Abstract
The dentin sialophosphoprotein (DSPP) gene on chromosome 4q21.3 encodes the major noncollagenous protein in tooth dentin. DSPP mutations are the principal cause of dentin dysplasia type II, dentinogenesis imperfecta type II, and dentinogenesis imperfecta type III. We have identified a DSPP splice junction mutation (IVS2-6T>G) in a family with dentin dysplasia type II. The primary dentition is discolored brown with severe attrition. The mildly discolored permanent dentition has thistle-shaped pulp chambers, pulp stones, and eventual pulp obliteration. The mutation is in the sixth nucleotide from the end of intron 2, perfectly segregates with the disease phenotype, and is absent in 200 normal control chromosomes. An in vitro splicing assay shows that pre-mRNA splicing of the mutant allele generates wild-type mRNA and mRNA lacking exon 3 in approximately equal amounts. Skipping exon 3 might interfere with signal peptide cleavage, causing endoplasmic reticulum stress, and also reduce DSPP secretion, leading to haploinsufficiency.
Keywords: Dentin, dentin dysplasia type II, dentinogenesis imperfecta, DSPP, pre-mRNA splicing
Pulpal obliteration can complicate the endodontic treatment of teeth, causing treatment failures and subsequent extraction of the involved teeth. Idiopathic or pathologic conditions can induce generalized or localized pulpal obliteration. Generalized pulpal pathosis is also a feature in certain genetic diseases that affect dentin and pulp tissue (1).
Hereditary dentin defects are separated into 2 main categories, dentinogenesis imperfecta (DGI) and dentin dysplasia (DD) (2). DGI type I is a syndromic phenotype and is synonymous with osteogenesis imperfecta (OI) (3). The nondental phenotype in OI is sometimes subtle (4), and dentists need to be vigilant to avoid making an incorrect diagnosis of DD type II or DGI type II (5). DGI type II is characterized by amber discoloration of the dentition, severe attrition of the teeth, bulbous crowns, and early pulpal obliteration. DGI type III, which is believed to be distinct from DGI type II, is now considered as a more severe form of DGI type II (6). DD type I is characterized by normal-looking crowns with short roots with crescent-shaped pulpal remnants (7). DD type II is similar to DGI type II in the deciduous dentition but minimally affects the permanent dentition. The crowns of the permanent teeth are normal-looking or discolored gray, with thistle-shaped pulp chambers and pulp stones that eventually obliterate the pulp chamber (8). DD type II is rare, compared with DGI type II.
Defects in the dentin sialophosphoprotein (DSPP) gene cause DGI type II and type III and DD type II (9-19). Only 4 mutations have been reported to cause DD type II, 1 missense mutation in the signal peptide and 3 frameshift mutations in the DPP coding region. Because of the rarity of DD type II and the lack of supporting data of known mutations, its molecular genetic etiology and proper genotype-phenotype correlations are still unknown. Here we report clinical findings and the results of mutational analysis of a Hispanic family with DD type II.
Materials and Methods
Identification of a Kindred and Enrollment of Human Subjects
This study was independently reviewed and approved by the Institutional Review Board at the University of Michigan and the Seoul National University Dental Hospital. The experiments were undertaken with the understanding and written consent of each subject according to the Declaration of Helsinki. The proband was a 4-year-old boy who presented with the dental features of DGI type II or DD type II. Five members of the family consented to participate in this study and contributed samples for genomic DNA analyses. All participating members received clinical and radiologic examinations.
Polymerase Chain Reaction and Sequencing
Genomic DNA was isolated from peripheral whole blood of participating family members (II-2, III-1, III-2, III-3, and III-4) by using the QIAamp DNA Blood Maxi Kit (Qiagen Inc, Valencia, CA). The purity and concentration of the DNA were quantitated by spectrophotometry, as measured by the OD260/OD280 ratio. Polymerase chain reaction (PCR) conditions and primer pairs used for PCR and sequencing were previously described (12). PCR amplification products were purified by QIAquick PCR Purification Kit and protocol (Qiagen Inc). DNA sequencing was performed at the DNA sequencing center (Macrogen, Seoul, Korea). All nucleotide numbering was determined by counting from the A of the ATG translation initiation codon of the human DSPP reference sequence (NM_014208.3).
