Abstract
This protocol describes tissue engineering of synchronously contractile cardiac constructs by culturing cardiac cell populations on porous scaffolds (in some cases with an array of channels) and bioreactors with perfusion of culture medium (in some cases supplemented with an oxygen carrier). The overall approach is ‘biomimetic’ in nature as it tends to provide in vivo-like oxygen supply to cultured cells and thereby overcome inherent limitations of diffusional transport in conventional culture systems. In order to mimic the capillary network, cells are cultured on channeled elastomer scaffolds that are perfused with culture medium that can contain oxygen carriers. The overall protocol takes 2–4 weeks, including assembly of the perfusion systems, preparation of scaffolds, cell seeding and cultivation, and on-line and end-point assessment methods. This model is well suited for a wide range of cardiac tissue engineering applications, including the use of human stem cells, and high-fidelity models for biological research.
INTRODUCTION
Myocardial infarction results in the substantial death of cardiomyocytes (CMs) in the infarct zone followed by a vigorous inflammatory response and removal of dead cells by marrow-derived macrophages. Over the subsequent weeks to months, fibroblasts (FBs) and endothelial cells proliferate forming granulation tissue and ultimately dense collagenous scar. Formation of the scar tissue reduces contractile function of the myocardium and leads to a pathological remodeling process that includes ventricular wall thinning, dilatation and ultimately heart failure, which affects >500,000 patients in the United States each year1. Tissue engineering of functional cardiac patches has a potential to provide replacement for damaged or diseased myocardium and ultimately lead to a novel treatment option.
Tissue engineering generally involves the presence of reparative cells (the actual ‘tissue engineers’) the use of biomaterial scaffolds (designed to provide a structural and logistic template for tissue development and biodegrade at a controlled rate) and bioreactors (designed to control cellular microenvironment, facilitate mass transport to the cells and provide the necessary biochemical and physical regulatory signals). Three-dimensional cardiac tissue constructs that express structural and physiological features characteristic of native cardiac muscle have been engineered in vitro using fetal or neonatal rat cardiac myocytes (CMs) on collagen fibers2, fibrous polyglycolic acid scaffolds3-7 and porous collagen scaffolds8-10.
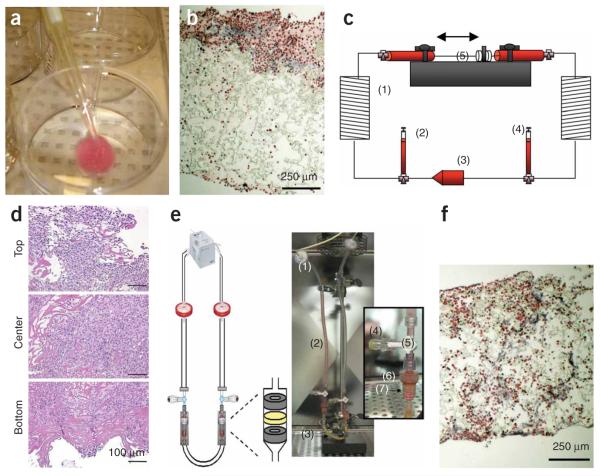
In early studies, cells were seeded onto scaffolds and cultivated in dishes4,7,8,11, in spinner flasks3,4,7 or in rotating vessels2,4,7. Inallof these systems, oxygen dissolved in culture medium was transported to the cells by molecular diffusion. Whereas human cardiac muscle is ~1 cm thick, diffusion alone can support only four to seven cell layers, that is, a 100-μm thick layer of viable and compact tissue3,4,7,12. We recently measured oxygen gradients in statically grown cardiac constructs and correlated them with the decrease in cell viability and density13 (Fig. 1, panels a–c). To overcome oxygen diffusional limitations during tissue culture, we employed perfusion bioreactors with interstitial medium flow (using porous collagen sponge scaffolds14) or flow through the channel array (using channeled poly(glycerol sebacate) (PGS) scaffolds15). We provide here a complete protocol for the assembly and operation of the perfusion system for engineering thick and compact cardiac tissue constructs.
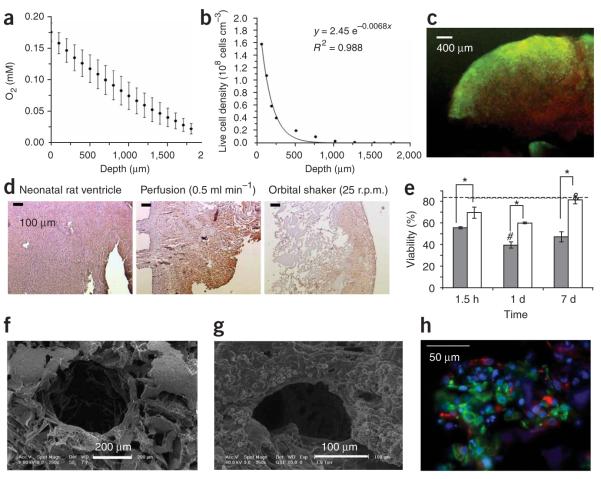
Figure 1.
Cultivation of cardiac tissue constructs. In (a–c) static and (d–h) perfusion culture. Neonatal rat cardiomyocytes after one pre-plating were seeded on collagen sponge scaffolds (~108 cells cm−3). (a) Owing to the high metabolic demand for oxygen and diffusion as the main mode of mass transfer within the interior of the scaffold dissolved oxygen concentration decreased linearly within the scaffold interior. (b) Cell density, as measured from histological cross-section, exhibited an exponential decay in the scaffold interior. (c) Live/dead staining indicated that most of the cells in the construct interior were dead (red). (d) Cardiac Troponin I staining of cardiomyocytes cultivated in collagen scaffolds in perfusion shows uniform and high cell density throughout the scaffold. (e) The viability of cells cultivated in perfusion (white bars) was significantly higher than those cultivated under static conditions (gray bars) at every time point tested. (f) A parallel channel array was laser-bored in poly(glycerol sebacate) scaffolds, seeded with cells and cultivated in perfusion in culture medium containing perfluorocarbon (PFC) oxygen carriers. (g) The channels remained open upon cultivation. (h) Double staining for cardiomyocytes (Troponin I, green) and fibroblasts (vimentin red) in constructs cultivated in the presence of PFC oxygen carriers on channeled PGS (poly (glycerol sebacate)) scaffolds. Nuceli are counterstained blue with 4,6-diamidino-2-phenylindole (DAPI).
Cultivation of cardiac tissue constructs with culture medium perfusion
Interstitial medium flow
Our group has used interstitial medium flow in conjunction with fibrous poly(glycolic acid) scaffolds5,6 and with porous collagen scaffolds14,16. In order to increase the thickness of viable tissue above ~100 μm, diffusional oxygen limitations have to be overcome during both cell seeding and tissue cultivation. The technique of cell seeding that was specifically developed for cardiac tissue engineering involves (i) rapid inoculation of cardiac cells into collagen sponges using Matrigel as a cell delivery vehicle, and (ii) transfer of inoculated scaffolds into perfused cartridges with immediate establishment of the interstitial flow of culture medium through the seeded scaffolds16,17. Forward-reverse flow was used for the initial period of 1.5–4.5 h in order to further increase the spatial uniformity of cell seeding16. Unidirectional flow of culture medium was then maintained for the duration of cultivation. In this system, cells were ‘locked’ into the scaffold during a short (10 min) gelation period, and supplied with oxygen at all times during culture.
Constructs seeded in perfusion had physiologically high and spatially uniform cell density throughout the perfused construct volume, whereas constructs seeded in dishes had most cells located in the ~100-μm thick layer at the top surface. Throughout the cultivation, the number of live cells in perfused constructs was significantly higher than in dish-grown constructs. Importantly, the final cell viability in perfused constructs (81.6 ± 3.7%) was not significantly different from the viability of the freshly isolated cells (83.8 ± 2.0) and it was markedly higher than the cell viability in dish-grown constructs (47.4 ± 7.8%)18 (Fig. 1). Consistently, the molar ratio of lactate produced to glucose consumed (L/G) was ~1 for perfused constructs, indicating aerobic cell metabolism. In dishes, L/G increased progressively from 1 to ~2, indicating a transition to anaerobic cell metabolism. Cell damage, assessed by monitoring the activity of lactate dehydrogenase (LDH) in culture medium, was at all time points significantly lower in perfusion than in dish cultures. Cells expressing cardiac-specific differentiation markers (sarcomeric α-actin, sarcomeric tropomyosin, cardiac troponin I, Fig. 1d) were present throughout the perfused constructs and only within a ~100-μm thick surface layer in dish-grown constructs. In response to electrical stimulation, perfused constructs contracted synchronously, had lower excitation thresholds (ETs) and recovered their baseline function levels following treatment with a gap junction blocker; dish-grown constructs exhibited arrhythmic contractile patterns and failed to recover their baseline levels.
Although interstitial medium flow enabled engineering of compact tissue that had physiologic density of viable aerobically metabolizing cells, most cells were round and mononucleated14 (Fig. 1d). This was likely due to the exposure of CMs to hydrodynamic shear, in contrast to the native heart muscle where blood is confined within the capillary bed and therefore not in direct contact with CMs. This motivated the design of scaffolds with arrays of channels that provide a separate compartment for medium flow.
Channeled scaffolds
We explored the use of an elastomer, PGS19, pretreated with cardiac FBs and seeded with neonatal rat heart cells (Fig. 1b). PGS is obtained by condensation of glycerol and sebacic acid, and formed by salt leaching into a 3D network with a desired pore size (e.g., ~100 μm), porosity (>95%) and thickness (1–5 mm). The cross-links and hydrogen bonds contribute to its unique elastic properties. PGS degrades by hydrolysis of its ester bond into glycerol (likely adsorbed in the body) and sebacic acid (secreted by urine either directly or metabolized into carboxylic acids). In vivo (5 weeks of subcutaneous implantation), PGS scaffold is biocompatible and biodegradable (linear loss of the scaffold mass to ~20% of initial over 5 weeks of culture), such that its shape and structural integrity were well maintained. The mechanical properties of PGS resemble vulcanized rubber: PGS is highly elastic and capable of up to 400% elongation before it yields.
In order to mimic the capillary network, neonatal rat heart cell populations were cultured on PGS scaffolds with a parallel array of channels made using a laser cutting/engraving system (Fig. 1f)and perfused with culture medium15. To mimic oxygen supply by hemoglobin, culture medium was supplemented by 5.4% vol/vol perfluorocarbon (PFC) emulsion (Oxygent, kindly donated by Alliance Pharmaceuticals), constructs perfused with unsupplemented culture medium served as controls. Constructs were subjected to unidirectional medium flow at a flow rate of 0.1 ml min−1 provided by a multichannel peristaltic pump (IsmaTec).
As the medium flowed through the channel array, oxygen was depleted from the aqueous phase of the culture medium by diffusion into the construct space where it was used for cell respiration. Depletion of oxygen in the aqueous phase acted as a driving force for the diffusion of dissolved oxygen from the PFC particles, thereby contributing to the maintenance of higher oxygen concentrations in the medium. Owing to the small size of PFC particles, molecular diffusion of dissolved oxygen from the PFC phase into the aqueous phase was very fast, and estimated not to be a rate-limiting step in this system. For comparison, in unsupplemented culture medium, oxygen was depleted faster because there is no oxygen carrier phase that acts as a reservoir20.
In PFC-supplemented medium, the decrease in the partial pressure of oxygen in the aqueous phase was only 50% of that in control medium (28 mmHg versus 45 mmHg between the construct inlet and outlet at the flow rate of 0.1 ml min−1). Consistently, constructs cultivated in the presence of PFC had higher amounts of DNA, troponin I and Cx-43, and significantly better contractile properties as compared to control constructs. In both groups, cells were present at the channel surfaces as well as within constructs (Fig. 1g,h). Improved constructs properties were correlated with the enhanced supply of oxygen to the cells within constructs.
