Abstract
Background
Recent studies have suggested that Cytotoxic T lymphocytes (CTL) play a key role in eliminating hepatitis B virus (HBV).
Objectives
We aimed to investigate the role of mutations in different immune epitopes of hepatitis B core antigen (HBcAg) among Iranians with HBeAg negative chronic hepatitis B (e-CHB), and asymptomatic carriers (ASCs).
Study design
Amino acids 1-150 of HBcAg were characterized for HBV strains from 29 e-CHB patients and 48 ASCs from Iran. All patients were infected with HBV genotype D and had previously been investigated for the presence of pre-core and basic core promoter (BCP) mutants.
Results
Amino acid mutations of core protein were observed more frequently in HBV strains from ASCs than e-CHB patients (p=0.014). Asn67 mutation was mutually exclusive to the combination Ile66 and Ser69 (P<0.001). Substitutions for Ser21 and Thr12Ser were associated with lower serum levels of HBV DNA (p<0.001). None of the patients with mutations in HLA-A2 CTL epitope, 18-27, had serum HBV DNA more than 105 copies/mL (p<0.001). By multivariate analysis, high level (> 105 copies/mL) of serum HBV DNA was inversely associated with the presence of mutations in CTL epitopes of HBc (OR:0.11 , p=0.015), while it was directly associated with presence of promoter double T1762A1764 mutations together with G1757 (OR:16.87, p=0.004)
Conclusion
The inverse correlation between serum levels of HBV DNA and CTL escape mutations of the core protein in HBeAg seroconverted patients, supports the notion that selection of CTL escape mutations consolidates the persistence of HBV infection despite reducing viral fitness.
Keywords: Hepatitis B, Tc, Immune, HBcAg, Iran, viral load
BACKGROUND
More than 300 million individuals worldwide are chronically infected with hepatitis B virus (HBV). Hepatitis B e antigen (HBeAg) negative chronic hepatitis B represents the predominant form of chronic hepatitis B (CHB) in several parts of world including Iran. This form of CHB, defined as HBeAg negative CHB (e-CHB) 1, is mostly associated with mutations in the basal core promoter (BCP) and pre-core (preC) regions of HBV. The HBV genome has four open reading frames, one of which is the core gene encoding the 183 or 185 amino acid (aa) long nucleocapsid, or core (HBc) protein which is preceded by the BCP and preC regions 2.
The major B-cell epitopes of HBV are localized around the most protruding HBc region (aa 71–87 3 involving the tip of the spike [residues 76-82]) 4, 5. The other B-cell epitope lies around aa 129–132 5, 6. Another epitope, corresponding to residues 107-118, was also suggested to be a B-cell epitope, although it does not seem to be on the surface of the complete virion 7.
HLA-class-II-restricted, T helper cell epitopes of the HBc protein have been mapped to peptides corresponding to aa positions 1–20, 28–47, 50–69, 72–90, 81–105, 90–99, 108–122, 111–125, 117–131, 120–139, 126–146, and 141–165 6, 8-11.
Special attention has been devoted to the search for HLA class I-restricted Cytotoxic T lymphocyte (CTL) epitopes within the HBc molecule. A single HLA-A2-restricted epitope HBc aa (18–27) has been identified, containing the predicted HLA-A2 binding motif with Leu at position 2 and Val at the C-terminus 8, 12
Although, the immunological basis for HBV persistence is not fully understood, CTLs are known as the main actors of the immune system in viral clearance 13. It is well known that the CTL response is less vigorous in chronically infected patients 11, and a robust T cell response against residues 18-27 of HBc protein 13 has been observed in patients who spontaneously clear the infection, but not in patients with CHB 8, 14, 15-16.However, the differences in immune responses among subgroups of patients with CHB patient have been less thoroughly investigated.
OBJECTIVES
In a recent study, we characterized the BCP and preC regions of HBV isolates of Iranian patients with e-CHB, and asymptomatic carriers (ASCs) 17. The present study investigates the prevalence of mutations in different HBc immune epitopes of HBV strains in the previously studied Iranian patients and its association to serum levels of HBV DNA, aminotransferase levels, and also BCP and preC mutational patterns.
