integrator complex component, very likely, Int11/RC-68 (Baillat et al. 2005; Dominski et al. 2005b). Int11/RC-68 interacts with the apparent homolog of CPSF-100, Int9/RC-74, forming a complex distinct from CPSF, as they do not associate with CPSF-160 (Dominski et al. 2005b). Depletion of Int11 by siRNA or inactivation of its catalytic activity by mutation revealed a defect in U1 and U2 3′-end processing (Baillat et al. 2005). However, inactivation of Int11 did not seem to interfere with termination of transcription.
Despite the presence of 3′-boxes, snRNA genes appear not to use a common termination process. While transcription of U1 genes terminates close to the 3′-box (Cuello et al. 1999), U2 transcription terminates ∼1 kb from its mature 3′-end (Medlin et al. 2003). An unidentified termination factor or complex appears to bind the U1 precursor RNA immediately downstream from the 3′-box at a G track, just upstream of a T stretch (Cuello et al. 1999). When this potential terminator-binding region is positioned downstream from the U2 3′-box, it leads to efficient transcription termination. However, such a sequence was not identified around the natural terminator region of U2. These experiments indicate that snRNA transcription termination might involve gene-specific factors and mechanisms, although details remain unclear.
The 3′-box can be compared with the poly(A) signal in mRNA encoding genes, as they are both required for efficient cleavage and termination. There is no evidence yet of an association of Xrn2 with snRNAs genes, but it is conceivable that termination occurs via degradation of the downstream product, at least in U2 termination. Termination of yeast snRNA transcription involves the same pathway used for snoRNA termination, which is discussed next.
Yeast sno/snRNAs
The majority of snoRNAs fall into two structurally and functionally well-defined classes. SnoRNAs carrying the conserved box C and D elements function mainly as guides in the site-specific 2′-O-methylation of target RNAs, whereas box H/ACA snoRNAs direct pseudouridylation. While some snoRNAs are required for pre-rRNA processing, the majority of snoRNAs guide modifications of pre-rRNAs (Kiss 2002). In mammals, these latter snoRNAs are encoded within introns of pre-mRNAs (Richard and Kiss 2006), while snoRNAs involved in pre-rRNA processing in humans, and most of the guide RNAs expressed in yeast, are transcribed by RNAPII from their own promoters.
Following cleavage by the endoribonuclease Rnt1 (yeast RNase III) (Chanfreau et al. 1998) or release from introns, the 3′-ends of snoRNA precursors are matured by exonucleolytic trimming via the exosome. In some cases, independently transcribed snoRNAs do not possess the RNA hairpin structures that are recognized by Rnt1 and may undergo endonucleolytic cleavage by the mRNA 3′-end formation machinery (Fatica et al. 2000; Morlando et al. 2002). After cleavage, cotranscriptional association of specific factors and formation of sn/snoRNPs protect the mature 3′-end of sn/snoRNAs from further exonucleolytic trimming (Morlando et al. 2004; Ballarino et al. 2005; Yang et al. 2005; Houalla et al. 2006).
Involvement of mRNA 3′-end processing factors in yeast snoRNA processing and termination
SnoRNAs and snRNAs are not polyadenylated. However, they require common factors from the cleavage/polyadenylation machinery (Fatica et al. 2000; Morlando et al. 2002), and thus cleavage must be uncoupled from polyadenylation. Cleavage factors are, in fact, required for correct termination and processing of sno/snRNAs (see Fig. 2; Morlando et al. 2002; Carneiro et al. 2008). Additionally, mutations in a subcomplex of the cleavage/polyadenylation machinery, the APT complex (associated with Pta1 [homolog of symplekin]), lead to read-through transcription of certain sn/snoRNA genes (Dheur et al. 2003; Ganem et al. 2003; Nedea et al. 2003; Steinmetz and Brow 2003; Kim et al. 2006; Ghazy et al. 2009). The APT factors implicated in sn/snoRNA termination include Pti1, a second yeast homolog of CstF-64 that also binds Rna14; Ref2, which interacts with the snoRNP-specific protein Nop1 (Morlando et al. 2004); the CTD phosphatase Ssu72 (Ganem et al. 2003; Steinmetz and Brow 2003; Kim et al. 2006); and Swd2, which is also a component of the Set1 complex (Cheng et al. 2004; Dichtl et al. 2004). Overexpression of Pti1 inhibits polyadenylation without affecting cleavage, suggesting that Pti1 may function in uncoupling cleavage and polyadenylation during snoRNA 3′-end formation (Dheur et al. 2003).
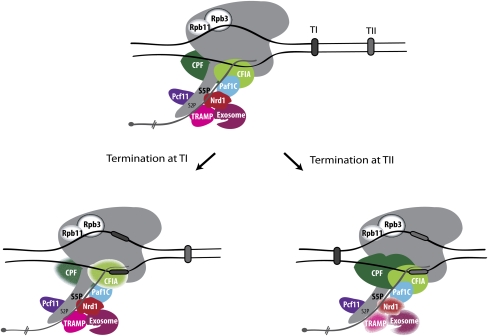
Figure 2.
Yeast RNAPII termination at snoRNA genes. Termination at terminator I (TI), the major termination site, is Nrd1-binding-dependent. The Nrd1 complex interacts with the Ser5-phosphorylated CTD and with the TRAMP and exosome complexes. The cleavage/polyadenylation machinery (CPF and CFIA) is not required for termination at TI (drawn as blurry complexes). Termination at terminator II (TII) involves cleavage/polyadenylation factors, while the Nrd1–TRAMP–exosome complex appears inactive in that process (blurry complexes). SnoRNA transcription couples termination with RNA surveillance. After release of the precursor, Trf4 and Pap1 adenylates the pre-snoRNA that will be processed by the exosome. Other factors shown to have a role in snoRNA termination are indicated. These include Pcf11, Paf1C, and the RNAPII subunits Rpb3 and Rpb11.
But the role of factors involved in 3′ processing/termination of pre-mRNA transcripts in snoRNA termination is not clear-cut. While ChIP experiments have shown that Rat1, mRNA cleavage factors including Rna15 and Rna14, as well as subunits of the APT complex are present at several snoRNA genes, surprisingly no termination defect was detected in RAT1 or RNA14 mutant strains (Kim et al. 2006). Normal snoRNA termination also occurred in strains lacking Rnt1 or the nuclear exosome component Rrp6 (Kim et al. 2006). Kim et al. (2006) also showed that cleavage by the cleavage/polyadenylation machinery is not required for snoRNA termination, as depletion of the CPSF-73 homolog Ysh1 or a mutation of PCF11 blocking cleavage did not affect snoRNA termination. However, transcription read-through was seen in strains expressing PCF11 mutants that are not capable of binding the CTD, and Garas et al. (2008) showed that termination at specific snoRNA genes is defective in a particular cold-sensitive YSH1 mutant strain. Interestingly, as shown in mRNA termination, Steinmetz et al. (2006a) observed that alteration of the heterodimer Rpb3/11 impairs snoRNAs termination. Another common point with mRNA termination is that in addition to proteins involved in 3′-end processing, the Paf1C is implicated in snoRNA termination. Defects in Paf1C lead to read-through transcription at snoRNA terminators (Sheldon et al. 2005). ChIP experiments suggested that Paf1C may facilitate snoRNAs 3′-end formation by recruiting Nrd1 (see below) (see Fig. 2; Sheldon et al. 2005).