Recently, we adapted the methods established by Wendt et al.21 for cell seeding onto PGS by perfusion of a cell suspension through the scaffold voids. We determined the proper flow rate to use in order to achieve a dense, uniform spatial distribution of cells throughout the scaffolds. This seeding technique does not involve the use of Matrigel; therefore, the method may serve as one of the important steps in moving toward a clinical application.
Selecting culture parameters
The two most important parameters that we attempt to control during cultivation in perfusion are oxygen supply and shear stress experienced by the cells. Overall, as the culture medium flow rate increases, so does the supply of oxygen and nutrients; however, the shear stress, which may have detrimental effects on heart cells, increases as well. In the native heart, CMs are shielded from direct contact with blood by endothelial cells. Low values of shear stress may induce phenotypic changes in cardiac cells, including elongation. However, higher values (e.g., ≥2.4 dyn cm−2 (ref. 22)) have detrimental effects on cardiac cells including cell death and apoptosis. When exposed to excessive shear stress, CMs round up and show signs of dedifferentiation5,6,23-25.
Recent studies with neonatal CMs cultivated in the porous alginate scaffolds indicate that the application of interstitial flow at shear stresses >2.4 dyn cm−2 resulted in p38 activation and initiation of apoptosis22. Importantly, the shear stress in this study was maintained <2.4 dyn cm−2. Perfusion of the macroporous scaffolds at low shear stresses and low average velocities had beneficial effects in many other tissue engineering systems. Shear stress of up to 1 dyn cm−2 (average velocity of up to 640 μm s−1) increased deposition of mineralized matrix by marrow stromal osteoblasts of a tissue engineered bone in a dose-dependent manner26,27. Similarly, perfusion cin the range from 1 to 170 μm s−1 increased the content of DNA, glycosaminoglycans and hydroxyproline of a tissue-engineered cartilage compared to the static controls28-30. The appropriate average velocities also depend on the cell type being cultivated23,25. Although perfusion at 5–110 μm s−1 had beneficial effect on the constructs based on MC3T3-E1 immature osteoblasts-like cells, an average velocity of 560 μm s−1 significantly reduced the viability in the same system31. For CM/polyglycolic acid constructs, perfusion at 140–710 μm s−1 increased uniformity of cell distribution and expression of cardiac markers compared to static controls5,6. In our previous work, perfusion in the range of 425–1,275 μm s−1 through the CM/collagen constructs improved cell viability compared to the static controls while maintaining high cell yield (~90%)10.
The minimum flow rate in perfused cartridges is determined from the overall mass balance of oxygen supply by culture medium and consumption by the cells for the high cell density:
| (1) |
where F (cm3 min−1) is the flow rate of culture medium, Cin 220 μmol 1−1 (at 37 °C and 20% O2 as in the incubator air) and Cout = 0 μmol 1−1 are inlet and outlet oxygen concentrations in culture medium, respectively, R is the oxygen consumption rate for the given cell type and N is the cell number. This calculation assumes that all of the oxygen is consumed in a single pass. Thus, for actual operation of the perfusion bioreactor, we increase the volumetric flow rate by two to five times as a safety factor. However, when doing so, we must check the shear stress experienced by the cells. For interstitial medium flow in porous and fibrous scaffolds, we use the following approaches.
- Porous scaffolds For the purpose of shear stress estimation in porous scaffolds, we assume that the pores are cylindrical following a tortuous pathway. For example, the tortuous pathway can be of length 2H where, H is the scaffold thickness. All calculations are based on the scaffold volume experiencing the interstitial flow (i.e., based on the inner diameter of the silicone gasket) and the total surface area of the pores in that volume. The average fluid velocity, U, though each pore can be determined according to the following equation:
where Q is volumetric flow rate through the cardiac patch, V is the scaffold volume and ε is the void fraction. The following equation, based on Poiseuille flow, can be used to estimate the shear stress:(2)
where η is viscosity and Rc is the pore radius. Reynolds (Re) number is usually calculated at this point. Low Re (Re < 1) is found for the flow of blood in capillaries, and that is what we usually try to reproduce in our bioreactor system.(3) - Fibrous scaffolds In order to calculate the shear stress acting on the cells in fibrous scaffolds, Carrier et al.6 equated the average energy dissipation with the drag force per unit surface area of a polymer fiber coated with cells:
where τd is the average shear stress on the cell surface, Fd drag force and S surface area of scaffold fiber. Drag force is equal to the pressure drop across a cell-polymer construct multiplied by its cross-sectional area, A, which can be calculated as:(4)
By substitution, equation (4) becomes:(5)
where ΔP is the pressure drop across the perfused tissue. The Re within the construct was estimated using:(6) (7)
where Uo is the superficial fluid velocity within the construct, df is diameter of polymer fibers coated with cells, ρ culture medium density and μ is the culture medium viscosity. If Re <10 at all conditions, the construct could be considered as an isotropic porous medium, and Darcy’s law could be applied:(8)
where k is permeability, a property which depends on the size, concentration and arrangement of the fibers in a fibrous medium. After substituting equation (9) into equation (6) and expressing S/V as a function of the construct void fraction, ε, and fiber diameter, df, equation (6) becomes:(9)
ε can be calculated from the volume fractions of cells and polymer fibers. The volume fraction of cells can be calculated by multiplying the cell number seeded onto the construct with a volume of each cell and dividing by the total scaffold volume.(10) - Channeled scaffolds with PFC oxygen carriers We have developed a mathematical model to study oxygen distribution in channeled scaffolds perfused with PFC emulsion supplemented culture medium. By setting ø, volume fraction of PFC droplets in the culture medium to 0, this model can also be applied to calculate oxygen distribution within a scaffold perfused with pure culture medium. A steady-state mathematical model based on the standard Krogh cylinder model was developed32. The model has to be solved numerically and it enables us to determine optimal channel spacing and flow rate that will yield high oxygen concentration in the entire tissue space. The construct was divided into an array of cylindrical domains, each representing a channel surrounded with a tissue space. The radial component of the velocity vector was assumed to be negligible in the channel lumen. Owing to the low hydraulic permeability of the tissue space, no convective transport in the tissue region was assumed32. Oxygen transfer in the channel lumen at the steady state is thus carried by convection in axial direction and by diffusion in axial and radial direction:
where Dm is the oxygen diffusion coefficient in the culture medium, w is the axial velocity of the culture medium through the channel and a function of the radial position, and Cm is the oxygen concentration in the culture medium. Depending on Re and the velocity entrance length, the flow within the channels will be either fully developed or plug flow. In our case, the flow was fully developed and the following equation was used to describe the profile:(11)
Uc is the average velocity through each channel determined as:(12)
where N is the number of channels in the scaffold.(13)
In the PFC-supplemented medium, oxygen release from PFC droplets has to be included as an additional step. PFC droplets were assumed to be small with uniform oxygen concentration (Cp) near equilibrium with the oxygen concentration in the aqueous phase (Ca), such that the total oxygen concentration in the emulsion, Ctot, is:
| (14) |
where ø is the fraction of PFC droplets. If the partition coefficient is defined as K = Cp/Ca, then equation (14) can be expressed as:
| (15) |
The governing equation for oxygen transfer in the channel lumen of the constructs perfused with PFC-supplemented medium is then:
| (16) |
where Deff is the effective diffusion coefficient of oxygen in the culture medium supplemented with PFC.
In the tissue region, both axial and radial diffusion were taken into account while the oxygen consumption rate was assumed to follow Michaelis–Menten kinetics:
| (17) |
where Dt is the oxygen diffusion coefficient in the tissue space, Ct is the local oxygen concentration in the tissue space, Qmax is the maximum oxygen consumption rate and Cm is the Ct at the half-maximal consumption rate.
For boundary conditions, the inlet and outlet oxygen concentrations in the channel and the tissue regions were set equal to those measured experimentally (Cin and Cout, respectively). In case model predictions are made before the actual experiment, the outlet oxygen concentration cannot be measured and the boundary conditions are calculated as follows. The oxygen concentration was assumed to vary only within the construct, that is, at the construct outlet oxygen concentration stops varying as a function of bioreactor length:
| (18) |
It was also assumed that the culture medium at the outlet was well mixed, with no variations in the radial direction, so that the mean value in the aqueous phase at the construct outlet was set as the outlet oxygen concentration for the tissue space as:
| (19) |
Symmetry conditions were applied at the channel axis (r = 0) and at the half distance between two channel centers (r = rt). Finally, at the channel-tissue interface (r = rc), oxygen concentrations in the aqueous phase and the tissues have to be equal and the oxygen diffusion flux across the interface has to be constant. Boundary conditions are summarized in Table 1.
TABLE 1.
| Boundary conditions for the porous tissue construct of the length L, channel radius rc, and half distance between centers of two channels rt
| Channel | Tissue annulus | |
|---|---|---|
| z = 0 | 0 ≤ r < rc Ca = Cin |
0 ≤ r < rt Ct = Cin (or ) |
| z = L | 0 ≤ r < rc Ca = Cout |
0 ≤ r < rt Ct = Cout (or) |
| 0 ≤ z ≤ L |
r = 0 ∂CL/∂r = 0 |
r = rt ∂Ct/∂r = 0 |
| 0 ≤ z ≤ L r = rc |
Ca = Ct |
The model was solved using the finite element method and a commercial software package FEMLAB 2.2 (ref. 32). Supplementation of the culture medium by PFC emulsion was predicted to improve mass transport by increasing convective term and effective diffusivity of culture medium, resulting in increased total oxygen concentration (Fig. 2). PFC particles served as oxygen reservoirs, replenishing oxygen in the culture medium, as it was depleted from the aqueous phase in the channel lumen by consumption in the tissue space. The presence of PFC emulsion increased both the axial transport by increasing the apparent convective term [by (K − 1)ø] and the radial transport by increasing the effective diffusivity. However, the increase in axial transport contributed ~98% to the increase in oxygen concentration in the tissue space32.
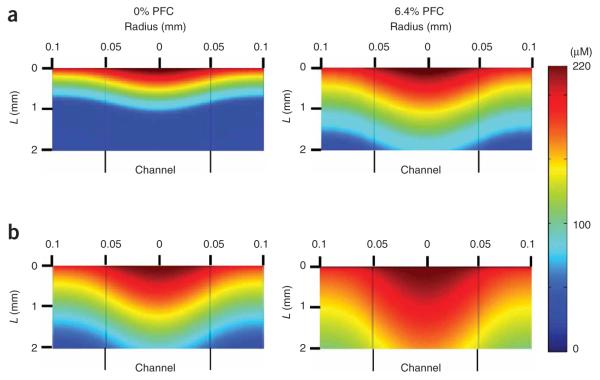
Figure 2.
Predictions of oxygen profiles (micromolar) in a channel and tissue space of a construct with cell density of 1 × 108 cell ml−1, 100-μm channel diameter and 100-μm wall-to-wall spacing. Perfused at a velocity of: (a) 490 μms−1 and (b) 1.35 mm s−1 with control medium (0% perfluorocarbon (PFC), left panels) and medium supplemented with 6.4% volume PFC emulsion (right panels); vertical lines designate channel walls. Adapted with modifications from ref. 32.
Owing to the low hydraulic permeability of the scaffold compared to the channels, the majority of the applied fluid flow goes through the channels. Thus, cells residing within the scaffold pores are shielded from the shear stress and only cells found immediately at the pore wall experience the fluid flow. The shear stress can then be calculated using equation (3), with Uc as the average culture medium velocity through the channel.