STUDY DESIGN
Patients
The study included 97 patients negative for HBeAg, and positive for anti-HBe referred to the Hepatology Clinic of Taleghani General Hospital, Tehran, Iran between March 2000 and April 2003. Based on clinical, biochemistry tests, and imaging findings, the patients were divided into two groups; Thirty-five patients were categorized as e-CHB while 62 were classified as ASCs. All patients were enrolled in the study after giving informed consent. Clinical characteristics of patients were previously described 17, and all patients were shown to be infected with HBV genotype D 17.
Methods
For core gene sequencing, HBV DNA was extracted from 200 μL of serum with QIAamp DNA mini kit (Qiagen Inc, Valencia, CA). Amplification was carried out with the primers hepA and hep66, and was nested with the primers hepA and hep68 18. The PCR amplicons were purified with GFX PCR DNA and gel band purification kit (Amersham, Uppsala, Sweden). Purified products were used as templates in the sequencing reaction using the dideoxynucleotide chain termination method with ABI PRISM Big dye TM terminator cycle sequencing reaction kit (Applied Biosystems, a Division of Perkin Elmer, Version 3) and the primers used in the PCR were used as sequencing primers. All amplificates were sequenced bi-directionally. The ABI PRISM 3100 Genetic Analyser (Applied Biosystems) was used for electrophoresis and data collection. The sequences obtained were edited using the SeqMan program in the LASERGENE package (DNA STAR Inc., Madison, WI). Three-dimensional predictions of the mutated HBc structures were performed on the basis of the X-ray structure of the HBc, genotype A 3, by a comparative modeling program 3D-JIGSAW 19 (http://bmm.cancerresearchuk.org/~3djigsaw/) and presented by Chimera software 20.
Statistical analysis
Statistical analysis was performed with Chi-square and Fisher’s exact tests for the absence or presence of mutations within different regions. Independent t test was used to compare the average mutation numbers within the same regions (SPSS Inc. Chicago, IL). Pearson’s correlation coefficient (cc) was used to calculate the correlation between each pair of regional mutations. We employed a multivariate logistic regression analysis in order to find the association between the serum HBV DNA, and a set of epitope mutations as independent variables. Correlation and regression analyses were done with the aid of Stata 11 (StataCorp, USA).
RESULTS
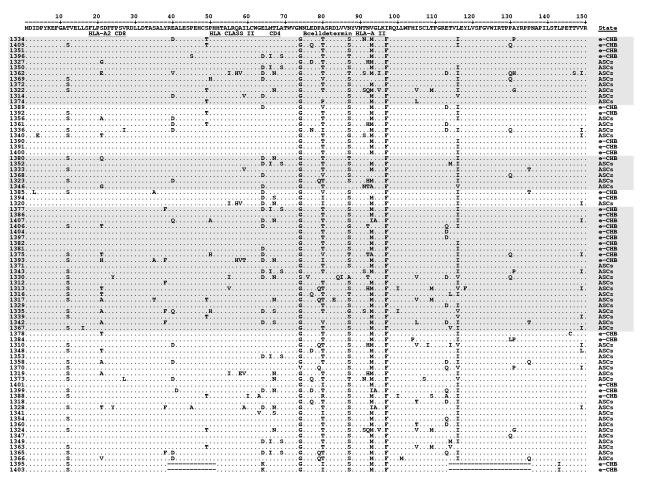
The core region of the infecting strain could be amplified from serum of 77 (79%) patients. Two e-CHB patients had strains with major deletions in core region and were excluded from statistical comparison. Sequencing data between amino acids 1-150 of core region in 48 ASCs and 27 e-CHB patients are presented in Fig 1. Data regarding HBV DNA and serum aminotransferase levels, preC, and core preC mutations are also available.17
Fig 1. Amino acid sequences of the HBV core protein (aa 1-150).
