Abstract
A molecular beacon that incorporates components of an artificially expanded genetic information system (Aegis) in its stem is shown not to be opened by unwanted stem invasion by adventitious standard DNA; this should improve the “darkness” of the beacon in real-world applications.
The desire to detect specific DNA and RNA sequences quantitatively and with high throughput has created a demand for biomolecular recognition probes that respond with high signal-to-noise ratios, low background, and few false positives in complex biological environments. These specifications are needed especially for molecular beacons (MBs), which are short oligonucleotide probes that carry a fluorescent molecule on one end and a quencher on the other. These come in close contact in a hairpin structure (Fig. 1(a)), but become separated when a target oligonucleotide analyte binds to their loop regions.1 Once the beacon is opened, its fluor can generate a fluorescence signal essentially free of quenching.
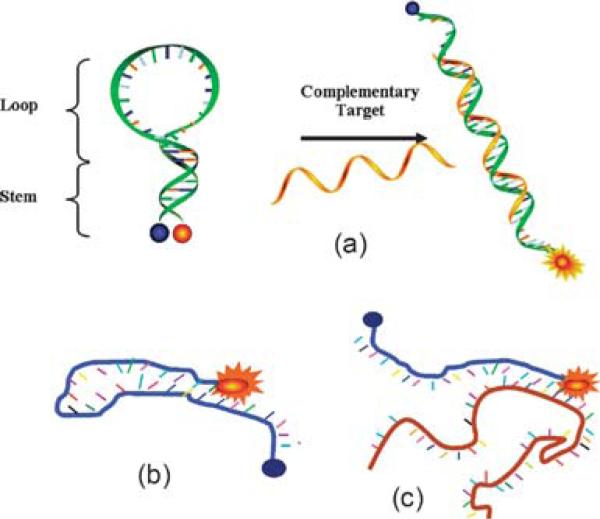
Fig. 1.
Molecular beacon design and interactions. (a) MB binding to complementary target; (b) MB mis-folds into non-hairpin structure through the hybridization of loop sequence with stem sequence; (c) MB opened by adventitious DNA that binds to MB stem.
Background fluorescence arises in MBs from various sources. First, quenching is never perfect, even when the fluor and quencher are in close proximity. Further, a class of false positives may arise when the beacon is digested with nucleases,2a or opened by binding to a protein.2b These issues may be managed using nuclease-resistant backbones (e.g. phosphorothioates or 2′-O-methyl RNA sugars),3a,b or by using sugars not recognized by DNA- and RNA-binding proteins (e.g. PNA,4a,b LNA,4c or HOMO DNA4d).
Another source of false positives arises when the MB has access in thermodynamic equilibrium to a different, non-hairpin intramolecular structure that pairs one of the stem segments with a segment in the loop (a conformational switch) (Fig. 1(b)); this generates background to the extent that the alternative conformation is present at equilibrium, and the distance is long between the fluor and the quencher in that conformation.5 While this might be managed by careful design of the beacon structure, rules that guide this design are imperfect, nearly always requiring a “winnowing” of alternative designs. False positives also arise if the MB is opened by binding to a standard oligonucleotide not in its loop region, but rather in its stem region (with perhaps some binding in the loop as well) (Fig. 1(c)). While this problem may be overcome using l-DNA in the stem,6 l-DNA phosphoramidites (Chem-Genes Corporation, Wilmington, MA) are quite expensive.
Accordingly, we examined an alternative approach to design MBs that relies less on design. This approach relies on an artificially expanded genetic information system (Aegis) (Fig. 2), which was developed some time ago in these laboratories.7 Pairing between Aegis components in a duplex retains an overall Watson–Crick geometry, and involves three hydrogen bonds. Aegis pairs therefore contribute to duplex stability to the same (or greater) extent as standard pairs. Because of non-standard arrangement of their hydrogen bonding functionality, however, Aegis components should not bind to standard nucleotides. Therefore, placing Aegis components into the stem of a MB should eliminate any opening of a beacon by adventitious natural DNA.