Overview of the procedure
A general outline of the procedure and the timing for all individual steps are shown as a flow diagram in Figure 3. Perfusion loop components and the perfusion chamber are prepared at least 2 d before the experiment. The components and the perfusion chamber are autoclaved and assembled under sterile conditions in the laminar flow hood. The complete loops are primed with culture media 1 d before the experiment to check for possible leaks (Fig. 4). The loops are placed in the incubator in order for the culture medium to equilibrate with the CO2 (turning the pump on is not required). Scaffold preparation occurs in parallel with the preparation of the perfusion loop. Commercially available scaffolds (collagen sponge, Ultrafoam) are punched out in the desired size 1 d before the experiment, placed in Petri dishes and incubated in culture medium for 24 h to ensure appropriate rinsing and conditioning of the scaffold. The custom-made scaffolds (such as PGS) are prepared at least 1 week before the experiment. Isolation of neonatal rat CMs requires 12–18 h. On the day of the experiment, cells are seeded onto the scaffolds using either rapid inoculation with Matrigel or cell suspension perfusion. Following cell seeding, the constructs are placed in the perfusion chamber and the entire perfusion loop is placed into the incubator for the duration of the experiment. The cell-scaffold constructs are maintained in perfusion for 3–14 d, followed by functional, biochemical and immunofluorescent evaluation as outlined below.
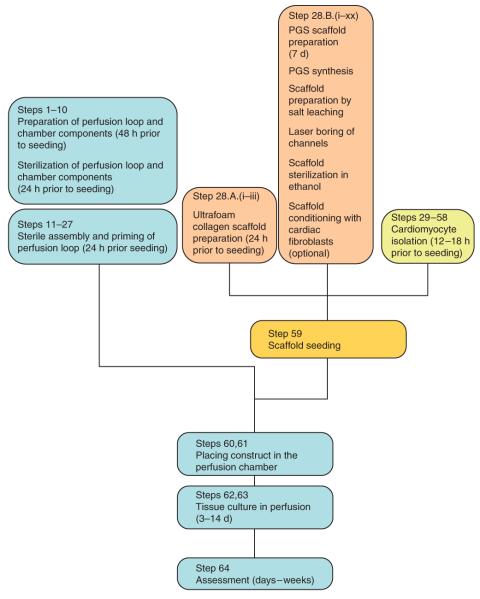
Figure 3.
Generalized protocol flowchart. The steps in this protocol can generally be divided into (i) preparation (Steps 1–58), (ii) scaffold seeding (Step 59), (iii) perfusion cultivation and (iv) construct assessment. During preparation steps, we would prepare, sterilize and assemble the perfusion loop (Steps 1–27), which can take up to 4 d. We would also prepare the appropriate scaffold (Step 28), which, depending on the scaffold type, can take up to 7 d. (Note that generalized loop preparation (Steps 1–27) and scaffold preparation (Step 28) can be performed in parallel, if desired). The cell isolation (Steps 29–58) should start ~12–18 h before scaffold seeding. It is required that Steps 1–28 be completed before cell isolation. Subsequently, the scaffolds are seeded with cells (Step 59), placed into the perfusion chamber of the generalized perfusion loop (Steps 60 and 61), and cultivated in perfusion for a desired time period (Steps 62 and 63) followed by assessment of the construct’s structural and functional properties (Step 64).
Figure 4.
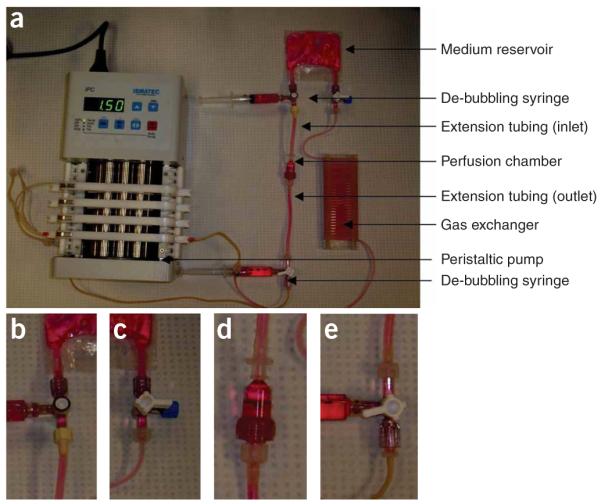
A perfusion loop for cultivation of cardiac tissue constructs. (a) Assembled perfusion loop. Luer connectors are inserted into the tubing before the assembly. (b) Connection between the medium reservoir and the inlet extension tubing of the perfusion chamber. (c) Connection between the medium reservoir and the gas exchanger. The three-way stopcock is capped with a male luer cap. (d) Connection between the perfusion chamber and the inlet/outlet extension tubing. (e) Connection between the outlet extension tubing of the perfusion chamber and the pump tubing.
MATERIALS
REAGENTS
Neonatal rat CMs and FBs ! CAUTION All studies must be conducted with an approved animal protocol from your institution and all animal experiments must comply with national regulations and US National Institutes of Health guidelines for the care and use of experimental animals.
0.25% (1 ×) Trypsin–EDTA solution in PBS (GIBCO, cat. no. 25200-072)
BioRad DC protein assay (Bio-Rad, cat. no. 500-0112)
Bleach, 5% sodium hypochlorite (wt/wt)
BSA
Calcium- and magnesium-free PBS, 10 × 0.067 M PO4 (HyClone, cat. no. SH30258.01)
Collagenase type II, (Worthington, cat. no. CLS-2)
4,6-Diamidino-2-phenylindole (DAPI) mounting medium (Vector Laboratories)
DNA standard(type I calf thymus, highly polymerized; Sigma, cat. no. D-1501)
DMEM, high glucose 4/g/ml,with L-GIn, without sodium pyruvate (GIBCO, cat. no. 11965)
Ethanol 100%, 95% and 70%
Ethidium monoazide bromide (EMA), powder (Molecular Probes)
Fluorescein-conjugated goat anti-rabbit IgG (Vector Laboratories)
Fluorescein-conjugated horse anti-mouse IgG (Vector Laboratories)
Formalin
Glycerol (Alfa Aesar, cat. no. 38988)
Hank’s balanced salt solution (HBSS), without calcium and magnesium (GIBCO, cat. no. 14170-112)
Hoescht dye 33258, powder (Polysciences)
LDH assay kit (Chiron Diagnostics)
Matrigel (Becton-Dickinson)
Ultrafoam scaffolds (Davol)
Mouse anti-β-myosin heavy chain (Chemicon)
Mouse anti-α-myosin heavy chain hybridoma supernatant (ATCC, cat. no. BA-G5)
Mouse anti-cardiac troponin I clone 23C6 (Biodesign)
Mouse anti-sarcomeric α-actin C5C (Sigma)
1 M HEPES (GIBCO, cat. no. 15630)
NaCl, powder
NaN3
NH4OH
Normal horse serum (NHS; Vector Laboratories)
Penicillin (GIBCO)
Propidium iodide (PI), powder (Molecular Probes)
Rabbit anti-Connexin-43 (Chemicon)
Reveal solution (Biocore Medical)
RNase A
Sebacic acid (>98% pure; Alfa Aesar, cat. no. L05051)
Sodium chloride (>99% pure; EMD Chemicals, cat. no. SX0420-3)
Tetrahydrofuran (THF, anhydrous, 99.9%; Fisher Scientific, cat. no. 610450010)
Texas Red–conjugated horse anti-mouse IgG (Vector Laboratories)
Tris-acid
Tris-base
Triton X-100
Trypsin, from bovine pancreas powder (Sigma, cat. no. T4665)
Tween 20
Tyrode’s salts (Sigma)
Culture medium (see REAGENT SETUP)
EQUIPMENT
30-, 10-, 5-ml syringes (Fisher, cat. no. 14-829-48, 14-823-2A, 14-823-35)
Three-way stopcock (Medex, cat. no. MX9311L)
BeadBeater (Cole-Parmer, cat. no. EW-36270-07)
Biopsy punch (10 mm; Acuderm, cat. no. P1025)
Blood gas analyzer (Model 1610;).
Caps, male and female for luer connectors
Deckloaking chamber (Biocare Medical, Concord CA)
Drill with bores of required sizes
Filters (0.45 and 0.2 μm; Millipore)
Fluorescence microscope (e.g., Olympus IX81 with ×10, ×20 and ×40 objectives and DAPI, FITC and Texas Red filter cubes)
Fluorometer
Glucose and L-lactate analyzer (Model 2300 STAT Plus; Yellow Springs Instruments)
Hemocytometer
Injection site rubber septum (Baxter, cat. no. 2N3399)
Kimwipes
Laser cutting/engraving system (model X-660; Universal Laser Systems)
Liquid nitrogen dewar
Luer connectors (McMaster-Carr)
Magnet (McMaster-Carr, cat. no. 5862K91)
Moira Spoon
Optical microscope
Perfusion chambers, polycarbonate ‘Apollo’, were kindly donated by the Advanced Tissue Sciences. (Millipore filter cartridges can be used in lieu of Apollo chambers.)
Peristaltic pump (Ismatec)
PharMed BPT tubing (Cole-Parmer, cat. nos. EW-96880-06 and EW-95809-32)
Plastic staining jars (Biocore Medical)
Platinum cured silicone tubing (1.6 mm i.d., 3.2 mm o.d.; Cole-Parmer)
Pump tubing, PharMed (Cole-Parmer, cat. no. 95706-26)
Refrigerator (2–8 °C)
Reservoir bag, 32-ml gas permeable VueLifetm bag (American Fluoroseal)
Set of USA standard testing sieves (VWR, cat. nos. 57334472 and 57334480)
Stainless steel beads 6.35 mm diameter for bead beater
Stainless steel screens, Millipore filter holders (85% open area)
Steel disk (McMaster-Carr, cat. no. 2895T56) coated with PTFE by Microsurfaces (Morris)
Steel ring (McMaster-Carr, cat. no. 97063A134), coated with PTFE by Microsurfaces
Stereomicroscope
Sterilization pouches
Syringe pump (Push/Pull; Harvard Apparatus or WPI Instruments)
Three-way stopcocks (Baxter Healthcare)
Tweezers (straight and curved)
Tygon tubing (McMaster, cat. no. 5103K13)
Ultrafoam collagen hemostat
Vacuum oven
ModFit software
REAGENTS SETUP
Culture medium
Our standard culture medium for cardiac tissue engineering is a high glucose DMEM containing 4.5 g l−1 glucose supplemented with 10% FBS, 10 mM HEPES, 2 mM L-GIn and 100 U ml−1 penicillin. In order to make this, take a 500-ml bottle of DMEM and remove 60 ml using a serological pipette. Add 50 ml of FBS. Add 5 ml of HEPES. Add 5 ml of penicillin.
PROCEDURE
Preparation and sterilization of a generalized perfusion loop
1| Measure 3 m of 1.6 mm i.d., 3.2 mm o.d. platinum-cured silicone tubing and coil around a holder to make a gas exchanger. Insert luer connectors at both ends. Loosely cap the ends with appropriate female or male caps for the luer connectors. Place the gas exchanger in a sterilization pouch.
2| Insert luer connectors at the ends of the peristaltic pump tubing. Loosely cap the ends with the appropriate female or male caps for the luer connectors. Place the tubing into a sterilization pouch.
3| Loosely cap the opening of the reservoir bag with the appropriate female or male caps for the luer connectors.
4| Autoclave the components (121 °C/2 bar) for 20 min followed by 20 min of drying.
Preparation and sterilization of perfusion chambers
5| Open the perfusion chamber and place a stainless steel screen into the upper half of the cartridge. Place one silicon gasket on top. We use polycarbonate Apollo cartridges for the perfusion chamber, although filter cartridges from Millipore can also be used.
6| Loosely close the cartridge.
7| Place 3-cm extension tubing, via luer connectors inserted into the tubing, on top of the cartridge. Place a 6-cm extension tubing, via luer connectors inserted into the tubing, at the bottom. The tubing is 3.2 mm i.d., 5 mm o.d. platinum-cured silicone (Fig. 4).
8| Loosely cap the openings of the extension tubing with the appropriate female or male caps for the luer connectors. Place the assembly into the sterilization pouch.
9| Sterilize five male and five female caps to be used in the assembly procedure.
10| Autoclave the components (121 °C/2 bar) for 20 min followed by 20 min of drying.
? TROUBLESHOOTING
Assembly and priming of the generalized perfusion loop
11| Place the sterilized pouches into the biosafety cabinet (laminar flow hood).
12| Use sterile gloves.