Sequences of HBV core protein of patients aligned with the genotype A sequence pHBV3200 21. The grouping of the shaded sequences corresponds to strains with different BCP mutants previously published. 17
Amino acid divergence in different regions of HBc protein
Ser21 and Thr80 were the most variable amino acid residues and could be substituted by seven and five different amino acids, respectively. Substitutions of Ser87 were found to be associated with substitutions of the Ser21 in most cases (p<0.002), while the latter were associated with Thr12 (p<0.02). Substitutions of Ser87 were also associated with Glu64 in most cases (p<0.01), while substitutions of Met93 were always accompanied with Asp64 (p<0.001). Asp64 was never accompanied with Thr49 (p<0.01), and was more frequently associated with substitutions of Thr67 (p=0.010), Tyr38 (p=0.010), and Thr80 (p=0.018). Double amino acid mutations Ile66 and Ser69 were always accompanied with Glu64Asp and never accompanied with Asn67 (p<0.001).
Seven regions were found to be more variable within the HBV core protein and corresponded to residues 18-27, 35-45, 49-69, 76-87, 91-95, 105-116, and 130-135. It was also possible to localize each of these regions within respective immune epitope (Th, CTL, and B-cell; Table 1). Region 49-69, which is a Th epitope, was the most variable, followed by region 105-116 which is a B-cell epitope. There were significant positive correlations among the amino acid substitutions when comparing different regions. Specifically, correlations were found significant between regions18-27 and 91-95 (cc=0.43, p=0.0001), regions 18-27 and 76-87 (cc=0.41, p=0.0001). The Thr12Ser mutation was also found to be correlated with substitutions within CTL epitopes 18-27 (cc=0.43, p=0.0001), and 91-95 (cc=0.40, p=0.0002).
Table 1.
Frequency of immune epitope mutations in HBV isolates of patients with different clinical categories
| Clinical States | Th Epitopes | CTL Epitopes | B-Cell Epitopes | ||||
|---|---|---|---|---|---|---|---|
| Regions | 35-45 | 49-69 | 18-27 | 91-95 | 76-87 | 105-116 | 130-135 |
| e-CHB (n=27) | |||||||
| Frequency | 6 | 33 | 5 | 10 | 13 | 14 | 6 |
| Mean* | 0.22 | 1.22 | 0.19 | 0.37 | 0.48 | 0.52 | 0.22 |
| Sum (Mean) | 39 (1.44) | 15 (0.56) | 33 (1.22) | ||||
| ASCs (n=48) | |||||||
| Frequency | 18 | 67 | 19 | 27 | 34 | 51 | 15 |
| Mean | 0.38 | 1.40 | 0.40 | 0.56 | 0.71 | 1.06 | 0.31 |
| Sum (Mean) | 85 (1.77) | 46 (0.96) | 100 (2.08) | ||||
| ALL (n=75) | |||||||
| Frequency | 24 | 100 | 24 | 37 | 47 | 65 | 21 |
| Mean | 0.32 | 1.33 | 0.32 | 0.49 | 0.63 | 0.87 | 0.28 |
| Sum (Mean) | 124 (1.65) | 61 (0.81) | 133 (1.77) | ||||
Mean (of mutations) was calculated as frequency of mutation in each epitope divided by number of patients in each clinical group.
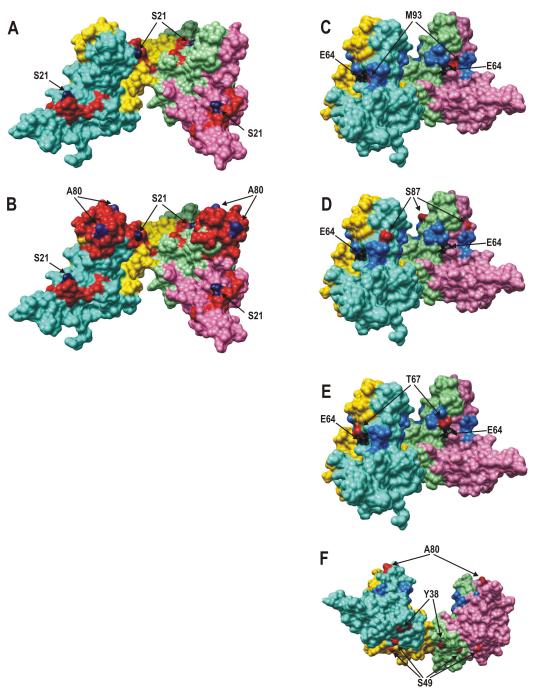
“Hot spot” amino acid exchanges were localized on the three-dimensional structure of HBc tetramers in the context of major CTL and B epitopes, and most variable HBc regions (Fig 2).