In Vitro Splicing Assay
Genomic DNA of a normal control without any dental and general health problems was amplified with pfu enzyme (Elpisbio, Seoul, Korea) by using PCR primers (amplicon size, 2767 base pairs; sense, GGGGGCGGCCGCGGGCAAATGCTTACACATCA; antisense, AGGATCCATGATTTCACATAAGAC) to include exons 2, 3, and 4 of DSPP gene. The amplification product was cloned into the pSPL3 splicing vector after double digesting with NotI and BamHI restriction endonucleases. PCR mutagenesis was performed to change T to G at the splicing acceptor site −6 position of intron 2. Sequences of normal and mutant pSPL3 vectors were confirmed by direct plasmid sequencing. Normal and mutant pSPL3 vectors were transfected into COS-7 cells, and total RNA was isolated after 36 hours. Reverse transcriptase–PCR (sense, TATTTTTGCATTTGGGCAGT; antisense, CCTACTTCTGCCCACTTAGAGC primers) was performed with HiPi PCR premix (Elpisbio). Amplification bands were excised from an agarose gel after electrophoresis, purified, and characterized by direct DNA sequencing as described above.
Results
Clinical Findings
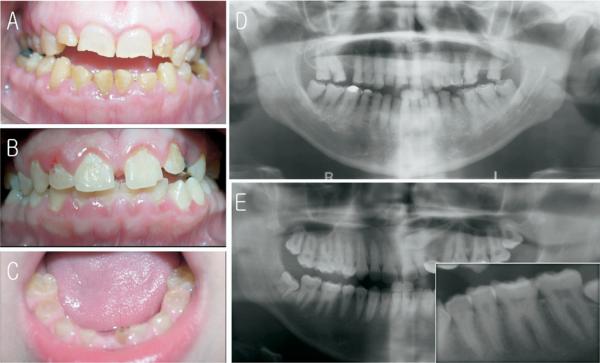
Physical examination of the family members revealed no obvious non-oral abnormalities such as bone fragility or progressive hearing loss (Fig. 1A). The proband (III-4) was a 4-year-old boy who presented with the dental features of DGI type II or DD type II. His primary teeth were discolored brown and showed severe attrition (Fig. 2C). The dentition of his affected brother (III-1) looked normal; however, the panoramic radiograph showed features characteristic of DD type II (Fig. 2B, E). The premolar and canine teeth had thistle-shaped pulp chambers with pulp stones. The coronal pulp chambers of the maxillary anterior teeth were obliterated, and the radicular pulp chambers of the molar teeth, especially those of the first molars, were obliterated. His mother (II-2) had a mildly discolored dentition, with minor attrition and fractures along the incisal edges of her anterior teeth (Fig. 2A). The panoramic radiograph showed almost complete pulpal obliteration except for small pulpal remnants in several posterior teeth (Fig. 2D).
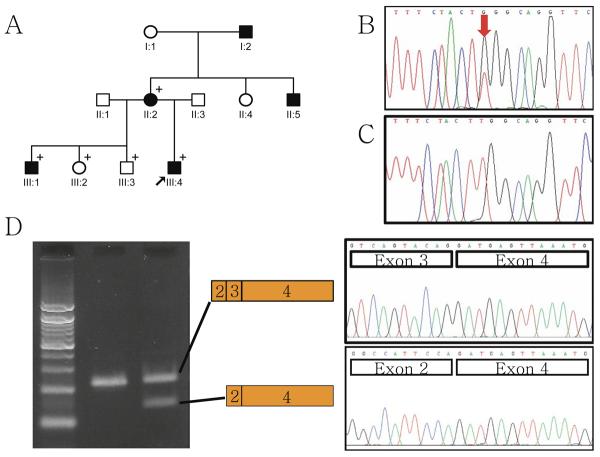
Figure 1.
Pedigree and mutational analyses of the kindred. (A) Pedigree of the kindred. Black arrow indicates proband. A + identifies members recruited in this study. (B) DNA sequencing chromatogram of the affected individual III-4. Red arrow indicates the position of the mutation. (C) DNA sequencing chromatogram of the unaffected individual III-3. (D) Agarose gel image of in vitro splicing assay. Left lane is DNA size marker. Middle lane is a result of pSPL3 vector with normal insert. Right lane is a result of pSPL3 vector with mutated insert. Upper band in the right lane is normally spliced mRNA transcript, and lower band is abnormally spliced mRNA with exon 3 deletion.
Figure 2.