13| Open the pouch with the medium reservoir, remove the cap from one tubing inlet and place the three-way stopcock.
14| Open the pouch with the gas exchanger, remove the cap from one end and connect to the three-way stopcock of the reservoir bag (Fig. 4c).
15| Open the pouch with the pump tubing. Remove the cap at the other end of the gas exchanger, remove the cap at one end the pump tubing and connect to the gas exchanger.
16| Open the pouch with the perfusion chamber. Remove the cap at one end of the perfusion chamber extension tubing and place the three-way stopcock. Repeat the same for the other end of the extension tubing.
17| Connect the inlet perfusion chamber extension tubing to the reservoir bag via the three-way stopcock (Fig. 4b). If alternating medium flow perfusion seeding is being used, do not do this step.
18| Connect the outlet perfusion chamber extension tubing to the pump tubing via the three-way stopcock (Fig. 4e). If alternating medium flow perfusion seeding is being used, do not do this step.
19| Close the access to the reservoir bag using the three-way stopcock placed between the inlet to the perfusion chamber and the reservoir bag. (The dial of the three-way stopcock should be pointing toward the culture medium reservoir.)
20| Take 30-ml syringe and fill it with 30 ml of warm culture medium.
21| Place the syringe at the stopcock between the gas exchanger and the reservoir bag. Close off the access to the reservoir bag and start priming the loop. The culture medium should slowly start filling the gas exchanger, followed by the pump tubing and finally the perfusion chamber. The air will be displaced from the tubing through the stopcock placed at the inlet to the perfusion loop.
22| Upon filling the gas exchanger, pump tubing and the perfusion chamber, close off the access to the gas exchanger, as well as the access to the perfusion chamber. Dispense the remainder of the culture medium into the medium reservoir. Air can be removed from the culture medium reservoir using the syringe placed between the gas exchanger and the reservoir bag. Remove the syringe and replace with a sterile luer cap.
23| Take two 5-ml syringes and fill each with 2.5 ml of culture medium.
24| Place the syringes at the stopcock flanking the perfusion chamber.
25| If there are any bubbles remaining in the perfusion chamber, perform de-bubbling by injecting from one syringe into the downstream syringe. (Elevate the downstream syringe as the bubbles go up.)
26| Check for leaks.
? TROUBLESHOOTING
27| If there are no leaks, place the loop inside the incubator. It is not necessary to turn the pump on at this time. Leak-free perfusion loop is anticipated at the end of this section. After placing the loop into the incubator overnight, gas bubbles may appear in the tubing as well as the perfusion chamber. The gas bubbles are due to the decreased solubility of CO2 at 37 °C compared to room temperature.
? TROUBLESHOOTING
Scaffold preparation
28| Prepare collagen scaffold or a PGS scaffold as outlined below in option A (collagen scaffold) or option B (PGS scaffold).
(A) Collagen scaffold preparation
Place the sheet of Ultrafoam collagen hemostat into a 100-mm Petri dish.
Punch out the scaffolds from the sheet using a 13-mm punch.
Place the scaffolds in the 60-mm Petri dish and hydrate with 10 ml of culture medium in a 37 °C/5% CO2 incubator for 24 h. Collagen should have dry dimensions of 13 mm diameter × 3 mm thickness and wet dimensions of 11 mm diameter × 1.5 mm thickness following hydration in cultu medium for 1 h in a 37 °C incubator. This allows the discs to fit tightly into the perfusion chamber.
(B) PGS scaffold preparation
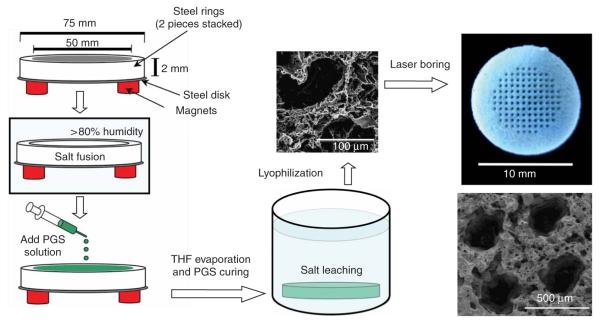
Machine a stainless steel ring and a plate of dimensions as in Figure 5. The mold, comprising Teflon-coated steel rings and a Teflon-coated steel plate, is cleaned with water, then 95% ethanol.
Allow molds to dry in air for 1 h.
Assemble the mold by stacking the polytetrafluoroethylene (PTFE)-coated steel ring on the PTFE-coated steel plate and use six strong magnets (neodymium–iron–boron disc magnet, 19 mm diameter, 6.4 mm thick, 14.3 lbs pull force) to hold the mold together33 (Fig. 5).
-
Add 8 g of salt particles of the desired size to the mold; gently tap the mold to ensure even distribution. ?
TROUBLESHOOTING
Scrape off the excess salt across the top of the mold using the side of a straight glass rod to ensure the thickness of the salt template matches that of the steel ring.
Transfer the mold into an incubator with >80% relative humidity and allow the salt to be fused into salt templates. The salt must be dried to stop salt fusion.
Remove the molds from the humidity chamber and place them in a vacuum oven. Dry the salt templates overnight at 100 mTorr and 60 °C.
Remove the molds from the vacuum oven and allow them to cool to room temperature (20–25 °C).
-
Synthesize PGS according to procedure published previously33. Add PGS into THF, vortex until the polymer dissolves to form a 7% (wt/vol) solution.
? TROUBLESHOOTING
-
Add appropriate amount of PGS solution evenly into the salt template in a fume hood. The weight ratio of PGS to salt should be 16:1.
? TROUBLESHOOTING
-
Allow the THF to evaporate for 1 h in a fume hood.
! CAUTION The fume hood should be certified and confirmed operational by your institution.
Cure the PGS in the vacuum oven for 24 h at 100 mTorr and 150 °C. Curing time and temperature can be varied to achieve softer (shorter time, lower temperature) or stiffer scaffolds.
Allow the scaffolds to cool to room temperature while in the mold.
Remove the magnets carefully to disassemble the mold and the scaffold/salt template should slide off easily.
Gently transfer the scaffold/salt template in a 3-l water bath placed on an orbital shaker. Set the shaking speed at 20–30 r.p.m. and leach the salt for 3 d at room temperature with complete water exchange once every 12 h.
After salt leaching, place the scaffolds on PTFE-coated aluminum sheet and transfer into a lyophilization flask, freeze at 20 °C, lyophilize, and store at 20 °C.
Bore parallel channels (e.g., 250 μm diameter and 250 μm spacing wall to wall) in a square array using a 120 W CO2 laser cutting/engraving system (model X-660; Universal Laser Systems), if desired.
Cut the scaffold into desired size and shape and sterilize the scaffolds by autoclaving at 121 °C/2 bar for 20 min.
Soak scaffolds in 70%, 50%, 25% ethanol and sterilized PBS (cell culture grade) each for 30 min with gentle shaking on an orbital shaker (20 r.p.m.).
Wash the scaffolds twice with culture medium and condition in cell culture medium overnight before cell seeding.
Figure 5.
Scaffold fabrication. The scaffolds were fabricated using a modified salt fusion-salt leaching method. The mold is held by magnets only for easy assembly and disassembly and to present a flush top surface to produce scaffolds of uniform thickness. The lyophilized scaffolds were bored using 120 W CO2 laser to generate a regular array of microchannels. Not drawn to scale.
Isolation of cells from heart ventricles
29| Put ice into a Styrofoam container and place the container into a biosafety cabinet. Place 60-mm Petri dishes on ice. Fill each dish with 10 ml of cold HBSS.
30| Upon isolation, place the ventricles in the 50-ml conical tube containing 25 ml of cold (2–8 °C) HBSS, and placed on ice.
! CAUTION All studies must be conducted with an approved animal protocol from your institution and all animal experiments must comply with national regulations. Cells should be obtained from ventricles of 2-d-old Sprague-Dawley rats. The volumes in the procedure are adjusted for isolation of cells from 10 to 20 hearts.
31| Prepare a solution of trypsin in HBSS (25 ml, 0.6 mg ml−1). Place the solution into a 100-ml plastic bottle on ice.
32| Using a 25-ml pipette, remove the HBSS from the conical tube.
! CAUTION Do not use vacuum aspiration because ventricles or cells may accidentally be aspirated into the vacuum line.
33| Add 25 ml of cold HBSS to the conical tube with ventricles and pipette the solution up and down to rinse the ventricles from blood. Repeat the procedure until the solution in the conical tube appears clear.
34| Using a 25-ml pipette, remove the HBSS from the conical tube. The ventricles should settle on the bottom of the tube. Open the lid on one of the Petri dishes placed on the ice. Transfer the ventricles from the conical tube into the Petri dish by slowly turning the conical tube upside down over the Petri dish.
35| Using sterile tweezers and scissors, remove any blood vessels and parts of atria that may be visible at the ventricle.
36| Using a Moira spoon, transfer the ventricles to the second Petri dish.
37| Using sterile forceps, hold the ventricles in place and cut them into equal pieces ~3 mm × 3 mm. Make sure to keep the ventricles submerged in the HBSS while cutting.
38| Using a Moira spoon, transfer the ventricles into the third Petri dish. Wash the pieces of the tissue by swirling them in the Petri dish.
39| Using a 25-ml pipette, transfer the ventricles to the plastic bottle with trypsin. This is accomplished easily by tilting the Petri dish to ~20° angle and applying the tip of the serological pipette to the region where the ventricles settled.
40| Place the bottle on an orbital shaker at 4 °C. Agitate overnight at 50 r.p.m.
41| Apply 6% bleach to the Petri dishes and conical tubes for 15 min. Remove the Styrofoam container and all plasticware from the biosafety cabinets.
! CAUTION You should adhere to your institutions regulations regarding the disposal of the bleached HBSS with the remaining heart parts. Wipe the surfaces of the biosafety cabinet using a paper towel and a spray bottle with 75% ethanol.
42| The next morning, place ice in a Styrofoam container and place the container in a biosafety cabinet.
43| Place four 50-ml conical tubes on ice: one containing 50 ml of cold HBSS (labeled ‘HBBS’), one containing 50 ml of collagenase type II (1 mg ml−1) solution (labeled ‘COLLAGENASE’) and two empty tubes (labeled ‘CELLS’, as cell suspension will be placed into these tubes).
44| Take the bottle with ventricles in trypsin solution and add 10 ml of culture medium to quench the action of trypsin. Place the bottle into a shaking water bath set to 37 °C and 50 r.p.m. for 4 min. In case a shaking water bath is not available, you can improvise as follows. A stand with a clamp is securely placed onto an orbital shaker. The entire setup is placed next to the regular water bath. The bottle is then fixed into the clamp and lowered into the water bath. The orbital shaker is set to 50 r.p.m.
45| Take an empty conical tube and place it into the Styrofoam container in the biosafety cabinet. This tube will serve as a ‘HELP’ tube to separate the tissues from the solution during collagenase digestions.
46| Using a 25-ml pipette, remove the tissues from the plastic bottle and transfer them into the ‘HELP’ conical tube. Remove the remaining solution from the bottle using a 50-ml serological pipette. Remove the remaining solution from the ‘HELP’ tube using a 5-ml serological pipette.
47| Using a 25-ml serological pipette, transfer 10 ml of collagenase solution from the ‘COLLAGENASE’ tube into the ‘HELP’ tube. Then, transfer the pieces of tissue together with the solution into the plastic bottle.
48| Place the bottle into the shaking water bath set at 75 r.p.m. for 4 min.
? TROUBLESHOOTING
49| Transfer the tissues from the plastic bottle into the ‘HELP’ conical tube. At this step, the solution contains CMs you are interested in collecting. Using a 5-ml pipette, transfer solution into the two conical tubes labeled ‘CELLS’. Make sure to split the volume equally between the two tubes.
50| Add 10 ml of cold HBSS (from the conical tube labeled ‘HBSS’, using 25-ml pipette). Pipette the tissues up and down gently for rinsing. Collect this solution using a 5-ml pipette and transfer it into the two conical tubes labeled ‘CELLS’. Make sure to split the volume equally between the two tubes.