Fig 2. Appearance of major CTL and B epitopes, the most variable regions, and “hot spot” aa exchanges on the surface of the HBc tetramer.
3D data are presented from the X-ray structure of the HBc, genotype A 3 by Chimera software.20 Chains A, B, C, and D of the HBc tetramer are colored pink, light green, gold, and light blue, respectively; three different viewing angles are used for HBc drawings: (i) A and B, (ii) C, D, and E, and (iii) F. A, CTL epitope 18-FLPSDFFPSV-27 is colored red, the “hot spot” S21 is blue; B, the same as A, but the B epitope 74-NNLEDPASRDLVVNYV-89 (red) and the “hot spot” A80 (blue) are added; C, two most variable regions 64-ELMTLAT-70 and 91-TNMGL-95 are colored blue, E64 — black, and M93 — red; D, the same as C, but the S87 is colored red; E, the same as C, but the T67 is red; F — the inner part of the HBc tetramer is exposed, “hot spots” of the inner HBc surface Y38 and S49 are red, A80 is colored red to mark the tips of the HBc spikes in the new presentation angle.
The overall frequency of amino acid mutations of HBc protein is higher in strains of ASCs than e-CHB patients (4.81 vs 3.22; p=0.014). HBV isolates of ASCs also had a higher frequency of mutation in their B-cell epitopes than isolates from e-CHB patients (2.08 vs 1.22; p=0.026, Table1). This difference was most significant in region aa 105-116 (p=0.013).
Immune epitope mutations and serum levels of HBV DNA
The overall amino acid variability of HBc protein was higher in strains from patients with lower serum levels of HBV DNA (<105 copies/mL) than in strains from patients with higher HBV DNA (>105 copies/mL) (4.49 vs 3.31; p=0.023, Table2), although this difference was only statistically significant in their CTL epitopes (0.93 vs 0.38, p=0.043).This difference was highly significant in region 18-27 (0.41 vs 0.00 p<0.001, Table 2). None of the HBV infected patients with HBV DNA greater than 105 copies/mL harbored amino acid mutations in region 18-27 of their isolates (p=0.002). Patients with HBV isolates containing Ser12 mutation also had lower HBV DNA than those whose isolates lacked this (p<0.001). The difference was also significant in region 76-87 (p=0.018).
Table 2.
Frequency of immune epitope mutations in HBV isolates of patients with different serum levels of HBV DNA
| Serum HBV DNA | Th Epitopes | CTL Epitopes | B-Cell Epitopes | ||||
|---|---|---|---|---|---|---|---|
| Regions | 35-45 | 49-69 | 18-27 | 91-95 | 76-87 | 105-116 | 130-135 |
| >105 copy/ml(n=16) | |||||||
| Frequency | 3 | 24 | 0 | 6 | 5 | 10 | 5 |
| Mean | 0.19 | 1.50 | 0 | 0.38 | 0.31 | 0.63 | 0.31 |
| Sum (Mean) | 27 (1.69) | 6 (0.38) | 20 (1.25) | ||||
| <105 copy/ml(n=59) | |||||||
| Frequency | 21 | 76 | 24 | 31 | 42 | 55 | 16 |
| Mean | 0.36 | 1.29 | 0.41 | 0.53 | 0.71 | 0.93 | 0.27 |
| Sum (Mean) | 97 (1.64) | 55 (0.93) | 113 (1.92) | ||||
HBc protein and preC/BCP mutants
Substitutions of Ser21 to Thr21 or Ala21 were never accompanied with T1762A1764 core promoter double mutation (p=0.005). Strains with BCP mutations or preC mutation had higher frequency of amino acid mutations within their immune epitopes of HBc protein compared to BCP wild type isolates, respectively, although these differences were not statistically significant (Table3). It was also found that strains containing preC mutants more frequently had a Ser12 (p=0.014).