The above results indicate that snoRNA genes appear to use different pathways to terminate transcription, as mutations or depletion in factors involved in mRNA termination have effects only on a subset of snoRNA genes. Further investigations will be needed to understand what underlies the differences among these genes.
The Nrd1 complex
Despite the apparent involvement of factors shared with pre-mRNA 3′-end processing, sno/snRNA termination relies heavily on a specific factor, the Nrd1 complex. Nrd1 (nuclear pre-mRNA down-regulation) is an essential nuclear RNA-binding protein that directs termination of snoRNA and some mRNA transcripts, in association with an interacting protein, the helicase Sen1 (Steinmetz and Brow 1996; Steinmetz et al. 2001; Ursic et al. 2004; Vasiljeva and Buratowski 2006). Nrd1 contains a CID (Meinhart and Cramer 2004; Meinhart et al. 2005) that interacts with CTD phosphorylated at Ser5 (Conrad et al. 2000; Gudipati et al. 2008; Vasiljeva et al. 2008a). Nrd1 also interacts with another essential RNA-binding protein, Nab3 (Yuryev et al. 1996).
Nrd1, Nab3, and Sen1 form the Nrd1 complex, which is detected by ChIP at snoRNAs genes (Kim et al. 2006) as well as some mRNAs genes, mainly at promoter regions (Nedea et al. 2003). Importantly, NRD1 and SEN1 mutations cause strong termination defects at some snoRNAs (Kim et al. 2006). Nrd1 and Nab3 recognize specific RNA sequences located downstream from the mature 3′-end of sn/snoRNAs (GUAA/G and UCUU, respectively) (Steinmetz and Brow 1996, 1998; Conrad et al. 2000; Steinmetz et al. 2001; Carroll et al. 2004, 2007), and mutations in these sites cause termination defects (Carroll et al. 2004). Several snoRNA terminators contain multiple potential binding sites for Ndr1 and Nab3, which form a heterodimer (Carroll et al. 2007). Nrd1 can also recruit the exosome complex, coupling the Nrd1 termination pathway to exosome-dependent trimming or degradation of transcripts (Fig. 2; Vasiljeva and Buratowski 2006).
Genome-wide analysis of RNAPII distribution in a SEN1 mutant strain confirmed the involvement of Sen1 in transcription termination of snoRNAs (Ursic et al. 1997; Rasmussen and Culbertson 1998) and other short transcripts (Steinmetz et al. 2006b). ChIP analysis showed RNAPII read-through at most snoRNAs genes in a SEN1 mutant strain or a strain defective for its helicase activity (Kim et al. 2006; Steinmetz et al. 2006b). Sen1 associates with the CTD and also coprecipitates with several snoRNAs and some snRNAs (Yuryev et al. 1996; Ursic et al. 1997, 2004). Nedea et al. (2008) showed that Glc7, the yeast PP1 (protein phophatase-1) catalytic subunit, is also a regulator of snoRNA termination, interacting directly with Sen1. Purification of TAP-tagged Sen1 identified Glc7, Nab3, Nrd1, and all the snoRNP core proteins as Sen1-associated proteins (Nedea et al. 2008). They propose that Sen1–Glc7–Nab3–Nrd1 form a stable complex in which Sen1 interacts directly with Nab3 and Glc7, which was shown to dephosphorylate Sen1 in vitro. Glc7 (He and Moore 2005) and PP1 in mammals (Shi et al. 2009) are known to function in mRNA 3′-end formation, and it will be of interest to elucidate the precise roles of dephosphorylation in all these processes.
Some snoRNA terminators appear to be bipartite, with a major (terminator I) Nrd1-binding-dependent terminator and a second, downstream terminator (terminator II) containing features similar to cleavage and polyadenylation signals (Fig. 2; Fatica et al. 2000; Morlando et al. 2002; Steinmetz and Brow 2003; Steinmetz et al. 2006a). Both terminators are required for efficient termination. Intriguingly, a snoRNA terminator can lead to polyadenylation when placed in the 3′ untranslated region (UTR) of a protein-coding gene, upstream of the natural poly(A) site (Steinmetz et al. 2006a). These data suggest that, depending on the gene context, snoRNA terminators possess the potential to direct polyadenylation. Previous data, indeed, reported polyadenylation of some sn/snoRNAs precursors in an exosome-defective yeast strain (van Hoof et al. 2000; Egecioglu et al. 2006). Accordingly, a strain deleted for RRP6 showed that one to four adenines are present at the 3′-end of C/D-box snoRNA precursors that had been incompletely processed (Grzechnik and Kufel 2008). Trf4/5 (PAPs from the TRAMP complex) add adenines at terminator I, while the canonical poly(A) polymerase, Pap1, polyadenylates snoRNA precursors at both terminators. SnoRNA read-through and defects in Nrd1 recruitment at terminator I in a TRAMP-defective strain indicate that the TRAMP complex is important for the appropriate association of Nrd1 with snoRNA genes and therefore for termination. Grzechnik and Kufel (2008) concluded that polyadenylation by Trf4 and Pap1 is a normal mechanism linked to snoRNA termination that stimulates 3′-end formation by the exosome (Fig. 2).
The regulation of transcription termination of snRNAs and snoRNAs involves a large number of factors, implicated in every step of transcription. Many factors are shared between mRNA and snoRNA termination pathways, and a tight regulatory network is necessary to correctly and efficiently discriminate between the two. An important feature of termination appears to be the distance traveled by RNAPII when it reaches the terminator. The Nrd1 complex is implicated in transcription of short transcripts (Steinmetz et al. 2006b). As explained below, this regulation reflects the different stages of CTD phosphorylation. On the other hand, the potential polyadenylation of snoRNAs adds another degree of complexity in possibly linking RNA surveillance, processing, and termination, and will need to be more fully explored.
A wider role for the Nrd1 complex in gene expression
Recent data indicate that the function of the Nrd1 complex is not restricted to snoRNA 3′-end formation and termination. The Nrd1 complex has wider functions in gene expression, especially in the synthesis/processing of CUTs. CUTs are short noncoding RNAs (300–600 nt) that were first detected by genome-wide microarray analysis in yeast strains defective for RNA nuclear degradation (Kapranov et al. 2005; Wyers et al. 2005; David et al. 2006; Davis and Ares 2006). In strains defective for the nuclear exosome protein Rrp6, CUTs with a defined 5′-end and heterogenous 3′-ends accumulate. These transcripts are estimated to originate from 10% of yeast intergenic regions (Wyers et al. 2005; although see Neil et al. 2009 and Xu et al. 2009). After Nrd1-dependent termination by RNAPII, CUTs are rapidly polyadenylated by the TRAMP complex, and then degraded by the nuclear exosome (Wyers et al. 2005; Arigo et al. 2006b; Houalla et al. 2006; Thiebaut et al. 2006). Recently, Gudipati et al. (2008) showed that Nrd1-dependent termination of CUTs is inhibited by CTD phosphorylation on Ser2 and promoted by Ser5 phosphorylation. They also showed that binding of Nrd1 to the nascent transcript, even if not sufficient, is necessary for efficient termination. Accordingly, Vasiljeva et al. (2008a) showed that Nrd1 has a high affinity for the CTD phosphorylated at Ser5 and Ser5/Ser2. Additionally, association of Nrd1 with a snoRNA or mRNA gene is not affected by inhibition of Ser2 phosphorylation.