51| Using a 25-ml pipette, transfer 10 ml of collagenase solution (from the ‘COLLAGENASE’ tube) into the ‘HELP’ tube. Transfer the tissues along with the solution into the plastic bottle.
52| Place the bottle into the shaking water bath set at 75 r.p.m. for 4 min.
53| Repeat Steps 49–52 four more times. The duration of second collagenase digest in the water bath can be increased to 8 min. Monitor the progression of digestion by observing the size of the tissue, as well as how cloudy the collagenase solution appears after digestion. Usually after the second cycle of collagenase digestion, the tissues are small enough to be transferred using a 10-ml pipette instead of the 25-ml pipette. Once the five cycles of the digestion are complete, the two ‘CELLS’ tubes will be full and the ‘HBSS’ and ‘COLLAGENASE’ tube will be empty.
? TROUBLESHOOTING
54| Centrifuge the ‘CELLS’ tubes at 121g for 4 min. Remove the supernatant using a 25-ml serological pipette. The red blood cells are not specifically removed during the cell isolation. Relatively low centrifugation speed ensures that most cells in the pellet are CMs and FBs.
▴ CRITICAL STEP The cell pellet is very fragile and it can easily be disturbed.
? TROUBLESHOOTING
55| Resuspend each pellet in 20 ml of culture medium. At this point, the cells can be used if enrichment for CMs is not required. If enrichment is required, proceed to the pre-plating steps described later in this protocol.
57| Place the flask into the laminar flow hood and gently tap it to remove the cells that are settled (rather than adhered). Transfer the cell suspension into a conical tube. Rinse the flask with 5 ml of culture medium. Transfer the culture medium to the conical tube. Repeat this procedure for the remaining pre-plating flasks. At this point, the cell suspension can be enumerated using a hemocytometer and trypan blue exclusion for cell viability and the cells can be used for experiments.
? TROUBLESHOOTING
58| Should further enrichment be required, repeat Steps 56 and 57 one more time (pre-plate 2) or two more times (pre-plate 2 and 3).
Cell seeding
59| There are three ways in which cells can be inoculated into scaffolds: using Matrigel (option A)16 (Fig. 6a,b); using alternating perfusion (option B) (Fig. 6c,d); or by scaffold seeding in perfusion without Matrigel inoculation (option C) (Fig. 6e,f).
Figure 6.
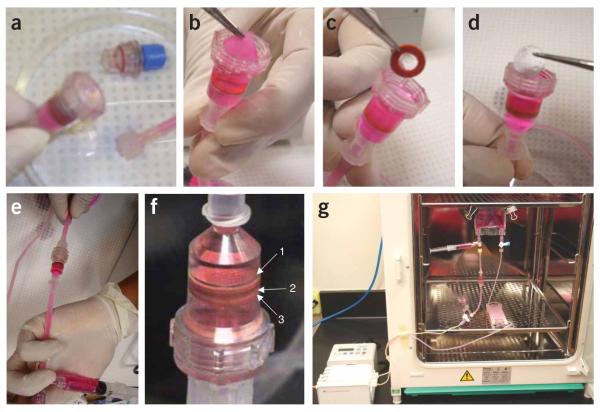
Seeding methods. (a) Cell inoculation using Matrigel. (b) H&E-stained cross-section of a poly(glycerol sebacate) (PGS) scaffold immediately following neonatal rat heart cell inoculation with Matrigel. (c) Set up for perfusion seeding using alternating culture medium flow. (1) Gas exchanger, (2,4) de-bubbling syringes, (3) perfusion chamber, (5) Syringe pump. (d) H&E-stained cross-section of a collagen scaffold inoculated with 12 million C2C12 cells using Matrigel and seeded under alternating flow perfusion at 1.5 ml min−1 for 4.5 h. (e) Perfusion loop for cell seeding consisting of a multichannel peristaltic pump, (1) filters, (2, 3) U shaped tubing, (4) injection sites, (5) three-way stopcocks, (6) Apollo perfusion chambers with tissue construct. In the perfusion chamber, the construct was squeezed between (7) two silicone gaskets to force the flow through the 5 mm diameter scaffold core. (f) H&E-stained cross-section of a construct after 2 h of perfusion seeding of cardiac cells.
(A) Inoculation using Matrigel
1d(~12 h) before cell seeding, place a vial with Matrigel on ice into a refrigerator (2–8 °C). This will enable Matrigel to defrost without gelling16.
Immediately before seeding, fill a small Styrofoam container with ice and place it into a biosafety cabinet. Place the vial with defrosted Matrigel on ice.
Place a sterile Kimwipe in a 60-mm Petri dish in a biosafety cabinet. (To sterilize the Kimwipe, fold it into quarters, place into a sterilization pouch and autoclave for 20 min at 121 °C/2 bar, followed by 20 min drying.)
-
On the basis of the desired cell seeding number, aliquot a volume of cell suspension into a 15-ml conical tube. Use one conical tube/scaffold, that is, fill the tube with the volume of cell suspension containing the exact number of cells to be seeded onto the scaffold. We usually use 12 million cells per scaffold. For a cell suspension containing 1 million cells/ml, we would use 12 ml of cell suspension/scaffold.
▴ CRITICAL STEP Do not combine the cells to be seeded on many different scaffolds into a single conical tube because you will not be able to measure the volume of a viscous cell suspension accurately in Matrigel during cell inoculation.
Centrifuge the conical tubes at 212g for 10 min.
Place the conical tubes on ice in the biosafety cabinet.
Remove the scaffold from the culture medium and gently blot dry on top of a Kimwipe. Use fast but gentle movement. The scaffold will shrink compared to its swollen volume.
Place the scaffold into a well of a 6– or 12– well plate. If folding of the scaffold occurs, gently unfold it and stretch to its original diameter.
Aspirate off the culture medium from the conical tubes. The cell pellet should be as dry as possible, that is, if there is residual culture medium, remove it immediately before inoculation by turning the conical tube gently upside down at an angle of ~150°.
-
Resuspend the cell pellet in Matrigel. Use 5 μl Matrigel/1 million cells to be inoculated, that is, 60 μl of Matrigel for 12 million cells. Instead of standard pipetting up and down, gently swirl the pipette tip inside the pellet. Pipette up and down slowly, releasing the piston on the pipette only half way.
▴ CRITICAL STEP Too vigorous pipetting at this point will introduce air bubbles that are very difficult to remove from the cell suspension and may cause the decrease in cell viability.
▴ CRITICAL STEP The cell suspension in Matrigel should be as uniform as possible, but at the same time should be bubble free. This critically determines the uniformity of initial cell distribution in the scaffold.
-
Apply the cell suspension in Matrigel into and on top of the scaffold by gently inserting the pipette tip at multiple locations at the scaffold face section. Some cell suspension may leak out. A layer of Matrigel may be visible on top of the scaffold (Fig. 6).
? TROUBLESHOOTING
Place the well plate into the 37 °C/5% CO2 incubator for 30 min.
If static or orbital mixing cultivation is required, add 4 ml of culture medium into the 6-well plate (for 12 million cells) and proceed with the cultivation. For orbital mixing seeding and cultivation, we use 25 r.p.m.
(B) Seeding of Matrigel inoculated scaffolds using alternating perfusion
Sterilize two gas exchangers (consisting of 80-cm-long coils of 1.6 mm i.d., 3.2 mm o.d. platinum-cured silicone tubing) and a perfusion chamber with extension tubing as described in Steps 1–10. Sterilize Kimwipes (Step 59A(iii)), is as well as female- and male-vented luer lock caps (Step 9)16.
Open the pouch with the perfusion chamber. Remove the cap at one end of the perfusion chamber extension tubing and place the three-way stopcock. Repeat the same for the other end of the extension tubing.
Open the pouch with the first gas exchanger, remove the cap from one end and connect to the three-way stopcock of the inlet extension tubing of the perfusion chamber.
Open the pouch with the second gas exchanger, remove the cap from one end and connect to the three-way stopcock of the outlet extension tubing of the perfusion chamber.
Remove the cap from the loose end of the first gas exchanger and connect it to a three-way stopcock. Connect the stopcock to a 10-ml syringe filled with 10.5 ml of culture medium. (To fill the syringe, dispense the culture medium into a Petri dish and apply the entire volume into the syringe.)
Repeat Step 59B(v) and connect the loose end of the second gas exchanger to the empty 10-ml syringe.
Take one 3-ml syringe filled with 3 ml of culture medium and attach it to the inlet three-way stopcock of the perfusion chamber.
Place an empty 3-ml syringe at the outlet stopcock from the perfusion chamber.
-
Hold the perfusion chamber above a Petri dish and open it.
! CAUTION The chamber is filled with culture medium, so you can expect some leaks. We chose to place scaffolds into the culture medium–filled perfusion chambers instead of into the empty ones to prevent entrapment of bubbles into the construct. That is, if the construct is placed into an empty chamber and the culture medium is subsequently applied, the displaced air tends to accumulate in the scaffolds. Place the perfusion chamber onto the sterile Petri dish slowly.
Disconnect the tubing from the perfusion chamber outlet. Cap the end of the cartridge with a sterile-vented male cap and place it onto a sterile Petri dish. Cap the free end of the outlet tubing with a sterile female cap and place it onto a sterile Petri dish (Fig. 7a).
Unscrew the bottom half of the cartridge and carefully place it into a sterile Petri dish (Fig. 7a).
-
Pick up the scaffold using sterile tweezers and place it into the top half of the perfusion chamber (Fig. 7b). Place the scaffold so that its top, covered with Matrigel, enters the chamber first. This will ensure that the culture medium flow forces the cells into the scaffold pores. Place a sterile silicone gasket over the scaffold (Fig. 7c), followed by placing of the stainless steel screen (Fig. 7d).
▴ CRITICAL STEP The cell-seeded scaffold should be placed into the perfusion chamber so that it lies perfectly flat over the stainless steel screen and the silicone gasket. If the construct is angled or folded, flow patterns will be disturbed and no perfusion through the construct interior will occur. At the same time, care must be taken to ensure that the cells are not displaced from the scaffold during this procedure by applying a minimum force.
Close the chamber.
Reconnect the outlet tubing.
-
De-bubble (Fig. 7e) if required, as described in Step 25. The perfusion chamber with the constructs should appear as in Figure 7f.
? TROUBLESHOOTING
Place the syringes onto a push/pull syringe pump.
Place the entire setup (Fig. 6c) into a 37 °C/5% CO2 incubator.
-
Program the pump to 0.5 ml min−1 or 1.5 ml min−1 with the reversal of flow direction after 2.5 ml was perfused in a given direction.
? TROUBLESHOOTING
Figure 7.
Tissue culture in a generalized perfusion loop. (a) Open perfusion chamber. (b) Insert a Matrigel-inoculated scaffold into a primed perfusion cartridge. (c) Insert a silicone gasket on top of the inoculated scaffold. (d) Insert the stainless steel screen into the perfusion chamber. (e) De-bubble by placing the perfusion chamber in vertical position such that a gas bubble is at a higher point than the scaffold. Apply the culture medium from the de-bubbling syringe below the scaffold. (f) Primed bubble-free perfusion chamber with cell-seeded scaffold in place (1) stainless steel screen, (2) cell-seeded scaffolds, and (3) silicone gasket. (g) Place the perfusion loop into the incubator. Note that these pictures were taken on a bench for better clarity; however, this procedure should be performed in a biosafety cabinet over a sterile Petri dish.
(C) Scaffold seeding in perfusion without Matrigel inoculation
Sterilize the following components as described in Steps 1–10: two perfusion chambers (open), four gaskets (made of platinum-cured silicone with 10 mm outer and 5 mm inner diameter and 4 mm thickness); and two pieces of Tygon tubing (3/16″ inner and 5/16″ outer diameter and length 35 cm) with luer connectors on each end (keep tubing open). One piece of PharMed BPT tubing (1/4″ outer and 1/8″ inner diameter and 12 cm length).