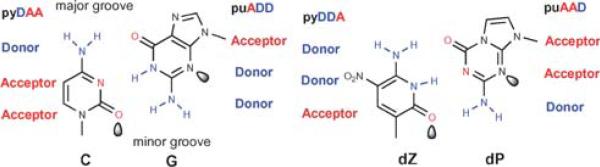
Fig. 2.
One example of an “artificially expanded genetic information system” (Aegis). Nucleobase pairs in this system have a Watson–Crick geometry, with large purines or purine analogs (indicated by “pu”) pairing with small pyrimidines or pyrimidine analogs (indicated by “py”) joined by hydrogen bonds. The hydrogen-bonding acceptor (A) and donor (D) groups are listed from the major to the minor groove as indicated. Lobes represent electron density in the minor groove. The nucleotides implementing the pyDDA:puAAD hydrogen bonding pattern (dZ:dP), the topic of this paper, are at the right.
To test this concept, we exploited 6-amino-5-nitro-3-(1′-beta-d-2′-deoxyribofuranosyl)-2(1H)-pyridone (dZ), and its complement 2-amino-8-(1′-beta-d-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (dP) as the Aegis pair.8 This has advantages with respect to chemical stability, enzymatic compatibility, and cost over another Aegis pair, 5-methylisocytidine (disoMeC) and isoguanosine (disoG), widely used today in FDA approved diagnostic assays that help manage the care of some 400 000 patients infected with HIV, hepatitis B and hepatitis C viruses.9 Hybridization of the dZ:dP Aegis pair is stronger than the dC:dG pair, and comparable to that of the disoMeC:disoG pair.8 In addition, the dZ:dP Aegis pair effectively discriminates against mismatches in short duplex DNA.8 Furthermore, the dZ:dP base pair is readily accepted by many native DNA polymerases.10 All these features make the dZ:dP pair a greater Aegis pair for future applications in developing advanced diagnostic tools.
To implement and benchmark this approach, four Aegis-containing MBs and a control (ZPMB-1, ZPMB-2C, ZPMB-2S, ZPMB-3 and Control MB, Table 1) were prepared by chemical synthesis from the appropriate phosphoramidites.8 Each MB sequence in the series was analogous to the previous, but had an additional dC:dG pair in its stem replaced by a dZ:dP pair. Standard conditions for automated DNA synthesis were used, except that extended coupling times were used for the dZ and dP nucleoside phosphoramidites (6 min) and the 6-fluorescein phosphoramidite (15 min). For deprotection, the modified MBs were first treated with DBU (1 M in acetonitrile, 12 h, room temperature) to remove the protecting group on dZ (p-nitrophenethyl (NPE)), then, incubated in NH3(aq) (29%) at 55 °C overnight. All molecular beacons were purified by preparative reverse-phase HPLC and their purities were checked by analytical ion-exchange HPLC.
Table 1.
Sequences of MBs and targets used in this study
| Name | Sequence |
|---|---|
| Control MB | 5'-FAM-CCTAGCTCTAAATCACTATGGTCGCGCTAGG-Dabcyl-3' |
| ZPMB-1 | 5'-FAM-CCTAGZTCTAAATCACTATGGTCGCPCTAGG-Dabcyl-3' |
| ZPMB-2Ca | 5'-FAM-CCTAPZTCTAAATCACTATGGTCGCPZTAGG-Dabcyl-3' |
| ZPMB-2Sb | 5'-FAM-CZTAPCTCTAAATCACTATGGTCGCGZTAPG-Dabcyl-3' |
| ZPMB-3 | 5'-FAM-CZTAPZTCTAAATCACTATGGTCGCPZTAPG-Dabcyl-3' |
| Loop Target | 3'-AGATTTAGTGATACCAGCG-5' |
| 6mer Target | 3'-CGATCC-5' |
| 8mer Target | 3'-CGCGATCC-5' |
| 10mer Target | 3'-AGCGCGATCC-5' |
| 12mer Target | 3'-CCAGCGCGATCC-5' |
| ZP-10mer Target | 3'-GPATZGAGAT-5' |
| ZP-12mer Target | 3'-GPATZGAGATTT-5' |
2C Indicates that the MB has two consecutively modified nucleobases in the stem.