Table 3.
Frequency of immune epitope mutations in HBV isolates of patients with different categories of BCP and pre-core mutations
| Th epitopes | CTL epitpes | B-cell epitopes | ||||
|---|---|---|---|---|---|---|
| Sum | Mean | Sum | Mean | Sum | Mean | |
| BCP | ||||||
| Mutants (n=58) | 134 | 2.31 | 51 | 0.88 | 107 | 1.84 |
| Wild-type (n=11) | 6 | 0.55 | 15 | 1.36 | 14 | 1.27 |
| Pre-core | ||||||
| Mutants* (n=66) | 113 | 1.71 | 56 | 0.85 | 121 | 1.83 |
| Wild-type (n=9) | 11 | 1.22 | 5 | 0.56 | 12 | 1.33 |
These consist of isolates with A1896 together with two other isolates with other mutations within pre-core region, one of which abolished the pre-core start codon, and the other made another stop codon in the second aa position of pre-core polypeptide.
Mutations in the context of multivariate analysis
In a multivariate analysis, with age, sex, preC mutation, core promoter mutation, and mutations within each immune epitope of HBc in the model (Table 4), the whole model was statistically significant (p=0.006), and high serum levels of HBV DNA (> 105 copies/mL) were inversely associated with presence of mutations in CTL epitopes of HBc (OR:0.11 , p=0.015), while they were positively associated with presence of promoter double T1762A1764 mutation together with G1757 (OR:16.87, p=0.004).
Table 4.
Mutivariate analysis for high levels of HBV DNA (> 105 copy/ml) as dependant variable
| Independent variable | Univariate odds ratio |
Multivariate odds ratio |
SEM | P-value | Confidence interval |
|---|---|---|---|---|---|
| Age | 1.02 | 0.98 | 0.02 | 0.048 | 0.93-1.04 |
| Sex | 1.93 | 2.86 | 2.21 | 0.172 | 0.63-12.99 |
| Pre-core mutation | 0.45 | 1.74 | 1.67 | 0.562 | 0.27-11.40 |
|
BCP mutation (G1757T1762A1764) |
5.57 | 16.87 | 16.44 | 0.004 | 2.50-113.90 |
|
Mutation of Th epitopes |
0.97 | 2.81 | 2.24 | 0.196 | 0.59-13.39 |
|
Mutation of CTL epitopes |
0.28 | 0.11 | 0.10 | 0.015 | 0.02-0.65 |
|
Mutation of B-cell epitopes |
0.43 | 0.40 | 0.32 | 0.245 | 0.08-1.90 |
DISCUSSION
The HBc protein is subjected to strong folding and self-assembly requirements in order to form the highly-symmetric icosahedric HBc particle, the observed mutations were analyzed from the structural point of view. Fig. 2 represents asymmetric unit of the HBc particle consisting of two such spikes formed by two HBc dimers consisting of four HBc molecule chains: A, B, C, and D.
The observed mutations appear predominantly on the outer surface of the HBc protein, since both “hot spot” HBc epitopes: CTL 18-27 and B-cell 74-89 are externally exposed (Fig. 2, A and B). The most variable position aa 21 within the CTL epitope is one of the most exposed positions. On the other hand, the most exposed region, HBc spike, contains position 80 of the B cell epitope which is the second most variable position. Position 21 is part of a short one-turn α-helix consisting of three aa residues which is also highly exposed on the HBc surface. Correlation of definite pairs of mutations at positions 64 and 93 (Fig. 2C), and 64 and 67 (Fig. 2E) could be explained by their close contact within the four-helix bundle. Other “hot spot” positions located on outer (Fig. 2D) or inner (Fig. 2F) surfaces of the HBc particle may influence immunological or packaging HBc properties, respectively, rather than its folding and self-assembly.