Cut two 10-mm diameter disks out of a 1-mm thick PGS sheet with a biopsy punch and sterilize as described in Step 28B(xix).
Place the sterilized components and the sterile PGS scaffolds into the hood.
Assemble the perfusion chambers using sterile gloves and sterile tweezers as follows: at the bottom of the cartridge place a gasket, the PGS scaffold and then a second gasket. The gaskets should squeeze the scaffold at its edges to hold it in place during perfusion. Repeat the same procedure for the second cartridge.
Close the cartridges.
Prepare two three-way stopcocks and attach one injection site to each of them.
Assemble the tubing, the three-way stopcocks and the cartridges in a U-tube configuration as shown in the scheme (Fig. 6e).
Attach one syringe filter at each open end of the tubing. From this point on, the system is closed and sterile.
Attach the assembled bioreactor to the lab stand using the clamps so that the U-tube and the cartridge inlet and outlet are aligned vertically. Using a 5-ml syringe with an attached cannula, inject 5 ml of cardiac medium through the injection site into one of the clear Tygon tubes.
-
With an empty 10-ml syringe attached to one of the filters, gently pull the medium until the two cartridges are filled. De-bubble the system by gently pushing or pulling on the medium. Be careful that the scaffolds are not damaged or displaced during this step.
? TROUBLESHOOTING
Prepare two 1-ml aliquots of cell suspension at a concentration of 3.3 ×106 cells ml−1. This corresponds to a seeding density of 1.35 × 108 cells cm−3 of scaffold because only the inner 5 mm of scaffold will be seeded.
Using a sterile syringe and attached cannula, inject through the injection site one of the cell suspension aliquots above each cartridge.
Transfer the lab stand and system into the incubator and attach pump tubing to the filters.
Apply a flow rate of 0.0176 ml s−1 (1000 μm s−1) in either direction.
When the descending medium level reaches one of the stopcocks, reverse the direction of flow. Repeat this process for the 2-h duration of the seeding process. We use a LabVIEW program to automate this process.
Placing the cell-seeded scaffold into the perfusion chamber
60| Place the assembled perfusion loop into the biosafety cabinet. Use sterile gloves.
61| Place scaffolds into the perfusion chamber. If Matrigel-inoculated scaffolds are used without the perfusion seeding, follow option A. If alternating medium flow perfusion seeding was used, follow option B. We found that it was the easiest way to transfer the entire perfusion chamber along with the extension tubing from the seeding loop into the perfusion cultivation loop. For this case, the generalized perfusion loop from Steps 1–27 can be assembled without the perfusion chamber (i.e., medium reservoir bag would connect directly through a three-way stopcock into the pump tubing, meaning Steps 17 and 18 would be omitted).
(A) If Matrigel-inoculated scaffolds are used without the perfusion seeding step
Follow Step 59B(ix–xv) as described earlier.
(B) If alternating medium flow perfusion seeding was used
Place the seeding loop perfusion chamber along the extension tubing into a sterile 60-mm Petri dish.
In the culture perfusion loop, close off the access to the reservoir bag via the three-way stopcock to prevent leaking. In the seeding loop, close off the three-way stopcocks toward the perfusion chamber.
In the perfusion loop, disconnect the medium reservoir from the pump tubing. In the seeding loop, disconnect the gas exchanger tubing and the three-way stopcock from the inlet of the perfusion chamber extension tubing.
Connect the inlet of the perfusion chamber extension tubing to the medium reservoir bag, via the three-way stopcock attached to the medium reservoir bag.
In the seeding loop, disconnect the second gas exchanger tubing from the three-way stopcock at the outlet of the perfusion chamber extension tubing. Connect the outlet of the perfusion chamber extension tubing, along with its three-way stopcock, to the peristaltic pump tubing.
Open three-way stopcocks.
-
Carefully de-bubble the perfusion chamber if required, according to Step 25.
▴ CRITICAL STEP It is critical to remove all of the bubbles at this point, but also make sure that no cells are displaced from the scaffolds, that is, de-bubbling should be performed slowly and carefully.
Place the loop into the incubator (Fig. 7g) in a vertical position.
Connect the pump tubing to the pump placed outside of the incubator (Fig. 7g). Draw the pump tubing through the incubator door and secure the tubing to the incubator using tape. Select the desired flow rate. Throughout the cultivation, check the loop for leakage and formation of bubbles in the perfusion chamber.
62| Place the loop into the incubator (Fig. 7g) in a vertical position.
63| Connect the pump tubing to the pump placed outside of the incubator (Fig. 7g). Draw the pump tubing through the incubator door and secure the tubing to the incubator using tape. Select the desired flow rate. Throughout the cultivation, check the loop for leakage and formation of bubbles.
? TROUBLESHOOTING
Assessments
64| From the culture medium samples, we determine the metabolism of glucose and lactate, the release of LDH and pO2, pH; and from the constructs, we determine: contractile properties, cell number, viability, cell cycle, protein content, DNA content, tissue and cell morphology by histology, the distribution and morphology of cells by immunofluorescent staining. Measurement of contractile force is an additional useful functional assay12. We provide a detailed description of methods that we developed or that required significant modification compared to the standard protocol. Methods that relied on commercially available kits are not discussed in detail here. See option A for assessment of culture medium samples. See option B for assessment of contractile properties. See option C for assessment of cell number, viability and cell cycle. Determine these parameters from constructs based on collagen scaffolds and rely on digestion of the scaffold using a solution of collagenase. See option D for protein and DNA content. This analysis can be performed on constructs based on either natural or synthetic scaffolds. See option E for histology and immunofluorescence.
(A) Culture medium samples
Sample culture medium and the constructs at timed intervals during cultivation and at the end of cultivation.
Measure glucose and lactate concentrations in culture medium samples using glucose and L-lactate analyzer Model 2300 STAT Plus (Yellow Springs Instruments).
Perform a LDH assay on samples of culture using a commercial kit (Chiron Diagnostics), according to the manufacturer’s instructions. Culture medium samples can be obtained from the reservoir bag through a three-way stopcock, located between the reservoir bag and the gas exchanger, to determine changes in medium composition with respect to days of cultivation.
To measure pO2 and pH, draw 1 ml of culture medium from the stopcock located upstream and downstream of the perfusion chamber. Stop the pump, remove the de-bubbling syringe from the stopcock in the incubator and draw the sample using a 1-ml syringe. Measure using a blood gas analyzer (Model 1610).
(B) Contractile properties
Prepare a solution of Tyrode’s salts (Sigma) according to the manufacturer’s instructions. Keep the solution at 37 °C throughout the experiment.
Prepare stimulating electrodes (1.5-mm diameter carbon rods). Prepare two polycarbonate electrode holders and insert the electrodes into the holes in the holder. The distance between the electrodes should be 1 cm.
Place 20 ml of Tyrode’s solution in a 60-ml Petri dish and place the dish onto a stage of an optical microscope. In one modification, a heating tape (manufacturer) can be placed on the bottom of the dish to maintain the temperature of 37 °C.
Place the electrodes into the solution, so that they are completely immersed.
-
Connect the electrodes via platinum wires to a cardiac stimulator (Grass s88x).
▴ CRITICAL STEP It is important that electrodes are completely immersed in the culture medium and that that applied voltage is maintained between the electrodes. Use an oscilloscope to ensure that the desired pulse duration and intensity is delivered to the system (e.g., 5 V cm−1 and 2 ms duration pulses between the electrodes; the current delivered at these conditions should be 200 mA).
Open the cartridge of the perfusion loop. Remove the screen and the silicone gasket using tweezers. Remove the constructs gently using tweezers.
Place the construct between the electrodes. Using a low magnification (×1 or ×4) objective, place the entire construct in the field of view.
Set the stimulation regime at square monophasic pulses of 2 ms duration, 1 Hz and start at 0.1 V.
Start stimulating and increase the voltage in 0.1-V increments until the entire face section of the construct is observed to beat synchronously. Continue observation for a minimum of 1 min. This voltage is the ET.
Set the stimulation voltage to 150% (or 200%) of ET. Increase the stimulation frequency in the interval of 0.2 Hz until the contractions become irregular or cease completely. This frequency is maximum capture rate.
(C) Cell number, viability and cell cycle
Prepare a stock solution of EMA at 50 μgml−1 per manufacturer’s instructions.
Remove the construct from the perfusion chamber, as described in Step 64B (vi).
Place the construct into 6-well plates.
Fill a Styrofoam container with ice and place the 6-well plates on ice.
Apply 100 μl of the EMA(10 μl of 50 μg ml−1 solution per 1 × 106 cells suspended in 100 μl PBS) solution on top of the construct. A meniscus held by the surface tension should form surrounding the construct.
Place the constructs 18 cm below fluorescent light for 10 min to allow for EMA to cross link to DNA of nonviable cells.
Transfer the constructs into 60-ml Petri dishes (one construct/well). Tear the constructs into pieces (~1 mm3)using tweezers.
Apply 10 ml of solution containing 0.6 mg ml−1 collagenase type II (282 U mg−1) and 1.2 U ml−1 of dispase in culture medium per construct. Maintain at 37 °C/5% CO2 incubator for 30 min, with periodic pipetting to dissociate cell aggregates.
Place the dishes on ice for 30 min.
Using a serological pipette, pipette the solution up and down, rinsing the bottom of the Petri dish to collect as many cells as possible. Transfer the cell suspension into a 15-ml conical tube. Rinse the bottom of the Petri dish with 5 ml of fresh culture medium, and then transfer the medium to the conical tube. Keep the conical tube on ice.
Count the cells in the tube using a hemocytometer, following a standard procedure.
Centrifuge the cell suspension at 212g for 10 min. Resuspend the cells at 106 cells ml−1 in PBS containing 5% of FBS.
To determine cell viability, perform flow cytometry, collecting the data in the FL3 (red) channel. Dead cells should have a higher level of fluorescence and should be clearly identified in the FL3 histogram. Use side scatter versus forward scatter data to exclude scaffold and cell debris from the analysis.
To determine cell cycle, take a separate construct and perform Steps 64C(vii–xii).
For permeabilization, resuspend the cells in 70% ethanol (we keep the ethanol solution at −20 °C) and place the cell suspension on ice for 30 min. We usually first resuspend the cell pellet in about 200 μl of PBS to prevent excessive damage and then add 2 ml of 70% ethanol.
Centrifuge the cell suspension for 10 min at 212g.
Resuspend the pellet in 1 ml solution of 50 μg ml−1 RNase A and 0.1% Triton X-100 in PBS (0.5 ml/10−6 cells) to digest double-stranded RNA that might interfere with staining.
Add 50 ml of PI stock solution. Prepare the stock solution at 1 mg ml−1 according to the manufacturer’s instructions. Subject the cells to flow cytometry to determine the fraction of cells in G0/G1,S and G2/M phases. We recommend that an experienced technician prepares the acquisition and analysis sheets for flow cytometry. In principle, the side, forward scatter and red fluorescence are measured. DNA peak deconvolution is performed using commercially available software (we used ModFit LT V2.0 for Macintosh). For more information on principles of cell cycle analysis, refer to ref. 34.
(D) Protein and DNA content
Prepare a homogenization solution consisting of 1 M NH4OH and 0.2% of Triton X-100. To make 50 ml of solution, add 3.45 ml of 14.5 N NH4OH solution to a 50-ml measuring flaks, then add 100 μl of Triton X-100 and fill the flask with dH2O.
-
At the end of the experiment, remove the construct from the perfusion chamber and place it into a 2-ml cryovial. Snap freeze the construct by placing the cryovial into a Dewar filled with liquid nitrogen.
! CAUTION Wear safety glasses and mittens.
■ PAUSE POINT Store the vials in a container with liquid nitrogen if they are not used immediately.
Before the homogenization, clean the beads with 70% ethanol and autoclave for 20 min followed by 20 min of drying. We use stainless steel beads, 6.35 mm in diameter, for homogenization. Place three beads in each vial.
Place 1 ml of homogenization solution into each vial.