2S Indicates that the MB has two separately modified nucleobases in the stem. Underlined regions are stems and their targets. The presence of Z in the synthetic oligonucleotides was confirmed by UV absorbance at 390 nm.
The Tm for opening of each beacon was first determined and compared. All MBs modified with dZ and dP showed higher Tms than the control MB (Tm = 57.8 °C). This was consistent with our previous work showing that dZ:dP base pair is stronger than dC:dG pair in duplex DNA.8 Melting temperatures indicate that the ZPMB-2S (Tm = 68.2 °C) and ZPMB-3 (Tm = 67.8 °C) form more stable hairpins than ZPMB-1 (Tm = 60.5 °C) and ZPMB-2C (Tm = 61.3 °C), suggesting that increased substitution of dC:dG pairs by dZ:dP pairs creates stepwise more stable hairpins. By comparing the Tms of ZPMB-3 with ZPMB-2S, and the Tms of ZPMB-2C with ZPMB-1, we found that the stability added by substituting two consecutive Aegis pairs (the dinucleotide dPdZ paired with the dinucleotide dZdP) approximately equals that added by substituting just one dP:dZ pair. This was unexpected given previous studies that showed that the thermal stability of two consecutive pairs in duplex DNA (dPdP:dZdZ) is more than additive, showing a degree of context dependence with Aegis components comparable to that found in standard DNA.
The signal enhancement observed after the AEGIS-containing MBs hydridize to their complementary fully matched target (binding in the loop), and the thermal denaturation profiles of MBs bound to its loop targets were then measured. Upon hybridization with their targets, all the Aegis-containing MBs show comparable signal enhancements with the control MB (lacking Aegis-components). Further, both modified and control MBs give consistently reasonable thermal denaturation profiles.
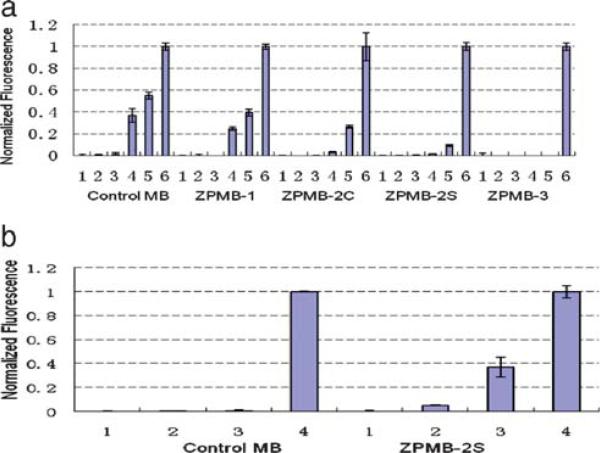
To show that MB stems containing dZ and dP resisted stem invasion and thereby reduced false-positive signals, MBs were challenged with various synthetic oligonucleotides with different lengths that had six nucleotides complementary to the stem of the MB, and a remaining sequence matched the loop region (Table 1). Fluorescence emission was recorded after separately incubating these with both modified and control MBs for 0.5 h. For the control MB, incubation with 10-fold excess of the 6mer and 8mer DNA target (Table 1) failed to open the hairpin structure (Fig. 3a, Control MB, lane 2 and 3). This is consistent with the design strategy; the intramolecular-binding constant between the stem sequences is far greater than the intermolecular interaction between one arm of the stem and the 6mer and 8mer DNA targets. On the other hand, in the presence of the 10mer and 12mer targets, one of the stem and part of loop of the control MB hybridizes with the target, disrupting the hairpin structure to give about 38% and 56% (assuming a linear fluorescence response) of opened MB, respectively (Fig. 3(a), Control MB, lane 4 and 5).
Fig. 3.
Comparison of selectivity of control MB and modified MBs. A final concentration of 50 nM MB was incubated with 500 nM of each target for 0.5 h and the fluorescence signal was measured. The experiment was repeated three times and the average value was calculated. (a) MBs (lane 1, normalized as zero); MBs incubate with the targets (6mer, 8mer, 10mer, and 12mer target, respectively, lane 2, 3, 4, and 5); MBs incubate with the loop targets (lane 6, normalized as one). (b) MBs (lane 1, normalized as zero); MBs incubate with the targets (ZP-10mer and ZP-12mer target, lane 2 and 3); MBs incubate with the loop targets (lane 4, normalized as one).