Clinical photographs and radiographs. (A) Frontal view of affected individual II-2 at age 39. Note mild discoloration with minor attritions. (B) Frontal view of affected individual III-1 at age 15. Clinically permanent teeth look normal. (C) Frontal view of affected individual III-4 at age 4. Note brown discoloration and attrition. (D) Panoramic radiograph of affected individual II-2 at age 39. Note almost complete pulpal obliterations. (E) Panoramic radiograph of affected individual III-1 at age 15. The premolar and canine teeth have thistle-shaped pulp chambers with pulp stones. The inset is a magnified view of left mandibular premolars and molars. Note thistle-shaped pulp chambers and pulp stones.
Mutation Analyses
Mutational analysis revealed a T to G change at the intron 2 splice acceptor site −6 position (g.1185T>G, IVS2-6T>G) of the DSPP gene (Fig. 1B, C). The identified mutation perfectly segregated with the disease phenotype in this family and furthermore was absent in 200 normal control chromosomes. An in vitro splicing assay showed that this mutation in the splicing acceptor site of intron 2 alters pre-mRNA processing and results in the deletion of exon 3 in the mutant mRNA, which is present in approximately equal amounts as the correctly spliced mRNA transcript (Fig. 1D).
Discussion
Currently 19 mutations in the DSPP gene, including 4 mutations causing DD type II, have been reported (Table 1). Several interesting patterns have emerged, but the genotype-phenotype correlationship is not yet known. Most of the mutations in the 5′ end of DSPP are single nucleotide substitutions near intron/exon junctions (15). It is difficult to predict with certainty what the effects of these mutations are in terms of DSPP expression. Retention of an intron would likely lead to degradation of the mRNA transcript by nonsense mediated mRNA decay (NMD), a mechanism that degrades mRNA harboring a premature translation termination codon (20). Degradation of the mRNA from the mutant DSPP allele would cause haploinsufficiency or a simple reduction in the quantity of normal mRNA and protein expressed. Mutations in the 3′ end of DSPP are deletions and/or insertions that shift the reading frame in a way that replaces the C-terminus of DSPP with a long chain of extraneous amino acids (18, 19). Interestingly, no mutations shift the reading frame in a way that simply truncates the protein (which would occur with shifts to the −2 reading frame), suggesting that such mutations do not cause a dominant dental phenotype.
TABLE 1.
Summary of Mutations in DSPP Resulting in Hereditary Dentin Defects
| cDNA* | Protein† | Diagnosis | Reference | |
|---|---|---|---|---|
| Exon 2 | c.16T>G | p.Y6D | DD-II | Rajpar et al. (11) |
| c.44C>T | p.A15V | DGI-II | Malmgren et al. (13) | |
| c.49C>A | p.P17T | DGI-II | Xiao et al. (9) | |
| c.49C>T | p.P17S | DGI-II | Hart and Hart (6), Zhang et al. (17) | |
| Intron 2 | c.52-6T>G | Normal + p.V18_Q45del | DD-II | This report |
| c.52-3C>G | p.V18_Q45del | DGI-II | Kim et al. (12) | |
| c.52-3C>A | p.V18_Q45del | DGI-II | Holappa et al. (15) | |
| Exon 3 | c.52G>T | p.V18_Q45del or p.V18F | DGI-II, DGI-III | Xiao et al. (9), Kim et al. (14), Song et al. (16), Holappa et al. (15) |
| c.133C>T | p.V18_Q45del or p.Q45X | DGI-II | Zhang et al. (10), Song et al. (16) | |
| Intron 3 | c.135 + 1G>A | p.V18_Q45del | DGI-II | Xiao et al. (9) |
| c.135 + 1G>T | p.V18_Q45del | DGI-II | McKnight et al. (18) | |
| Exon 5 | c.1870_1873delTCAG | p.S624fsX1312 | DD-II | McKnight et al. (18) |
| c.1918_1921delTCAG | p.S640fsX1312 | DD-II | McKnight et al. (18) | |
| c.2040delC | p.S680fsX1313 | DD-II | Song et al. (19) | |
| c.2272delA | p.S758fsX1313 | DGI-II | McKnight et al. (18) | |
| c.2525delG | p.S842fsX1313 | DGI-II | McKnight et al. (18) | |
| c.2593delA | p.S865fsX1313 | DGI-II | Song et al. (19) | |
| c.2684delG | p.S895fsX1313 | DGI-II | Song et al. (19) | |
| c.3438delC | p.D1146fsX1313 | DGI-II | Song et al. (19) | |
| c.3546_3550delTAGCAinsG | p.D1182fsX1312 | DGI-II | Song et al. (19) |
Numbering assumes the A of the ATG start codon as nucleotide 1 on the basis of reference sequence NM_014208.3.
These are mostly predictions.