The studies of Gudipati et al. (2008) and Vasiljeva et al. (2008a) provide an explanation of why termination depends on the distance traveled by RNAPII, and how RNAPII chooses between the 3′ cleavage and polyadenylation pathway for mRNA termination and the Nrd1 complex for noncoding RNAs, short ORFs and CUT termination. Short transcripts are mostly associated with Ser5 phosphorylation, which recruits the Nrd1 complex. In contrast, longer transcripts (most mRNAs) will be largely associated with Ser2 phosphorylation at the 3′-end of the gene, which recruits Pcf11 and Rtt103, facilitating Rat1 function (Rondon et al. 2008). Additional studies will be needed to understand how the cleavage/polyadenylation factor pathway is regulated at terminator II of snoRNA genes. Even when this terminator is at a greater distance from the promoter, this distance is usually <100 bp from terminator I.
Some CUTs play a regulatory function by controlling adjacent gene activity (Martens et al. 2004; Steinmetz et al. 2006b). For example, the regulatory region of the SER3 gene produces a CUT that represses transcription of SER3 (Martens et al. 2004). Nrd1 complex-dependent termination was shown to recognize regulatory terminators called “attenuators” located in the 5′-UTR of three characterized protein-coding genes—NRD1, HRP1 (or NAB4), and IMD2 (Steinmetz et al. 2001, 2006b; Arigo et al. 2006a; Kuehner and Brow 2008). Termination of the noncoding RNAs initiated upstream of the mRNA start site of the protein-coding genes regulates their expression. For instance, binding of Nrd1 and Nab3 to the 5′-UTR and upstream coding region of NRD1 directs premature termination, resulting in negative autoregulation of Nrd1 levels (Steinmetz et al. 2001; Arigo et al. 2006a). Likewise, termination of CUTs initiated upstream of the IMD2 mRNA start site regulates IMD2 mRNA expression (Jenks et al. 2008; Kuehner and Brow 2008; Thiebaut et al. 2008). All three attenuators are sensitive to mutations in SEN1 and RPB11, consistent with previous studies showing the importance of the Rpb3/11 heterodimer in termination (Steinmetz et al. 2006a).
The Nrd1–TRAMP/exosome complex appears to have a broad role in gene expression. CUTs provide an example of RNAs controlled by the Nrd1 complex that appear important for regulation of mRNA expression. Despite the existence of human homologs of exosome and TRAMP subunits, there is still only limited evidence for such an RNA quality and gene expression control pathway in humans (Houseley et al. 2006; Kapranov et al. 2007; Vanacova and Stefl 2007). However, West et al. (2006a) showed that the 5′ products of β-globin transcription resulting from cleavage at the CoTC frequently contain a short A-tail and accumulate upon siRNA-mediated depletion of the Rrp6 human homolog. But more importantly, new noncoding RNAs with unknown function were recently detected in the human genome (Kapranov et al. 2007). Further studies will be needed to elucidate the role of these RNAs and to reveal whether or not they function in gene expression.
RNAPII termination and chromatin remodeling factors
Chromatin architecture plays an important role in gene expression (Li et al. 2007), and mechanisms that remodel chromatin coordinate elongation with termination and 3′-end processing. Most commonly, chromatin remodeling occurs via histone modifications (Berger 2002) or ATP-dependant DNA remodeling complexes (Narlikar et al. 2002). Chromatin remodeling factors, grouped into three different families (SWI/SNF, ISWI, and CHD/Mi2), play crucial roles in transcription initiation (Xue et al. 2000). However, evidence has accumulated that these factors may also function in transcription termination. A genetic screen in Schizosaccharomyces pombe identified Hrp1, the homolog of the Saccharomyces cerevisiae chromatin remodeling protein Chd1, as a termination factor (Alen et al. 2002). Transcription run on (TRO) analysis revealed a termination defect in both hrp1Δ S. pombe and in chd1Δ S. cerevisiae strains, correlating with an alteration in chromatin structure at the 3′-end of the tested genes.
Another remodeling complex, ISWI, has also been implicated in remodeling at the 3′-ends of genes. ChIP analysis confirmed the presence of the CHD/Mi2 and ISWI complexes at the terminator regions of yeast protein-coding genes (Alen et al. 2002). Additionally, an ATPase-deficient version of Isw1 resulted in a termination defect at the 3′-end of protein-coding genes and a defect in Rna15 recruitment that correlated with a drop in CTD Ser2 and Ser5 phosphorylation (Morillon et al. 2003).
The Set1-containing complex is responsible for the H3-K4 trimethylation that is associtated with actively transcribed genes (Santos-Rosa et al. 2003). Mutation or depletion of Swd2, a component of both the Set1 and APT (see above) complexes, leads to transcriptional read-through at certain genes, including mRNAs and snoRNAs; however, this is independent of a defect in methylation at H3-K4 (Cheng et al. 2004; Dichtl et al. 2004).
Although heterochromatin is thought to constitute a transcriptionally silent region, a large analysis of RNAPII distribution throughout the entire genome of S. cerevisiae showed that RNAPII is present in heterochromatin (Steinmetz et al. 2006b). Genome-wide analysis of human cells suggests that most protein-coding genes, including transcriptionally inactive genes, are occupied by RNAPII, and evidence of transcription initiation is frequently detected (Barski et al. 2007; Guenther et al. 2007). Two recent papers (Houseley et al. 2007; Vasiljeva et al. 2008b) provide evidence that the Nrd1/exosome complex represses transcription within heterochromatin. In yeast, IGS1 (intergenic spacer region 1) in the rDNA locus (Fig. 3; and see below) produces CUTs, whose transcriptional repression prevents recombination and loss of rDNA repeats. This rDNA silencing appears mostly to be due to the histone deacetylase Sir2. However, inactivation of the Nrd1 complex or the exosome led to termination defects of CUTs within the rDNA loci (Vasiljeva et al. 2008b). This is correlated with rDNA chromatin changes and higher rDNA recombination. While histone deacetylation by Sir2 plays an important role in this process by preventing RNAPII access to the template, the Nrd1 complex might ensure termination of transcripts generated by RNAPII that did manage to access to the template, by recruiting the TRAMP and exosome complexes (Houseley et al. 2007; Vasiljeva et al. 2008b). Interestingly, this mechanism might also be used at repressed chromatin at telomeres (Houseley et al. 2007).
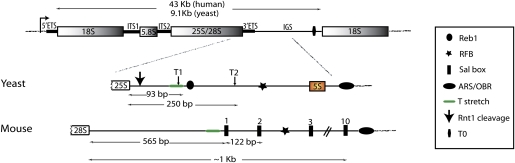
Figure 3.
Organization of the rDNA genes in yeast and mouse. The rDNA transcription unit is 9.1-kb long in yeast and 43-kb long in human. It consists of a 35S precursor in yeast and 45S in human. The precursor contains the sequences of the mature 18S, 5.8S, and 25S/28S rRNAs separated by two ITSs, ITS1 and ITS2, and is flanked by two ETSs, the 5′ETS and 3′ETS. In yeast, the IGS is interrupted by the 5S rDNA gene, which is transcribed by RNAPIII in the opposite orientation. In yeast, RNAPI terminates predominantly at the T1 terminator, apparently dependent on Reb1 and located ∼90 bp downstream from the 25S sequence, in IGS1 (IGS upstream of the 5S gene). The “fail-safe” terminator (T2) is located ∼250 bp downstream from the 25S sequence. The RFB that binds Fob1 is located downstream from T2. In mouse, the major terminator (Sal box 1) is located ∼550 nt downstream from the 28S RNA. The origin of bidirectional replication (OBR) in mouse and the autonomously replicating sequence (ARS) in yeast are indicated, as well as the Rnt1 cleavage and the T0 terminator.