Take an ice bucket and place the vials on ice. Homogenize each sample for six 10-s cycles in the BeadBeater. Use the speed setting of 42. Return the vial to the ice when the sample is not in the BeadBeater.
Transfer the homogenate to a new cryovial and save for protein and DNA analysis at −20 °C.
For protein analysis, take 100 μl of homogenate and pipette up and down several times to shear the DNA.
Centrifuge for 10 min at 2,500g and 4 °C.
Proceed with the BioRad DC protein assay kit according to the manufacturer’s instructions.
For DNA analysis, prepare a DNA standard by dissolving DNA standard (Sigma D-1501, type I calf thymus, highly polymerized) at 1 mg ml−1 in PBE buffer. PBE buffer is PBS containing 1.0% BSA, 1.0 mM EDTA, and 1.5 mM NaN3 at pH7.6. Let the standard dissolve for 12–18 h at 60 °C. Filter sterilize the solution with a 0.45-μm filter and store at 4 °C. Check OD at 260 nm; it should be 1.
-
Prepare a stock solution of Hoescht dye 33258 (Polysciences) at 1 mg ml−1 in dH2O.
! CAUTION Hoescht dye is toxic.
Prepare 10 × assay buffer consisting of 1.0 M NaCl, 10 mM EDTA and 100 mM Tris at pH 7.4. Filter sterilize and store at 4 °C. Before use, dilute ten times.
Construct a standard curve by diluting a desired amount of DNA stock solution in the 1× assay buffer.
Dissolve 100 μl of construct homogenate (after Step 64D(vi)) in 1.9 ml of assay buffer.
Pipette 100 μl of DNA standard curve solution or sample into a 5-ml acrylic cuvette. Add 2 ml of dye and read the fluorescence on a fluorometer (excitation 350 nm and emission 450 nm). (Calibrate and operate the fluorometer according to the operating manual for your model.)
(E) Histology and immunofluorescence
At the end of cultivation, fix the constructs in formalin for 1 h at room temperature.
Subsequently, transfer the constructs in PBS. Paraffin embed and section the constructs. We usually use the services of an in-house pathology laboratory for paraffin embedding and sectioning into 5-μm thick sections. For those researchers who wish to perform paraffin embedding and H&E staining in their own laboratories, there are protocols described elsewhere. For example, the online National Cancer Institute protocol for tissue dehydration and embedding is available at http://cgap-mf.nih.gov/Protocols/Tissues/TissueProtocols/ParaffinEmbedding.html. Another example is a kit for H&E staining, available from Dako (AR 157).
Before staining, incubate the slides for 30 min at 58 °C.
For deparaffinization, we use a deckloaking chamber and the reveal solution available from BioCoreMedical. The deckloaking chamber is similar to a pressure cooker and the main component of the reveal solution is a citrate buffer. Fill a plastic staining jar (also available from Biocore Medical) with reveal solution and place the slides in the jar. Place the jar with the slides and a staining jar with dH2O in the deckloaking chamber. Close the chamber and set the chamber to 20 min at 95 °C.
Let the chamber cool down for 20 min, then open it and transfer the slides into the jar with dH2O. Let the chamber cool down for additional 10 min.
Take the jars out and place them on the bench for 10 min.
-
Rinse with dH2O two times for 2 min.
▴ CRITICAL STEP Pour water out before taking the slides out to make sure paraffin does not stick to the sections.
For blocking, wipe the slides carefully around the section. Apply 200 μl of 10% NHS in PBS. Place the slides in a humidified chamber and incubate for 40 min at room temperature. (A humidified chamber can be improvised by placing a moist paper towel into a plastic slide box.)
Apply 200 μl primary Ab in a solution consisting of 0.5 vol% Tween 20 and 1.5 vol% of NIHS in PBS. We usually apply primary antibodies for 1 h at 37 °C or overnight at 4 °C. We used the following primary antibodies and their corresponding dilution factors: mouse anti-cardiac troponin I (clone 23C6, Biodesign, 1:150 (ref. 35)) and mouse anti-sarcomeric α-actin (C5C, 1:500 Sigma35), mouse anti-α-myosin heavy chain (hybridoma supernatant, full strength35), mouse anti-β-myosin heavy chain (clone 5B9, Chemicon, full strength35), and rabbit anti-Connexin-43 (Chemicon, 1:50 (ref. 35)).
Rinse slides four times, each time for 3 min. Perform rinsing by replacing PBS in the staining jar.
Apply 200 μl of secondary Ab in a solution consisting of 0.5 vol% Tween 20 and 1.5 vol% of NHS in PBS for either 30 or 40 min at room temperature. We used the following secondary antibodies, all from Vector Laboratories: Texas Red-conjugated horse anti-mouse IgG (1:100), fluorescein-conjugated horse anti-mouse IgG (1:100) and fluorescein-conjugated goat anti-rabbit IgG (1:200).
Rinse as described in Step 64E(x).
Mount the slide by applying one drop of mounting medium with DAPI (Vector Laboratories).
Proceed with fluorescence microscopy.
● TIMING
Steps 1–4, 2 h
Steps 5–10, 1 h
Steps 11–27, 0.5 h per perfusion loop
Step 28A, 15 min for scaffold punching, 24 h for soaking
Step 28B, 24 h for PGS synthesis, 72 h for PGS scaffold preparation
Steps 29–41, 2 h
Step 42, 8–10 h
Steps 43–55, 2.5 h
Steps 56–58, 1 h per pre-plating step
Step 59A(i–v), 15 min; Step 59A(vi–xi), 3 min per scaffold; Step 59A(xii), 30 min; Step 59A(xiii), 0.5–4 h
Step 59B(i–xvii), 15 min per construct (1 construct/loop); Step 59B(xviii), 0.5–4 h
Step 59C(i–xii), 20 min per loop; Step 59C(xiii–xv), 15 min to set up and 2 h for cell seeding
Step 60, 3 min per loop
Step 61A, 10 min per construct
Step 61B(i–vii), 15 min per construct
Step 62, 10 min per loop
Step 63, 3-7 days
Step 64A(ii), 3 min per sample; Step 64A(iii), 7 min per sample; Step 64A(iv), 3 min per sample Step 64B(i–x), 10 min per construct
Step 64C(i–ix), 2 h; Step 64C(x–xvi), 2 h
Step 64D(i–iii), 2 h; Step 64D(iv–vi), 5 min/sample; Step 64D(vii–ix), 2 h; Step 64D(x–xv), 4 h
Step 64E(i–iii), 24–48 h; Step 64E(iv–viii), 2 h; Step 64E(ix), 1 h or 12 h; Step 64E(x–xiii), 1 h; Step 64E(xiv), 10–20 min per slide
? TROUBLESHOOTING
Step 10
Cartridges may become more fragile with repeated autoclaving and the screw-top cap’s threading may not provide a tight seal, leading to fluid leaking from the system. In this case, we recommend that before sterilizing the cartridge, the threading be wrapped with Teflon tape.
Step 26
Leaks in the perfusion loop usually occur in the perfusion chamber and the extension tubing, and can be treated by thoroughly fastening the connectors. However, if persistent leaks occur, it may be due to the material defects caused by autoclaving. We recommend that additional perfusion chambers with extension tubing be prepared and sterilized so that the chamber can be replaced before the experiment if leaks persist. If the pump will be placed far from the incubator, we recommend that a piece of PharMed tubing of identical diameter as the pump tubing and desired length be used to extend the pump tubing at either end.
Step 27
The gas bubbles from the perfusion chamber can be removed using two syringes. The chamber should be placed in the vertical position and the fluid should be forced from the lower syringe into the higher syringe. The bubbles will end up in the higher syringe, moving by a combination of buoyancy and culture medium flow.
Step 28B(iv)
The mold is designed to ensure the thickness of the salt template matches that of the steel ring. To achieve this, it helps to overfill the mold with salts and scrape the top of the mold flush.
Step 28B(ix)
PGS is a colorless highly viscous polymer. It will swell before it dissolves in THF. Make sure a homogeneous solution is formed instead of transparent PGS xenogels floating in THF.
Step 28B(x)
It is important to keep the PGS solution from leaking into the seam of the mold. This can be achieved using strong magnets that match or exceed the specification and steel rings and plates that are flat and scratch free at the coupling surfaces.
Step 48
Do not use a glass bottle, as it cannot heat up fast enough during the collagenase digestion steps in the water bath, thus decreasing cell yield.
Step 53
If the pieces of undigested tissue are still too big after the fifth collagenase digestion, try to make smaller and more even tissue pieces next time. There is no point in proceeding with further collagenase digestions (e.g., 6th), as cell viability will decrease dramatically.
Step 54
If a cell pellet does not appear, increase the centrifugation time to 5 min.
Step 57
If cell viability appears too low (≤70%), decrease the enzyme concentration, the time tissues spend in trypsin and the agitation speed during the overnight trypsin digestion, as well as the serial collagenase digestions.
Step 59A(xi)
For cell inoculation using Matrigel, if significant leaking of Matrigel occurs after seeding, collect the leaked solution with a pipette and gently apply on top of the scaffold (rather than inside). The flow of culture medium will force the cells to enter the scaffold pores.
Steps 59B(xv) and 59C(x)
De-bubbling of the system could cause displacement or damage of the scaffolds. This problem is easily visualized because the cartridge is transparent. If the problem occurs, the only solution is to open the cartridge and replace or substitute the scaffold. Leaks in the perfusion loop may occur at the tubing connections or at the cartridge inlet, outlet and screw-top cap. Assure that all the connections are tight.
Step 59B(xviii)
If a bubble is observed in the perfusion chamber during the seeding procedure, stop the pump and de-bubble.
Step 63
If a bubble is observed in the perfusion chamber during cultivation, stop the pump and de-bubble. Occasionally, the cultivation with the interstitial medium flow (on collagen scaffolds or non-channeled PGS scaffolds) may result in the cells clogging the pores of the scaffold. Also, the culture medium flow may apply sufficient pressure to the scaffold, so that the pores collapse and the flow of culture medium is stopped. The first sign of this is formation of persistent bubbles below the construct, as well as yellowing of the culture medium. If that occurs, the experiment unfortunately needs to be stopped. That was one additional motivation for us to develop channeled scaffolds where such clogging does not occur.
ANTICIPATED RESULTS
The PGS scaffold should have a porosity of ~90% and pore size as determined by the selection of salt particles (usually 100–200 μm in diameter). The scaffold will appear white and should be elastic (i.e., revert to its original position upon application of gentle pressure with tweezers).
After cell isolation, the cell viability should be ~80% (as determined by trypan blue exclusion, as well as FACS in conjunction with EMA labeling). Before pre-plating, we usually obtain ~5 million cells per heart used. As the heart has a heterogeneous cell population, we performed flow cytometry to determine cellular composition after one and two pre-plating steps. We used cardiac troponin I to identify CMs and prolyl-4-hydroxylase to identify FBs. After one pre-plating step, the cell suspension consisted of 64 ± 5% CM and 36 ± 16% FB. After two pre-plating steps, the cell suspension consisted of 81 ± 14% CM and 16 ± 3% FB36.