In our kindred with DD type II, sequence analysis of the proband's DNA revealed a mutation (g.1185T>G, IVS2-6T>G) in the sixth nucleotide from the end of intron 2. This nucleotide is part of the 3′ polypyrimidine tract that is necessary for proper pre-mRNA splicing (21). A polypyrimidine (C or T) tract longer than 6 nucleotides is necessary for splicing in mammalian species such as human, mouse, and dog (22). Computer splicing algorithms (www.fruitfly.org/seq_tools/splice.html) decreased the splice acceptor site prediction value of the mutant compared with the wild-type polypyrimidine tract from 0.97 to 0.74. The results of an in vitro splicing assay supported this prediction that the IVS2-6T>G alteration would diminish recognition of the end of intron 2 as a splice junction. RNA analysis showed normally spliced mRNA as well as mRNA with exon 3 deleted (Fig. 1D). This finding suggests that disruption of the splice acceptor site by the IVS2-6T>G mutation is not complete but partial and does not result in intron retention. Skipping of exon 3 during pre-mRNA splicing deletes 28 amino acids (from Val18 through Gln45; p.V18_Q45) from the N-terminal region of DSPP. Such an alteration could secondarily interfere with either the secretion of DSPP or cleavage of its signal peptide after Ala15. A percentage of the pre-mRNA from the diseased allele, however, would splice normally and combine with expression from the wild-type DSPP allele to produce more than half of the normal amount of DSPP protein in affected individuals. Previously, a point mutation in the DSPP signal peptide coding region (c.16T>G, p.Y6D) that was manifested as a DD type II phenotype was shown to reduce the secretion of DSPP as a result of partial failure of endoplasmic reticulum (ER) translocation (11). Perhaps the reduction of the normal amount of DSPP protein (about 25% reduction according to this study) results in the relatively mild phenotype of DD type II, whereas a larger reduction, as would be expected in haploinsufficiency, results in the more severe forms of inherited dentin defects (DGI type II and type III).
The data, however, are also consistent with a competing theory. Deletions or point mutations in or near the signal peptide coding region might affect normal signal peptide cleavage and interfere with protein folding and secretion, inducing an ER stress response and general cell pathology (23). It has even been suggested that all DSPP mutations yet reported cause their DD-II, DGI-II, and DGI-III phenotypes through dominant negative effects and not haploinsufficiency (18). The findings of our study indicate that exon skipping rather than intron retention is the more likely outcome of DSPP splice junction mutations, which is more consistent with the dominant negative explanation for the dental pathology. The effects of exon 3 deletion on the efficiency of signal peptide cleavage causing possible ER stress need to be studied further.
A better understanding of the pathogenesis of inherited diseases affecting dentin might improve the ability of dental practitioners to choose among treatment options in patients with DSPP mutations. Pathologic pulp chamber obliteration related to genetic conditions like those seen in our family with DD type II can be predicted at early stages before the development of pulp stones or pulp canal obliteration. Furthermore, mutational analysis would reveal genotype-phenotype correlations and lead us to a better understanding of dentin mineralization and homeostasis. The functional role of DSPP and its relationship to various transcription factors are critical to the success of regenerative treatment or gene therapy for dentin-pulp complex (24, 25).
In conclusion, we have identified a novel mutation in the splice acceptor site of intron 2 of the DSPP gene in a family with DD type II. The partial disruption of splicing efficiency resulted in normal mRNA transcripts along with an equal quantity of exon 3 deleted transcripts. The functional role of the DSPP protein as well as the effect of the DSPP mutations on protein expression needs to be further studied to better understand the role of DSPP in normal and pathologic dentin formation.
Acknowledgments
We would like to thank all the family members for their cooperation. We thank Dr Alessandro Stella, University of Bari, Italy, for the pSPL3 splicing vector used in this study. This research was supported by grant 03-2007-0080 from the Seoul National University Dental Hospital Research Funds (J.-W. Kim) and NIDCR/NIH grant DE015846 (J .P. Simmer).