All these data show that changes in chromatin are important for termination and suggest that RNAPII requires a specific chromatin structure to terminate efficiently. In addition, it is possible that chromatin remodeling factors facilitate the recruitment of factors that are directly involved in termination. This is illustrated by a recent study in S. pombe showing that Swi6 (human HP1) recruits cohesin to the intergenic region of convergent genes, which forces RNAPII to terminate by blocking RNAPII elongation (Gullerova and Proudfoot 2008). While ChIP analysis in HeLa cells showed an association of several proteins involved in chromatin remodeling with the entire transcription unit, including the region downstream from the poly(A) site (Venkataraman et al. 2005), detailed studies of the possible role of chromatin remodeling factors in mammalian termination are necessary to better understand the exact function and generality of chromatin in termination.
RNAPI termination
RNAPI synthesizes >50% of total cellular RNAs in actively growing cells. RNAPI is composed of five subunits common to all RNAPs, two subunits shared with RNAPIII, and seven unique subunits (Reeder 1999). Human cells contain ∼400 copies of ribosomal DNAs repeats distributed over five chromosomes, while yeast cells contain 150–200 copies of rDNAs, concentrated on one chromosome. Each repeat is transcribed in the nucleolus by RNAPI to synthesize the 35S precursor RNA (45S in human) that is processed into mature 18S, 5.8S, and 25S (28S in mammals) rRNAs (Fig. 3). Chromatin structure influences the active or silent status of rDNA genes, and only a portion of these genes are transcribed at a time (Grummt and Pikaard 2003; Birch and Zomerdijk 2008). Most ribosomal gene units consist of a transcribed sequence containing the mature rRNA sequences, internal and external transcribed spacer sequences (ITS and ETS), and the IGS. The yeast IGS is interrupted by the 5S RNA gene, which is transcribed independently by RNAPIII in an opposite polarity. Termination of transcription of the 35S/45S precursor occurs in the IGS region at a series of terminator elements downstream from the 3′-end of the 35S transcript, and upstream of the 5S gene (see Fig. 3; van der Sande et al. 1989; Johnson and Warner 1991; El Hage et al. 2008).
RNAPI termination: not that simple
RNAPI termination involves significantly fewer factors than RNAPII termination but is still a complex process. Three major termination sites have been identified in the IGS sequence of yeast rDNA. Ninety percent of yeast RNAPI terminates at the first terminator (T1), situated upstream of a pause-inducing factor-binding site that is recognized in vitro by Reb1 (Ju et al. 1990; Lang and Reeder 1995). In mammals, the major terminator, called the Sal box (Grummt et al. 1985; Kuhn et al. 1988), is recognized by TTF-I (transcription termination factor for polI) (Bartsch et al. 1988; Evers et al. 1995), and like the Reb1-binding site, it must be correctly oriented to cause termination (Grummt et al. 1985; Lang and Reeder 1993). The mouse Sal box is an 18-bp element repeated 10 times in the IGS, while humans contain a shorter Sal box (11 bp) that is recognized by the human TTF-I (Bartsch et al. 1987; Pfleiderer et al. 1990; Evers and Grummt 1995). Several mammalian rDNAs contain a Sal box (T0) upstream of the transcription start site (Grummt et al. 1986; Labhart and Reeder 1986; Pfleiderer et al. 1990). Such terminators seem to prevent transcriptional interference by terminating transcripts initiated in the IGS (Bateman and Paule 1988; Henderson et al. 1989) and have been shown to stimulate transcription as well (Grummt et al. 1986). Yeast rDNA also contains a T0 terminator, which is, however, in the wrong orientation to mediate termination (Fig. 3).
Another sequence element situated upstream of the Reb1-binding site in yeast and the Sal box in mammals, which is necessary for termination, acts as a transcript release element (Lang et al. 1994; Jeong et al. 1995; Lang and Reeder 1995). This sequence is composed of a stretch of Ts. The weakness of the resulting A·U heteroduplex might destabilize RNAPI pausing at the Reb1/TTF-I-mediated pause site (Kuhn and Grummt 1989; Lang and Reeder 1995). Consequently, rRNA precursors terminated at T1 are formed ∼12–20 bp upstream of the Reb1-binding site and within the T stretch, 93 nt downstream from the mature 3′-end of 25S RNA (Lang et al. 1994; Jeong et al. 1995).
A release factor is additionally needed for rRNA 3′-end formation and RNAPI release in mammals (Mason et al. 1997). The factor, designated as PTRF (Pol I and transcript release factor), is associated with RNAPI and TTF-I. Addition of PTRF to an in vitro transcription assay containing RNAPI is sufficient to release the transcript and RNAPI from the DNA template. PTRF function is dependant of the T-rich stretch located upstream of the terminator T1 and most likely binds the U stretch of the 3′-end of the precursor RNA (see Figs. 3, 4; Jansa et al. 1998; Jansa and Grummt 1999).
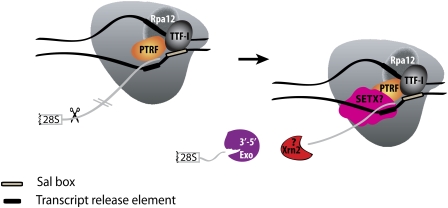
Figure 4.
Mammalian RNAPI termination. Transcription terminates at the transcript release element composed of a stretch of Ts, upstream of the TTF-I-mediated pause site. The release factor PTRF, which interacts with RNAPI and TTF-I, recognizes the transcript release element and most likely binds the stretch of Us on the nascent RNA. The RNAPI-specific subunit Rpa12 also plays a role in yeast RNAPI termination that is possibly conserved in mammals. After cleavage of the precursor downstream from the 28S gene, Xrn2 degrades the downstream RNA, “catches up” with RNAPI, and participates in RNAPI release, possibly in conjunction with SETX. An unidentified 3′–5′ exonuclease might be involved in processing of the rRNA precursor.
Ten percent of yeast RNAPI transcripts do not terminate at T1 and read-through to a second terminator (T2 or “fail-safe” terminator) located ∼250 nt downsteam from the 25S RNA 3′-end (Reeder et al. 1999; Prescott et al. 2004) or to the replication fork barrier (RFB). The RFB, located roughly 300 bp downstream, recruits the protein Fob1, which blocks DNA replication forks (see Fig. 3; Takeuchi et al. 2003; Huang et al. 2006). TTFI also acts as an RFB when it is bound to the second Sal box and flanking regions of the IGS (Gerber et al. 1997). Unexpectedly, the RFB was found to act as an additional RNAPI transcription terminator in a xrn1Δ strain depleted of Rat1 (El Hage et al. 2008). Absence of Rat1 and the mainly cytoplasmic exonuclease Xrn1 causes read-through, mostly at T1 but also at T2 and RFB. The additional absence of Fob1 increased read-through at T2 and RFB, leading to the accumulation of very large transcripts extending through the IGS1 region. The role of Rat1/Xrn2 in RNAPI termination is discussed further below.