In contrast, the Aegis-containing MBs cannot be opened by any of the stem targets tested (Fig. 3(a), ZPMB-3). Indeed, results of the experiments shown in Fig. 3(a) suggested that the ability of an Aegis-containing MB to avoid the stem invasion issue is directly related to both the number of Aegis components in the stem and the melting temperature of the stem. In general, three Aegis substitutions are better than two, and two are better than one. As for ZPMB-3 and ZPMB-2S, the thermal stability of each stem is roughly the same, however, the ability of ZPMB-3 to resist stem invasion is superior to that of ZPMB-2S. The selectivity of hybridization is presumably due to the orthogonality of Aegis pairing, which is directly related to the number of Aegis-components in the stem. On the other hand, with two substitutions, the modified MB with a higher Tm, ZPMB-2S, generates a better signal-tto-noise ratio than the one with lower Tm, ZPMB-2C.
To show that the modified MBs eliminate stem invasion mainly because of the orthogonality of Aegis pairing, and not because of the higher Tm associated with dZ:dP pairs in the stem, two sequences were designed to have complementarity to one stem in the Aegis-containing MBs (Table 1). Specifically, the ZP-10mer target and ZP-12mer target, replaced one dC and one dG with one dZ and one dP, respectively. As shown in Fig. 3(b), the control MB is not opened by either the ZP-10mer or the ZP-12mer target (lane 2 or 3). However, the ZPMB-2S beacon is partially opened by ZP-12mer target (land 3), even though the Tm of the modified MB is about 10 °C higher than that of the control MB.
In conclusion, we have shown that the use of Aegis-components in the stems of MB can eliminate unwanted stem invasion by adventitious DNA. The selectivity of the modified MBs arises mainly from the orthogonality of dZ:dP pair over natural base pairs. The stability of the hairpin stem increase stepwise 3.2 °C in average, after an additional dC:dG pair in the stem replaced by a dZ:dP pair. This should further mitigate any background by alternative beacon conformations involving stem–loop interactions.
Acknowledgments
This work was supported by the National Institutes of Health Grants R01GM081527 (S. A. B.) and GM66137 (W. T.).
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Notes and references
- 1.Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.a Li J, Fang XH, Schuster SM, Tan WH. Angew. Chem., Int. Ed. 2000;39:1049. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]; b Tan WH, Fang XH, Li J, Liu XJ. Chem.–Eur. J. 2000;6:1107. [Google Scholar]
- 3.a Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. Nucleic Acids Res. 2005;33:1902. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Molenaar C, Marras SA, Slats JCM, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Nucleic Acids Res. 2001;29:e89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Seitz O. Angew. Chem., Int. Ed. 2000;39:3249. doi: 10.1002/1521-3773(20000915)39:18<3249::aid-anie3249>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]; b Xi CW, Balberg M, Boppart SA, Raskin L. Appl. Environ. Microbiol. 2003;69:5673. doi: 10.1128/AEM.69.9.5673-5678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH. J. Am. Chem. Soc. 2005;127:15664. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]; d Crey-Desbiolles C, Ahn DR, Leumann CJ. Nucleic Acids Res. 2005;33:e77. doi: 10.1093/nar/gni076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santangelo PJ, Nix B, Tsourkas A, Bao G. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y, Yang CYJ, Tan WH. Nucleic Acids Res. 2007;35:7279. doi: 10.1093/nar/gkm771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benner SA. Acc. Chem. Res. 2004;37:784. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Hutter D, Sheng P, Sismour AM, Benner SA. Nucleic Acids Res. 2006;34:6095. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbeik T, Surtihadi J, Destree M, Gorlin J, Holodniy M, Jortani SA, Kuramoto K, Ng V, Valdes R, Valsamakis A, Terrault NA. J. Clin. Microbiol. 2004;42:563. doi: 10.1128/JCM.42.2.563-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]