In our study, mutations were found in Th/CTL epitopes of strains of both ASCs and e-CHB patients: however, contrary to our expectation; mutations were more frequent in HBV strains from ASCs. Although longitudinal studies could be more informative to correlate between HBc mutation and outcome of chronic hepatitis B, the higher frequency of these escape mutations in ASCs than in patients with liver disease emphasizes the role of these immune escape mutations for the persistence of virus rather than their role, if any, for causing liver disease.
Residues 18-27 of the HBc protein constitute an HLA-A2 restricted CTL epitope, and mutants with substitutions of amino acid 21 in the HLA-A2 restricted CTL epitope have been suggested to act as T cell receptor antagonists leading to inhibition of the CTL response to the wild-type HBV CTL epitope 22. We found that patients having isolates with mutations within residues 18-27 never had HBV DNA exceeding 105 copies/mL. This level has been proposed 23 as a cut point for differentiating ASCs from e-CHB patients.
It was recently shown that replacing many positions within HLA-A2 CTL epitopes of HCV replicons severely decreased viral replication, and this might be the reason that these mutations are rarely seen in the nature 24. In our patients acquisition of similar mutations but with a more conservative effect in lowering viral replication, like Ser21Ala might considered as a cost to viral fitness that paid by HBV to permit survival by mitigating CTL response . Another recent study also showed HBV reactivation despite reduced viral fitness due to HBsAg immune escape mutations in an HIV-coinfected patient 25. Besides, HBV mutants that reduce viral replication rate might be compensated with other mutations that restore viral fitness like preC and BCP mutations.
In conclusion, our research corroborates the idea that selection of CTL escape mutants consolidates the persistence of HBV infection despite reducing viral fitness because these mutations are more frequently seen in the context of lower serum levels of HBV DNA. We speculate that similar mechanisms for selection of immune escape mutations occur in other chronic viral diseases as well.
ACKNOWLEDGMENT
This study was funded by scholarship from the Swedish Institute 00475/2004-2005 for Dr Hossein Sendi, grant from the Swedish Research Council: VR521-2006-2573, and also NIH grant R01-DK38825 and contracts N01-DK92326, U01-DK065193, and U01-DK065201 [to HLB]
Abbrevations
- aa
amino acid(s)
- ASCs
asymptomatic carriers
- BCP
basal core promoter
- cc
correlation coefficient
- CHB
chronic hepatitis B
- CTL
cytotoxic T lymphocyte
- e-CHB
HBeAg-negative CHB
- HBc
Hepatitis B core
- HBcAg
Hepatitis B core antigen
- HBeAg
Hepatitis B e antigen
- HBV
Hepatitis B virus
- preC
pre-core
- Th
T helper
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hadziyannis SJ. Hepatitis B e antigen negative chronic hepatitis B. From clinical recognition to pathogenesis and treatment. Vir Hep Rev. 1995;1:7–36. [Google Scholar]
- 2.Hur GM, Lee YI, Suh DJ, Lee JH, Lee YI. Gradual accumulation of mutations in precore core region of HBV in patients with chronic active hepatitis: implication of clustering changes in a small region of the HBV core region. J Med Virol. 1996;48:38–46. doi: 10.1002/(SICI)1096-9071(199601)48:1<38::AID-JMV6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 4.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sallberg M, Ruden U, Magnius LO, Harthus HP, Noah M, Wahren B. Characterisation of a linear binding site for a monoclonal antibody to hepatitis B core antigen. J Med Virol. 1991;33:248–252. doi: 10.1002/jmv.1890330407. [DOI] [PubMed] [Google Scholar]
- 6.Sallberg M, Pushko P, Berzinsh I, Bichko V, Sillekens P, Noah M, et al. Immunochemical structure of the carboxy-terminal part of hepatitis B e antigen: identification of internal and surface-exposed sequences. J Gen Virol. 1993;74:1335–1340. doi: 10.1099/0022-1317-74-7-1335. [DOI] [PubMed] [Google Scholar]
- 7.Colucci G, Beazer Y, Cantaluppi C, Tackney C. Identification of a major hepatitis B core antigen (HBcAg) determinant by using synthetic peptides and monoclonal antibodies. J Immunol. 1998;141:4376–4380. [PubMed] [Google Scholar]