Coupling of rRNA processing and termination
The 3′-end of the primary rRNA transcript is cleaved by the dsRNA-specific RNase III, Rnt1 (Elela et al. 1996; Kufel et al. 1999; Henras et al. 2004). Inactivation of Rnt1 causes read-through at T1, indicating that, as with protein-coding genes, cotranscriptional cleavage is important for efficient termination (Reeder et al. 1999; Prescott et al. 2004; Kawauchi et al. 2008). Recently, Catala et al. (2008) showed that Rnt1 interacts with two RNAPI-specific subunits, Rpa12 and Rpa34. Rpa12 shares sequence similarity with the small RNAPII subunit Rpb9 and with C11 of RNAPIII. While C11 is implicated in RNAPIII reinitiation (see below), Rpb9 might regulate arrest of elongating RNAPII at pause sites (Awrey et al. 1997). TRO analysis and electron microscopy (EM) visualization of Miller spreads demonstrated the involvement of Rpa12 in RNAPI termination (Prescott et al. 2004; Kawauchi et al. 2008). These experiments link transcription termination and processing of pre-rRNA. Deletion of Rnt1 or mutation disrupting its interaction with RNAPI altered the chromatin conformation of rDNA by increasing the number of open rRNA gene repeats. However, deletion of RPA12 alone had no effect on chromatin opening and pre-rRNA processing (Catala et al. 2008). Interestingly, as shown for RNAPII termination (Alen et al. 2002), chromatin remodeling factors are also involved in RNAPI termination. Deletion of CHD1, ISW1, and ISW2 had no effect on transcription initiation or on accumulation of rRNA, but caused an RNAPI termination defect (Jones et al. 2007). It is thus possible that normal RNAPI termination requires a specialized chromatin structure directed by these remodeling factors.
RNAPI termination: also torpedoed?
Recent data have unexpectedly implicated Rat1 in RNAPI termination. ChIP assays showed the presence of Rat1 in the IGS1 region of rDNA, and increased RNAPI occupancy in the IGS1 region was detected in a strain lacking active Rat1 (El Hage et al. 2008). Additionally, deletion of the gene encoding the Rat1 cofactor Rai1 caused the same RNAPI termination defect. Extending these results, Kawauchi et al. (2008) showed that Sen1 is also detected at the terminator region of rDNA and cooperates with Rat1 to promote efficient RNAPI termination. However, contrary to what was previously shown in vitro (see above), Reb1 was not detected at the terminator region in vivo, and a fourfold reduction of Reb1 did not affect RNAPI termination in TRO analysis. These results question the necessity of Reb1 in termination at T1 and indicate that the mechanism of RNAPI termination is more complicated than expected (Kawauchi et al. 2008). On the other hand, it is possible that Reb1 protein levels were not sufficiently reduced enough to cause a termination defect. El Hage et al. (2008) and Kawauchi et al. (2008) proposed that after cotranscriptional cleavage of the rRNA precursor by Rnt1, the downstream RNA is bound by Rat1/Rai1, which “catches up” with RNAPI paused at T1. Rat1, in association with the helicase Sen1, most likely recruited cotranscriptionally by Rnt1 (Ursic et al. 2004), might then be able to disrupt the EC and release the transcript and RNAPI from the DNA template.
The above data indicate that RNAPI termination shares common features with RNAPII termination, at least in yeast. No RNase III cleavage site has been identified at the 3′-end of the human 45S precursor, even though the precursor is trimmed by an exonuclease at its 3′-end (Kuhn and Grummt 1989). RNAPI termination in mammals requires more investigation in order to determine whether Xrn2, and perhaps SETX, is also involved (Fig. 4).
RNAPIII termination
RNAPIII transcribes a variety of short, nuclear, and cytoplasmic noncoding genes (100–150 bp) including those encoding 5S rRNA, U6 spliceosomal snRNA, tRNAs, and the RNAse P, RNAse MRP, adenovirus-associated (VA), and 7SK RNAs (for review, see Dieci et al. 2007). RNAPIII is the largest of the three RNAPs; yeast RNAPIII has an overall mass of 700 kDa and is composed of 17 subunits, all essential for cell viability. Nine subunits define the structural core, evolutionarily related to the other nuclear RNAPs, with only five subunits specific to RNAPIII (Jasiak et al. 2006). RNAPIII is a very efficient polymerase not only because of the short transcription units, but also because of rapid reloading of RNAPIII on the same transcription unit (Dieci and Sentenac 1996; Ferrari et al. 2004). RNAPIII can terminate efficiently with no apparent need of other factors.
RNAPIII: a terminator on its own?
Three classes of RNAPIII-transcribed genes exist (tRNAs/VA RNAs, 5S rRNA, and U6 snRNA), which each uses a characteristic promoter. Despite the use of different promoters, RNAPIII terminates transcription at a simple common consensus sequence at the 3′-end of genes. More than 25 years ago, Bogenhagen and Brown (1981) identified a nucleotide sequence composed of a cluster of T residues in the coding DNA strand, required for transcription termination of the Xenopus laevis oocyte 5S RNA gene. Later, Cozzarelli et al. (1983) showed that a purified RNAPIII unable to carry out transcription initiation of a 5S DNA was able to terminate accurately and efficiently by recognizing the simple T-rich consensus. This experiment provided early evidence that RNAPIII itself possesses transcription termination activity without requirement of associated factors.
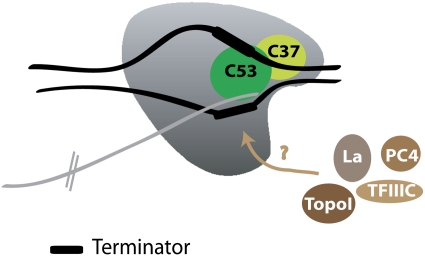
In vitro transcription using RNAPIII lacking the RNAPIII-specific subunits C11, C37, and C53 (RNAPIIIΔ) can support transcription initiation but fails to terminate properly (Landrieux et al. 2006). Adding back the heterodimer C37/C53 was sufficient to allow recognition of the terminator and to correct the termination defect, but not for transcription reinitiation. It was proposed that the C37/C53 complex reduces the elongation rate of RNAPIIIΔ, allowing for an increased pausing time at the terminator elements, which leads to release of the transcript and RNAPIII (Fig. 5). C11 is an essential subunit that mediates the intrinsic RNA cleavage activity of RNAPIII (Whitehall et al. 1994) and shows homology with the RNAPII subunit Rpb9 and to RNAPII elongation factor TFIIS (Chedin et al. 1998). Landrieux et al. (2006) showed that C11 RNA cleavage activity is not required for correct recognition of the RNAPIII terminator elements and release of the newly synthesized transcript. However, C11 itself, but not its cleavage activity, is required for reinitiation (Chedin et al. 1998; Landrieux et al. 2006). These experiments strongly suggest that termination and cleavage of the RNA precursor are not coupled in the RNAPIII transcription process. Consistent with its role in termination, structural analysis of RNAPIII showed that the C37/C53 heterodimer is positioned at the outer end of the DNA-binding cleft, toward the front of the EC, allowing the complex to sense the incoming DNA (Fernandez-Tornero et al. 2007).
Figure 5.
RNAPIII termination. Most of the termination activity is triggered by RNAPIII itself. The subunits C37 and C53 are essential for termination. The heterodimer C37–C53 might play a role in reducing the elongation rate of RNAPIII, allowing for an increased pausing time at the terminator, composed of a stretch of four to five Ts in the coding DNA strand. Several auxiliary factors, such as La, PC4, Topoisomerase I, or TFIIIC, are proposed to participate in RNAPIII termination in mammals.