Previously, we used two cell types for the cell-seeding studies outlined in the PROCEDURE above: C2C12 cells (a murine myo-blast cell line) and neonatal rat CMs. Cells were seeded at densities corresponding to those normally present in the adult rat heart (0.5–1 × 108 cells cm−3) into collagen sponges (10 mm × 3 mm discs), using Matrigel as a vehicle for rapid cell delivery. Scaffolds inoculated with cell-gel suspension (Step 59A) were seeded either with alternating medium flow (Step 59B) or in orbitally mixed (25 r.p.m.) Petri dishes. The effects of seeding time (1.5 or 4.5 h), initial cell number (6 or 12 million cells per scaffold) and seeding setup (medium perfusion at 0.5 and 1.5 ml min−1; orbitally mixed dishes) were investigated using a randomized three-factor factorial experimental design with two or three levels and three replicates. The seeding cell yield was consistently high (over 80%) and appeared to be determined by the rapid gel inoculation. The decrease in cell viability was markedly lower for alternating medium flow than for orbitally mixed dishes (e.g., 8.8 ± 0.8% and 56.3 ± 4%, respectively, for 12 million cells at 4.5 h after seeding). Spatially uniform cell distributions were observed in perfused constructs (Fig. 6d, Step 59B), whereas cells were mainly located within a thin (100–200 μm) surface layer of dish-seeded constructs (Fig. 6b). In Step 59C, seeding efficiency for cells seeded using a high flow rate (0.0176 ml s−1) was higher than either when a low flow rate (0.00176 ml s−1) perfusion was used or when cells were inoculated with Matrigel into the PGS scaffolds (respectively 72 ± 9%, 58 ± 15% and 49 ± 23). The lowest s.d. of the cell seeding method with the high flow rate highlights the reliability and the reproducibility of the seeding procedure. Histological assessments also indicated that only a high flow rate for Step 59C (0.0176 ml s−1) yielded constructs with cells that are uniformly distributed throughout the entire cross-section of the scaffold (Fig. 6f).
Perfusion cultivation previously yielded cardiac tissue constructs that were able to contract synchronously in response to electrical field stimulation with ET of ~3.3 V cm−1 and MCR as high as 7 Hz. After 7 d of perfusion cultivation, the cell viability was ~80% and the live cell number was ~5 million. In addition, 80% of cells were in the G0/G1 state. The cell metabolism was aerobic, as measured by the molar ratio of lactate produced to glucose consumed, which was maintained between 1 and 2 during the entire cultivation. The cells within the constructs should be uniformly distributed throughout the cross-section and express cardiac markers, such as cardiac Troponin I and sarcomeric α-actin.
An alternative method to enhance mass transport within in vitro-cultured cardiac constructs is mechanical stretching, as elegantly demonstrated by Zimmermann et al.12. Following implantation in vivo, vascularization is the mechanism for increasing the thickness of viable and functional tissue. Two representative examples include the implantation of stacked monolayer sheets of CM36 and the use of a vascularized chamber37.
Ongoing developments
We are currently establishing methods of perfusion seeding to achieve a dense, uniform cell distribution in channeled scaffolds. This approach is designed to provide efficient local supply of oxygen while protecting the cells from hydrodynamic shear associated with perfusion, and to maximize cell yield, spatial uniformity and viability by seeding in perfusion. We are also combining the medium perfusion and electrical field stimulation of cultured constructs to provide a truly ‘biomimetic’ environment for the cells to express their differentiated phenotype and assemble into highly functional tissue constructs. The functional assessments are being extended to the measurements of contractile forces in vitro and pumping function in vivo, following implantation in a model of cardiac ischemia. These methods are currently being extended to tissue engineering of cardiac grafts using human stem cells.
ACKNOWLEDGMENTS
The authors gratefully acknowledge research support of the work described in this protocol by the National Institutes of Health (R01 HL076485 and P41-EB002520 to G.V.-N.), National Science and Engineering Council (Discovery Grant to M.R.) and Canada Foundation for Innovation (Leaders Opportunity Fund to M.R.). The authors also thank Melissa A.N. Brown for help with taking pictures for Figures 4 and 7.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/ reprints and permissions
References
- 1.Kirkpatrick JN, Vannan MA, Narula J, Lang RM. Echocardiography in heart failure: applications, utility, and new horizons. J. Am. Coll. Cardiol. 2007;50:381–396. doi: 10.1016/j.jacc.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 2.Akins RE, et al. Cardiac organogenesis in vitro: Reestablishment of three-dimensional tissue architecture by dissociated neonatal rat ventricular cells. Tissue Eng. 1999;5:103–118. doi: 10.1089/ten.1999.5.103. [DOI] [PubMed] [Google Scholar]
- 3.Bursac N, et al. Cardiac muscle tissue engineering: toward an in vitro model for electrophysiological studies. Am. J. Physiol. Heart Circ. Physiol. 1999;277:H433–H444. doi: 10.1152/ajpheart.1999.277.2.H433. [DOI] [PubMed] [Google Scholar]
- 4.Carrier RL, et al. Cardiac tissue engineering: cell seeding, cultivation parameters and tissue construct characterization. Biotechnol. Bioeng. 1999;64:580–589. doi: 10.1002/(sici)1097-0290(19990905)64:5<580::aid-bit8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 5.Carrier RL, et al. Effects of oxygen on engineered cardiac muscle. Biotechnol. Bioeng. 2002;78:617–625. doi: 10.1002/bit.10245. [DOI] [PubMed] [Google Scholar]
- 6.Carrier RL, et al. Perfusion improves tissue architecture of engineered cardiac muscle. Tissue Eng. 2002;8:175–188. doi: 10.1089/107632702753724950. [DOI] [PubMed] [Google Scholar]
- 7.Papadaki M, et al. Tissue engineering of functional cardiac muscle: molecular, structural and electrophysiological studies. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H168–H178. doi: 10.1152/ajpheart.2001.280.1.H168. [DOI] [PubMed] [Google Scholar]
- 8.Li R-K, et al. Survival and function of bioengineered cardiac grafts. Circulation. 1999;100:II63–II69. doi: 10.1161/01.cir.100.suppl_2.ii-63. [DOI] [PubMed] [Google Scholar]
- 9.Li R-K, et al. Construction of a bioengineered cardiac graft. J. Thorac. Cardiovasc. Surg. 2000;119:368–375. doi: 10.1016/S0022-5223(00)70193-0. [DOI] [PubMed] [Google Scholar]
- 10.Radisic M, et al. High density seeding of myocyte cells for tissue engineering. Biotechnol. Bioeng. 2003;82:403–414. doi: 10.1002/bit.10594. [DOI] [PubMed] [Google Scholar]
- 11.Leor J, et al. Bioengineerred cardiac grafts: A new approach to repair the infarcted myocardium? Circulation. 2000;102:III56–III61. doi: 10.1161/01.cir.102.suppl_3.iii-56. [DOI] [PubMed] [Google Scholar]
- 12.Zimmermann WH, et al. Three-dimensional engineered heart tissue from neonatal rat cardiac myocytes. Biotechnol. Bioeng. 2000;68:106–114. [PubMed] [Google Scholar]
- 13.Radisic M, et al. Oxygen gradients correlate with cell density and cell viability in engineered cardiac tissue. Biotechnol. Bioeng. 2006;93:332–343. doi: 10.1002/bit.20722. [DOI] [PubMed] [Google Scholar]
- 14.Radisic M, et al. Medium perfusion enables engineering of compact and contractile cardiac tissue. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H507–H516. doi: 10.1152/ajpheart.00171.2003. [DOI] [PubMed] [Google Scholar]
- 15.Radisic M, et al. Biomimetic approach to cardiac tissue engineering: oxygen carriers and channeled scaffolds. Tissue Eng. 2006;12:2077–2091. doi: 10.1089/ten.2006.12.2077. [DOI] [PubMed] [Google Scholar]
- 16.Radisic M, et al. High-density seeding of myocyte cells for cardiac tissue engineering. Biotechnol. Bioeng. 2003;82:403–414. doi: 10.1002/bit.10594. [DOI] [PubMed] [Google Scholar]
- 17.Vunjak-Novakovic G, Radisic M. Cell seeding of polymer scaffolds. Methods Mol. Biol. 2004;238:131–146. doi: 10.1385/1-59259-428-x:131. [DOI] [PubMed] [Google Scholar]
- 18.Radisic M, et al. Medium perfusion enables engineering of compact and contractile cardiac tissue. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H507–H516. doi: 10.1152/ajpheart.00171.2003. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Ameer GA, Sheppard BJ, Langer R. A tough biodegradable elastomer. Nat. Biotechnol. 2002;20:602–606. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 20.Radisic M, Deen W, Langer R, Vunjak-Novakovic G. Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H1278–H1289. doi: 10.1152/ajpheart.00787.2004. [DOI] [PubMed] [Google Scholar]
- 21.Wendt D, Marsano A, Jakob M, Heberer M, Martin I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol. Bioeng. 2003;84:205–214. doi: 10.1002/bit.10759. [DOI] [PubMed] [Google Scholar]
- 22.Dvir T, Levy O, Shachar M, Granot Y, Cohen S. Activation of the ERK1/2 cascade via pulsatile interstitial fluid flow promotes cardiac tissue assembly. Tissue Eng. 2007;13:2185–2193. doi: 10.1089/ten.2006.0364. [DOI] [PubMed] [Google Scholar]
- 23.Kretzmer G, Schugerl K. Response of mammalian cells to shear stress. Appl. Microbiol. Biotechnol. 1991;34:613–616. doi: 10.1007/BF00167909. [DOI] [PubMed] [Google Scholar]
- 24.Smith CG, Greenfield PF, Randerson D. Shear sensitivity of three hybridoma cell lines in suspension culture. In: Spier RE, Griffith JB, editors. Modern Approaches to Animal Cell Technology. Butterworth; Kent, UK: 1987. pp. 316–327. [Google Scholar]
- 25.Stathopoulos NA, Hellums JD. Shear stress effects on human embryonic kidney cells in vitro. Biotechnol. Bioeng. 1985;27:1021–1026. doi: 10.1002/bit.260270713. [DOI] [PubMed] [Google Scholar]
- 26.Sikavitsas VI, Bancroft GN, Holtorf HL, Jansen JA, Mikos AG. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc. Natl. Acad. Sci. USA. 2003;100:14683–14688. doi: 10.1073/pnas.2434367100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bancroft GN, et al. Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc. Natl. Acad. Sci. USA. 2002;99:12600–12605. doi: 10.1073/pnas.202296599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pazzano D, et al. Comparison of chondrogenesis in static and perfused bioreactor culture. Biotechnol. Prog. 2000;16:893–896. doi: 10.1021/bp000082v. [DOI] [PubMed] [Google Scholar]
- 29.Davisson TH, Sah RL, Ratcliffe AR. Perfusion increases cell content and matrix synthesis in chondrocyte three-dimensional cultures. Tissue Eng. 2002;8:807–816. doi: 10.1089/10763270260424169. [DOI] [PubMed] [Google Scholar]
- 30.Raimondi MT, et al. The effect of media perfusion on three-dimensional cultures of human chondrocytes: Integration of experimental and computational approaches. Biorheology. 2004;41:401–410. [PubMed] [Google Scholar]
- 31.Cartmell SH, Porter BD, Garcia AJ, Guldberg RE. Effects of medium perfusion rate on cell-seeded three-dimensional bone constructs in vitro. Tissue Eng. 2003;9:1197–1203. doi: 10.1089/10763270360728107. [DOI] [PubMed] [Google Scholar]
- 32.Radisic M, et al. Biomimetic approach to cardiac tissue engineering. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007;362:1357–1368. doi: 10.1098/rstb.2007.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Crapo PM, Wang Y. Macroporous elastomeric scaffolds with extensive micropores for soft tissue engineering. Tissue Eng. 2006;12:917–925. doi: 10.1089/ten.2006.12.917. [DOI] [PubMed] [Google Scholar]
- 34.Ormerod MG. Flow Cytometry. Oxford University Press; New York: 2000. [Google Scholar]
- 35.Nuccitelli R. Endogenous ionic currents and DC electric fields in multicellular animal tissues. Bioelectromagnetics. 1992;(Suppl 1):147–157. doi: 10.1002/bem.2250130714. [DOI] [PubMed] [Google Scholar]
- 36.Iyer RK, Chiu LL, Radisic M. Microfabricated poly(ethylene glycol) templates enable rapid screening of tri-culture conditions for cardiac tissue engineering. J. Biomed. Mater. Res. A. doi: 10.1002/jbm.a.32014. (in press) [DOI] [PubMed] [Google Scholar]
- 37.Morritt AN, et al. Cardiac tissue engineering in an in vivo vascularized chamber. Circulation. 2007;115:353–360. doi: 10.1161/CIRCULATIONAHA.106.657379. [DOI] [PubMed] [Google Scholar]