- 8.Bertoletti A, Ferrari C, Fiaccadori F, Penna A, Margolskee R, Schlicht HJ, et al. HLA class I-restricted human cytotoxic T cells recognize endogenously synthesized hepatitis B virus nucleocapsid antigen. Proc Natl Acad Sci USA. 1991;88:10445–10449. doi: 10.1073/pnas.88.23.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari C, Bertoletti A, Penna A, Cavalli A, Valli A, Missale G, et al. Identification of immunodominant T cell epitopes of the hepatitis B virus necleocapsid antigen. J Clin Invest. 1991;88:214–222. doi: 10.1172/JCI115280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakita T, Kakumu S, Tsutsumi Y, Yoshioka K, Machida A, Mayumi M, et al. Gamma-interferon production in response to hepatitis B core protein and its synthetic peptides in patients with chronic hepatitis B virus infection. Digestion. 1990;47:149–155. doi: 10.1159/000200490. [DOI] [PubMed] [Google Scholar]
- 11.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Ann Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 12.Bertoletti A, Chisari FV, Penna A, Guilhot S, Galati L, Missale G, et al. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J Virol. 1993;67:2376–2380. doi: 10.1128/jvi.67.4.2376-2380.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol. 2006;87:1439–1449. doi: 10.1099/vir.0.81920-0. [DOI] [PubMed] [Google Scholar]
- 14.Missale G, Redeker A, Person J, Fowler P, Guilhot S, Schlicht HJ, et al. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J Exp Med. 1993;177:751–762. doi: 10.1084/jem.177.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penna A, Chisari FV, Bertoletti A, Missale G, Fowler P, Giuberti T, et al. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J Exp Med. 1991;174:1565–1570. doi: 10.1084/jem.174.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nayersina R, Fowler P, Guilhot S, Missale G, Cerny A, Schlicht HJ, et al. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J Immunol. 1993;150:4659–4671. [PubMed] [Google Scholar]
- 17.Sendi H, Mehrab-Mohseni M, Zali MR, Norder H, Magnius LO. T1764G1766 core promoter double mutants are restricted to Hepatitis B virus strains with an A1757 and are common in genotype D. J Gen Virol. 2005;86:2451–2458. doi: 10.1099/vir.0.81023-0. [DOI] [PubMed] [Google Scholar]
- 18.Arauz-Ruiz P, Norder H, Visona KA, Magnius LO. Genotype F prevails in HBV infected patients of hispanic origin in Central America and may carry the preC stop mutant. J Med Virol. 1997;51:305–312. [PubMed] [Google Scholar]
- 19.Bates PA, Kelley LA, MacCallum RM, Sternberg MJE. Enhancement of protein modelling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins: Structure, Function and Genetics. 2001;5(Suppl):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 20.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 21.Valenzuela P, Quiroga M, Zaldivar J, Gray P, Rutter WJ. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. In: Fields BN, Jaenisch R, Fox CF, editors. Animal Virus Genetics. Academic Press; New York: 1980. pp. 57–70. [Google Scholar]
- 22.Bertoletti A, Sette A, Chisari FV, Penna A, Levrero M, De Carli M, et al. Natural variants of cytotoxic epitopes are T cell receptor antagonists for anti-viral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 23.Lok AS, Heathcote J, Hoofnagle JH. Management of hepatitis B. summary recommendations. Gastroenterology. 2001;120:1828–1853. doi: 10.1053/gast.2001.24839. [DOI] [PubMed] [Google Scholar]
- 24.Soderholm J, Ahlen G, Kaul A, Frelin L, Alheim M, Barnfield C, et al. Relation between viral fitness and immune escape within the hepatitis C virus protease. Gut. 2006;55:266–274. doi: 10.1136/gut.2005.072231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henke-Gendo C, Amini-Bavil-Olyaee S, Challapalli D, et al. Symptomatic hepatitis B virus (HBV) reactivation despite reduced viral fitness is associated with HBV test and immune escape mutations in an HIV-coinfected Patient. J Infect Dis. 2008;198:1620–1624. doi: 10.1086/592987. [DOI] [PubMed] [Google Scholar]