References
- 1.Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;86:392–9. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 2.Shields ED, Bixler D, el-Kafrawy AM. A proposed classification for heritable human dentine defects with a description of a new entity. Arch Oral Biol. 1973;18:543–53. doi: 10.1016/0003-9969(73)90075-7. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell AC, Marini JC. Evaluation of oral problems in an osteogenesis imperfecta population. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1999;87:189–96. doi: 10.1016/s1079-2104(99)70272-6. [DOI] [PubMed] [Google Scholar]
- 4.Pallos D, Hart PS, Cortelli JR, et al. Novel COL1A1 mutation (G559C) [correction of G599C] associated with mild osteogenesis imperfecta and dentinogenesis imperfecta. Arch Oral Biol. 2001;46:459–70. doi: 10.1016/s0003-9969(00)00130-8. [DOI] [PubMed] [Google Scholar]
- 5.Teixeira CS, Felippe MC, Felippe WT, Silva-Sousa YT, Sousa-Neto MD. The role of dentists in diagnosing osteogenesis imperfecta in patients with dentinogenesis imperfecta. J Am Dent Assoc. 2008;139:906–14. doi: 10.14219/jada.archive.2008.0277. [DOI] [PubMed] [Google Scholar]
- 6.Hart PS, Hart TC. Disorders of human dentin. Cells Tissues Organs. 2007;186:70–7. doi: 10.1159/000102682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright T. The molecular control of and clinical variations in root formation. Cells Tissues Organs. 2007;186:86–93. doi: 10.1159/000102684. [DOI] [PubMed] [Google Scholar]
- 8.Beattie ML, Kim JW, Gong SG, Murdoch-Kinch CA, Simmer JP, Hu JC. Phenotypic variation in dentinogenesis imperfecta/dentin dysplasia linked to 4q21. J Dent Res. 2006;85:329–33. doi: 10.1177/154405910608500409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao S, Yu C, Chou X, et al. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–4. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Zhao J, Li C, et al. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet. 2001;27:151–2. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 11.Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet. 2002;11:2559–65. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- 12.Kim JW, Nam SH, Jang KT, et al. A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum Genet. 2004;115:248–54. doi: 10.1007/s00439-004-1143-5. [DOI] [PubMed] [Google Scholar]
- 13.Malmgren B, Lindskog S, Elgadi A, Norgren S. Clinical, histopathologic, and genetic investigation in two large families with dentinogenesis imperfecta type II. Hum Genet. 2004;114:491–8. doi: 10.1007/s00439-004-1084-z. [DOI] [PubMed] [Google Scholar]
- 14.Kim JW, Hu JC, Lee JI, et al. Mutational hot spot in the DSPP gene causing dentino-genesis imperfecta type II. Hum Genet. 2005;116:186–91. doi: 10.1007/s00439-004-1223-6. [DOI] [PubMed] [Google Scholar]
- 15.Holappa H, Nieminen P, Tolva L, Lukinmaa PL, Alaluusua S. Splicing site mutations in dentin sialophosphoprotein causing dentinogenesis imperfecta type II. Eur J Oral Sci. 2006;114:381–4. doi: 10.1111/j.1600-0722.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Wang C, Peng B, et al. Phenotypes and genotypes in 2 DGI families with different DSPP mutations. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:360–74. doi: 10.1016/j.tripleo.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Chen L, Liu J, et al. A novel DSPP mutation is associated with type II dentinogenesis imperfecta in a Chinese family. BMC Med Genet. 2007;8:52. doi: 10.1186/1471-2350-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKnight DA, Hart PS, Hart TC, et al. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Hum Mutat. 2008 doi: 10.1002/humu.20783. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song YL, Wang CN, Fan MW, Su B, Bian Z. Dentin phosphoprotein frameshift mutations in hereditary dentin disorders and their variation patterns in normal human population. J Med Genet. 2008;45:457–64. doi: 10.1136/jmg.2007.056911. [DOI] [PubMed] [Google Scholar]
- 20.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. Embo J. 2008;27:471–81. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coolidge CJ, Seely RJ, Patton JG. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–96. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura M. Endoplasmic reticulum stress in the kidney. Clin Exp Nephrol. 2008 doi: 10.1007/s10157-008-0060-7. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24.Jadlowiec J, Koch H, Zhang X, Campbell PG, Seyedain M, Sfeir C. Phosphophoryn regulates the gene expression and differentiation of NIH3T3, MC3T3-E1, and human mesenchymal stem cells via the integrin/MAPK signaling pathway. J Biol Chem. 2004;279:53323–30. doi: 10.1074/jbc.M404934200. [DOI] [PubMed] [Google Scholar]
- 25.Jadlowiec JA, Zhang X, Li J, Campbell PG, Sfeir C. Extracellular matrix-mediated signaling by dentin phosphophoryn involves activation of the Smad pathway independent of bone morphogenetic protein. J Biol Chem. 2006;281:5341–7. doi: 10.1074/jbc.M506158200. [DOI] [PubMed] [Google Scholar]