Most RNAPIII termination signals occur within the first 40 bp downstream from the mature 3′-end of RNAs. In yeast, the most frequent T stretch length is six or seven with no stretch shorter than five Ts (Allison and Hall 1985; Hamada et al. 2000). In mammals, a stretch of four Ts is the most frequent terminator, and a stretch of more than five Ts is rare (Braglia et al. 2005). RNAPIII pauses not only at the terminator region but also at several other T stretches during transcription (Campbell and Setzer 1992; Matsuzaki et al. 1994). Campbell and Setzer (1992) proposed that RNAPIII release at the terminator is dependent on nascent RNA strand displacement during transcription elongation and can be uncoupled from termination signal recognition. RNAPIII recycling is also dependent on a natural termination signal (Dieci and Sentenac 1996; Ferrari et al. 2004). Termination can be influenced by sequences surrounding the T tract (Bogenhagen and Brown 1981; Mazabraud et al. 1987; Gunnery et al. 1999; Braglia et al. 2005). Braglia et al. (2005) proposed that the surrounding residues and, in particular, downstream residues of the yeast terminator influence the pausing time or the processivity of RNAPIII. However, no consistent pattern for this context effect has been elucidated.
Does RNAPIII need auxiliary factors to terminate?
Even though RNAPIII appears to be sufficient for accurate termination, some studies have demonstrated factor-dependent mechanisms for efficient termination and reinitiation in mammals (Fig. 5). The general RNAPIII transcription factor TFIIIC, the RNAPII transcription coactivator PC4, and Topoisomerase I interact with the terminator region of VA1 and tRNA genes and appear to facilitate RNAPIII termination and reinitiation (Wang and Roeder 1996, 1998). Purification of VA1 terminator-binding factors identified nuclear factor 1 (NF1) transcription factor as a stimulatory factor for RNAPIII termination and recycling (Wang et al. 2000). However, the NF1-binding sequence is not present at the terminator regions of many RNAPIII-transcribed genes and might only be stimulatory for specific genes. Additionally, ChIP assays showed that NF1 is not detectable at RNAPIII-transcribed genes (Fairley et al. 2005).
The La protein is associated with newly transcribed RNAPIII RNAs via their common 3′-U termini and functions to prevent their degragation by exonucleases (Maraia and Intine 2001). Twenty years ago, Gottlieb and Steitz (1989) proposed a role of La protein in transcript release and RNAPIII termination/reinitiation. In vitro studies (Maraia et al. 1994; Maraia and Intine 2001) and the fact that La is found at RNAPIII-transcribed genes in vivo (Fairley et al. 2005) reinforced the idea that La is a transcription termination factor. On the other hand, La-depleted X. laevis extracts are capable of efficient RNAPIII termination, and nascent transcripts are released from the DNA template (Lin-Marq and Clarkson 1998). Other in vitro studies show no role for La protein in RNAPIII transcription (Pannone et al. 1998; Weser et al. 2000; Hu et al. 2003). Recent work from French et al. (2008) might reconcile both hypotheses. Using EM to visualize RNAPIII on the yeast 5S rRNA, a higher average RNAPIII density was detected in a strain deleted for the yeast La protein, and ChIP assays demonstrated an association of La protein with the 5S gene, apparently mediated by the nascent RNA. French et al. (2008) showed that La has no effect on the rate of synthesis and size of 5S rRNA but proposed that the extra RNAPIII visualized at the 5S gene in the yeast laΔ strain reflects a defect in RNAPIII release.
The above data point to possible differences in requirement for yeast and mammalian RNAPIII termination. The yeast terminator may be less stable than the human terminator, as it possesses a longer U·A heteroduplex, and might therefore not require the action of accessory factors to disrupt the EC. In contrast, the shorter mammalian terminators might require the addition of accessory factors to terminate transcription efficiently. More work will be needed to elucidate the requirement and role of accessory factors in RNAPIII termination.
Conclusions
Transcription termination is a complex mechanism that appears to cooperate with essentially every other step in transcription. Termination, in particular, RNAPII termination, involves a large number of factors and a variety of cis-acting elements to coordinate the process. Yeast RNAP termination, and again, RNAPII termination, in particular, is better understood than is termination in mammals. It will be important to determine what features of termination now only observed in yeast can be transposed to mammalian systems. Even in yeast, there are significant questions, some concerning apparent differences in RNAPII termination even in similar classes of genes. For instance, why do some snoRNAs require only the Nrd1 complex, while others also require components of the cleavage/polyadenylation machinery? Why do some mRNAs require the Nrd1 complex for termination, but most do not? And what precisely is the role of Rat1/Xrn2, and what fraction of genes require it? Currently, transcription termination appears to involve multiple mechanisms and pathways. More work will be needed to elucidate not only the details of these mechanisms, but also what distinguishes one gene from another.
Even though the three RNAPs terminate transcription differently, some similarities exist. Significantly, these might in part be explained by RNAP subunits. Apart from the shared subunits, similarities exist between Rpa12 (RNAPI), Rpb9 (RNAPII), and C11 (RNAPIII), and all have a direct or indirect role in termination. Rpa12 has been implicated directly in termination, while C11 has a role in nascent RNA cleavage and RNAPIII reinitiation, and Rpb9 in RNAPII pausing. Pausing at the 3′-end of genes is, indeed, a feature of termination shared by all three RNAPs. Two other RNAP subunits, Rbp3 and Rbp11, also function in RNAPII termination. RNAPs themselves therefore seem to have intrinsic functions in termination, which is, of course, most evident with RNAPIII.
Even though considerable progress has recently been made in understanding RNAP termination, many questions remain to be answered. Studies in mammalian systems should now be a central focus for new investigations.
Acknowledgments
We thank Emanuel Rosonina for his helpful corrections and comments on the manuscript and his support. P.R. is supported by a grant from the Muscular Dystrophy Association and relevant work in our laboratory is supported by grants from the NIH.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1792809.
References
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Alen C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- Allison DS, Hall BD. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985;4:2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes & Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell. 2006a;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006b;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ashfield R, Patel AJ, Bossone SA, Brown H, Campbell RD, Marcu KB, Proudfoot NJ. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 1994;13:5656–5667. doi: 10.1002/j.1460-2075.1994.tb06904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae . Mol Cell Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae . Proc Natl Acad Sci. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartsch I, Schoneberg C, Grummt I. Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribsomal genes. Mol Cell Biol. 1987;7:2521–2529. doi: 10.1128/mcb.7.7.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch I, Schoneberg C, Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988;8:3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E, Paule MR. Promoter occlusion during ribosomal RNA transcription. Cell. 1988;54:985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- Bentley D. The mRNA assembly line: Transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Lee BA, Hansen K, Proudfoot NJ. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Blair IP, Bennett CL, Abel A, Rabin BA, Griffin JW, Fischbeck KH, Cornblath DR, Chance PF. A gene for autosomal dominant juvenile amyotrophic lateral sclerosis (ALS4) localizes to a 500-kb interval on chromosome 9q34. Neurogenetics. 2000;3:1–6. doi: 10.1007/pl00022976. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Bond UM, Yario TA, Steitz JA. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes & Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-β-lactamase fold within nucleic acids processing enzymes: The β-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- Calvo O, Manley JL. Strange bedfellows: Polyadenylation factors at the promoter. Genes & Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- Campbell FE, Jr, Setzer DR. Transcription termination by RNA polymerase III: Uncoupling of polymerase release from termination signal recognition. Mol Cell Biol. 1992;12:2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M. Inactivation of cleavage factor I components Rna14p and Rna15p induces sequestration of small nucleolar ribonucleoproteins at discrete sites in the nucleus. Mol Biol Cell. 2008;19:1499–1508. doi: 10.1091/mbc.E07-10-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala M, Tremblay M, Samson E, Conconi A, Abou Elela S. Deletion of Rnt1p alters the proportion of open versus closed rRNA gene repeats in yeast. Mol Cell Biol. 2008;28:619–629. doi: 10.1128/MCB.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: Dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes & Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Hashemi SH, Anderson SK, Huang Y, Moreira MC, Lynch DR, Glass IA, Chance PF, Bennett CL. Senataxin, the yeast Sen1p orthologue: Characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol Dis. 2006;23:97–108. doi: 10.1016/j.nbd.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes & Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N, Levine BJ, Sprecher C, Skoultchi AI, Marzluff WF. Expression of mouse histone genes: Transcription into 3′ intergenic DNA and cryptic processing sites downstream from the 3′ end of the H3 gene. Mol Cell Biol. 1987;7:1039–1047. doi: 10.1128/mcb.7.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy N, Pandey NB, Marzluff WF. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol Cell Biol. 1991;11:497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley JL. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell. 1989;57:561–571. doi: 10.1016/0092-8674(89)90126-8. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annual Review of Biophysics. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- Cuello P, Boyd DC, Dye MJ, Proudfoot NJ, Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 1999;18:2867–2877. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Allen MA, Larsen A, Macmorris M, Han M, Blumenthal T. Genes involved in pre-mRNA 3′-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc Natl Acad Sci. 2008;105:16665–16670. doi: 10.1073/pnas.0807104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae . Proc Natl Acad Sci. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar HE, Lund E, Dahlberg JE. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur S, Vo LTA, Voisinet-Hakil F, Minet M, Schmitter JM, Lacroute F, Wyers F, Minvielle-Sebastia L. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 2003;22:2831–2840. doi: 10.1093/emboj/cdg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Aasland R, Keller W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA. 2004;10:965–977. doi: 10.1261/rna.7090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Erkmann JA, Yang X, Sanchez R, Marzluff WF. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005a;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol. 2005b;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem Soc Trans. 2008;36:590–594. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- Elela SA, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes & Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P, Levitt N, Briggs D, Proudfoot NJ. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991;10:1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers R, Grummt I. Molecular coevolution of mammalian ribosomal gene terminator sequences and the transcription termination factor TTF-I. Proc Natl Acad Sci. 1995;92:5827–5831. doi: 10.1073/pnas.92.13.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers R, Smid A, Rudloff U, Lottspeich F, Grummt I. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 1995;14:1248–1256. doi: 10.1002/j.1460-2075.1995.tb07108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc Natl Acad Sci. 2005;102:18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Morlando M, Bozzoni I. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 2000;19:6218–6229. doi: 10.1093/emboj/19.22.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, Sentenac A, Muller CW, Schoehn G. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol Cell. 2007;25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Ferrari R, Rivetti C, Acker J, Dieci G. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc Natl Acad Sci. 2004;101:13442–13447. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bentley DL. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Schneider DA, Sikes ML, Fernandez CF, Copela LA, Misra VA, Nomura M, Wolin SL, Beyer AL. Visual analysis of the yeast 5S rRNA gene transcriptome: Regulation and role of La protein. Mol Cell Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garas M, Dichtl B, Keller W. The role of the putative 3′ end processing endonuclease Ysh1p in mRNA and snoRNA synthesis. RNA. 2008;14:2671–2684. doi: 10.1261/rna.1293008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio M, Chavez S, Huertas P, Tous C, Jimeno S, Luna R, Aguilera A. Different physiological relevance of yeast THO/TREX subunits in gene expression and genome integrity. Mol Genet Genomics. 2008;279:123–132. doi: 10.1007/s00438-007-0301-6. [DOI] [PubMed] [Google Scholar]
- Gerber JK, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: Polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Ghazy M, He X, Singh BN, Hampsey M, Moore C. The essential N-terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol Cell Biol. 2009;29:2296–2307. doi: 10.1128/MCB.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Fan R. Derailing the locomotive: Transcription termination. J Biol Chem. 2008;283:661–664. doi: 10.1074/jbc.R700032200. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Steitz JA. Function of the mammalian La protein: Evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Aranda A, Proudfoot N. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae . Proc Natl Acad Sci. 2000;97:8415–8420. doi: 10.1073/pnas.140217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Grummt I, Maier U, Ohrlein A, Hassouna N, Bachellerie JP. Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell. 1985;43:801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I, Kuhn A, Bartsch I, Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986;47:901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Marzluff WF. 3′ Processing and termination of mouse histone transcripts synthesized in vitro by RNA polymerase II. Nucleic Acids Res. 1996;24:3797–3805. doi: 10.1093/nar/24.19.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Wind M, Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc Natl Acad Sci. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe . Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Gunnery S, Ma Y, Mathews MB. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J Mol Biol. 1999;286:745–757. doi: 10.1006/jmbi.1998.2518. [DOI] [PubMed] [Google Scholar]
- Hamada M, Sakulich AL, Koduru SB, Maraia RJ. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J Biol Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Prakash L, Prakash S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol Cell Biol. 2005;25:10183–10189. doi: 10.1128/MCB.25.22.10183-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Moore C. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol Cell. 2005;19:619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes & Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SL, Ryan K, Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes & Dev. 1989;3:212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- Henras AK, Bertrand E, Chanfreau G. A cotranscriptional model for 3′-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA. 2004;10:1572–1585. doi: 10.1261/rna.7750804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast. 2006;23:439–454. doi: 10.1002/yea.1369. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N. A minimal RNA polymerase III transcription system from human cells reveals positive and negative regulatory roles for CK2. Mol Cell. 2003;12:699–709. doi: 10.1016/j.molcel.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes & Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EY, Ogiwara I, Weiner AM. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol Cell Biol. 2004;24:846–855. doi: 10.1128/MCB.24.2.846-855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa P, Grummt I. Mechanism of transcription termination: PTRF interacts with the largest subunit of RNA polymerase I and dissociates paused transcription complexes from yeast and mouse. Mol Gen Genet. 1999;262:508–514. doi: 10.1007/s004380051112. [DOI] [PubMed] [Google Scholar]
- Jansa P, Mason SW, Hoffmann-Rohrer U, Grummt I. Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. EMBO J. 1998;17:2855–2864. doi: 10.1093/emboj/17.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiak AJ, Armache KJ, Martens B, Jansen RP, Cramer P. Structural biology of RNA polymerase III: Subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol Cell. 2006;23:71–81. doi: 10.1016/j.molcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Jenks MH, O'Rourke TW, Reines D. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol Cell Biol. 2008;28:3883–3893. doi: 10.1128/MCB.00380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SW, Lang WH, Reeder RH. The release element of the yeast polymerase I transcription terminator can function independently of Reb1p. Mol Cell Biol. 1995;15:5929–5936. doi: 10.1128/mcb.15.11.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SP, Warner JR. Termination of transcription of ribosomal RNA in Saccharomyces cerevisiae . Mol Cell Biochem. 1991;104:163–168. doi: 10.1007/BF00229816. [DOI] [PubMed] [Google Scholar]
- Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju QD, Morrow BE, Warner JR. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990;10:5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes & Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes & Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Choe J, Seo YS. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999;38:14697–14710. doi: 10.1021/bi991470c. [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004a;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004b;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes & Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 2008;9:1013–1018. doi: 10.1038/embor.2008.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Grummt I. 3′-End formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes & Dev. 1989;3:224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Normann A, Bartsch I, Grummt I. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 1988;7:1497–1502. doi: 10.1002/j.1460-2075.1988.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P, Reeder RH. Characterization of three sites of RNA 3′ end formation in the Xenopus ribosomal gene spacer. Cell. 1986;45:431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang WH, Reeder RH. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae . Mol Cell Biol. 1993;13:649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang WH, Reeder RH. Transcription termination of RNA polymerase I due to a T-rich element interacting with Reb1p. Proc Natl Acad Sci. 1995;92:9781–9785. doi: 10.1073/pnas.92.21.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang WH, Morrow BE, Ju Q, Warner JR, Reeder RH. A model for transcription termination by RNA polymerase I. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, Snyder M, Weissman SM. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome Res. 2008;18:1224–1237. doi: 10.1101/gr.075804.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Lutz CS. p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA. 2006;12:111–121. doi: 10.1261/rna.2213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Lin-Marq N, Clarkson SG. Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopus extracts depleted of La protein. EMBO J. 1998;17:2033–2041. doi: 10.1093/emboj/17.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J, Falck-Pedersen E, Darnell JE, Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc Natl Acad Sci. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes & Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Intine RV. Recognition of nascent RNA by the human La antigen: Conserved and divergent features of structure and function. Mol Cell Biol. 2001;21:367–379. doi: 10.1128/MCB.21.2.367-379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SW, Sander EE, Grummt I. Identification of a transcript release activity acting on ternary transcription complexes containing murine RNA polymerase I. EMBO J. 1997;16:163–172. doi: 10.1093/emboj/16.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H, Kassavetis GA, Geiduschek EP. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- Mazabraud A, Scherly D, Muller F, Rungger D, Clarkson SG. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis . J Mol Biol. 1987;195:835–845. doi: 10.1016/0022-2836(87)90488-8. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–934. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes & Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Natl Acad Sci. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes & Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- Morlando M, Greco P, Dichtl B, Fatica A, Keller W, Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol Cell Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes & Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Kazerouninia A, Martinson HG. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA. 2006;12:1534–1544. doi: 10.1261/rna.103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Martinson HG. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol. 2007;14:662–669. doi: 10.1038/nsmb1253. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli . Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- Orozco IJ, Kim SJ, Martinson HG. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J Biol Chem. 2002;277:42899–42911. doi: 10.1074/jbc.M207415200. [DOI] [PubMed] [Google Scholar]
- Osheim YN, Proudfoot NJ, Beyer AL. EM visualization of transcription by RNA polymerase II: Downstream termination requires a poly(A) signal but not transcript cleavage. Mol Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- Osheim YN, Sikes ML, Beyer AL. EM visualization of Pol II genes in Drosophila: Most genes terminate without prior 3′ end cleavage of nascent transcripts. Chromosoma. 2002;111:1–12. doi: 10.1007/s00412-002-0183-7. [DOI] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Tsao DC, Martinson HG. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol Cell Biol. 2004;24:4092–4103. doi: 10.1128/MCB.24.10.4092-4103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1–RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleiderer C, Smid A, Bartsch I, Grummt I. An undecamer DNA sequence directs termination of human ribosomal gene transcription. Nucleic Acids Res. 1990;18:4727–4736. doi: 10.1093/nar/18.16.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant KE, Dye MJ, Lafaille C, Proudfoot NJ. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Gγ-globin gene. Mol Cell Biol. 2005;25:3276–3285. doi: 10.1128/MCB.25.8.3276-3285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Osheim YN, Jones HS, Alen CM, Roan JG, Reeder RH, Beyer AL, Proudfoot NJ. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc Natl Acad Sci. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rasmussen TP, Culbertson MR. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae . Mol Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- Reeder RH, Guevara P, Roan JG. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol Cell Biol. 1999;19:7369–7376. doi: 10.1128/mcb.19.11.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Kiss T. Integrating snoRNP assembly with mRNA biogenesis. EMBO Rep. 2006;7:590–592. doi: 10.1038/sj.embor.7400715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Mischo HE, Proudfoot NJ. Terminating transcription in yeast: Whether to be a ‘nerd’ or a ‘rat.’. Nat Struct Mol Biol. 2008;15:775–776. doi: 10.1038/nsmb0808-775. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Manley JL. From transcription to mRNA: PAF provides a new path. Mol Cell. 2005;20:167–168. doi: 10.1016/j.molcel.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Kaneko S, Manley JL. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Ng SB, Cloute JP, Brow DA. Cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol. 2006a;26:2688–2696. doi: 10.1128/MCB.26.7.2688-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006b;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Suraweera A, Becherel OJ, Chen P, Rundle N, Woods R, Nakamura J, Gatei M, Criscuolo C, Filla A, Chessa L, et al. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J Cell Biol. 2007;177:969–979. doi: 10.1083/jcb.200701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne IA, Meyer CA, Liu XS, Silver PA, Brodsky AS. Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 2006;16:912–921. doi: 10.1101/gr.5211806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes & Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A, Tahiri-Alaoui A, West S, Thomas B, Ramadass A, Martianov I, Dye M, James W, Proudfoot NJ, Akoulitchev A. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: A role for the nrd1–nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae . Mol Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Uguen P, Murphy S. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 2003;22:4544–4554. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, DeMarini DJ, Culbertson MR. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol Gen Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Chinchilla K, Finkel JS, Culbertson MR. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004;32:2441–2452. doi: 10.1093/nar/gkh561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sande CA, Kulkens T, Kramer AB, de Wijs IJ, van Heerikhuizen H, Klootwijk J, Planta RJ. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989;17:9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008a;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008b;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes & Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TN, Wittop Koning TH, Schumperli D, Muller B. A 5′ 3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA. 1998;4:1034–1046. doi: 10.1017/s1355838298971771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Bai L, Hsieh YJ, Roeder RG. Nuclear factor 1 (NF1) affects accurate termination and multiple-round transcription by human RNA polymerase III. EMBO J. 2000;19:6823–6832. doi: 10.1093/emboj/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J. hClp1: A novel kinase revitalizes RNA metabolism. Cell Cycle. 2007;6:2133–2137. doi: 10.4161/cc.6.17.4642. [DOI] [PubMed] [Google Scholar]
- Weser S, Bachmann M, Seifart KH, Meissner W. Transcription efficiency of human polymerase III genes in vitro does not depend on the RNP-forming autoantigen La. Nucleic Acids Res. 2000;28:3935–3942. doi: 10.1093/nar/28.20.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res. 2008;36:905–914. doi: 10.1093/nar/gkm1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ. Transcriptional termination enhances protein expression in human cells. Mol Cell. 2009;33:354–364. doi: 10.1016/j.molcel.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell. 2006a;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- West S, Zaret K, Proudfoot NJ. Transcriptional termination sequences in the mouse serum albumin gene. RNA. 2006b;12:655–665. doi: 10.1261/rna.2232406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Proudfoot N. α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α 2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehall SK, Bardeleben C, Kassavetis GA. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J Biol Chem. 1994;269:2299–2306. [PubMed] [Google Scholar]
- Winey M, Culbertson MR. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae . Genetics. 1988;118:609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5′ → 3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol. 2009;29:31–42. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M, Proudfoot NJ. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- Yonaha M, Proudfoot NJ. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 2000;19:3770–3777. doi: 10.1093/emboj/19.14.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Henderson AJ, Gilmour DS. Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes & Dev. 2007;21:1609–1614. doi: 10.1101/gad.1542707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]