Summary
Protein-DNA interactions are fundamental to core biological processes including transcription, DNA replication, and chromosomal organization. We have developed In vivo Protein Occupancy Display (IPOD), a technology that reveals protein occupancy across an entire bacterial chromosome at the resolution of individual binding sites. Application to Escherichia coli reveals thousands of protein occupancy peaks, highly enriched within and in close proximity to non-coding regulatory regions. In addition, we discovered extensive (>1 kilobase) protein occupancy domains (EPODs), some of which are localized to highly-expressed genes, enriched in RNA-polymerase occupancy. However, the majority are localized to transcriptionally-silent loci dominated by conserved hypothetical ORFs. These regions are highly enriched in both predicted and experimentally determined binding sites of nucleoid proteins and exhibit extreme biophysical characteristics such as high intrinsic curvature. Our observations implicate these transcriptionally-silent EPODs as the elusive organizing centers, long proposed to topologically isolate chromosomal domains.
Keywords: IPOD, Protein-DNA interactions, transcriptional networks, nucleoid proteins, chromosomal organization, E. coli
Introduction
Replication, maintenance, and expression of genetic information are processes that are orchestrated through precise interactions of hundreds of proteins with chromosomal DNA. For decades, research has focused on the behavior and functional consequences of DNA-protein interactions at individual loci. However, understanding systems-level behaviors, such as chromosomal organization, genome replication, and transcriptional network dynamics requires observations at the scale of the entire system. Microarray-based chromatin immunoprecipitation (ChIP-chip) allows global measurements of chromosomal occupancy for individual proteins (Ren et al., 2000). In another global approach, methylase protection, a fraction of all occupied sites are monitored in vivo, independent of the identity of the bound proteins (Tavazoie and Church, 1998). However, there currently exists no comprehensive approach for simultaneous, high-resolution monitoring of all in vivo protein-DNA interactions across the genome. We have developed such a technology and used it to profile protein occupancy of the E. coli chromosome at the resolution of individual binding sites.
Results
In vivo Protein Occupancy Display (IPOD)
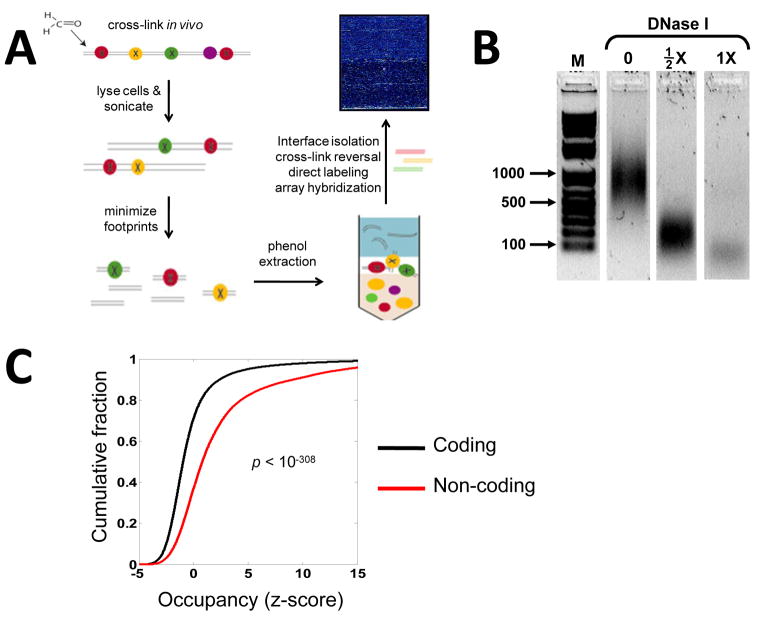
In order to globally profile the occupancy of all proteins on chromosomal DNA, we first stabilize in vivo protein-DNA interactions through covalent cross-linking with formaldehyde (Fig. 1A). After cell-lysis and sonication, protein footprints are minimized to a mode of ~ 50 bp through DNase I digestion (Fig. 1B). Phenol extraction is then used to trap amphipathic protein-DNA complexes at the interface between the organic and aqueous phases. Following interface isolation and cross-link reversal, short DNA fragments are end-labeled and hybridized to a high-density tiling array containing 25-mer oligonucleotides at the resolution of one every four base pairs across the entire genome. After scanning and data normalization, a high-resolution global protein occupancy profile is achieved. For each probe on the chip, protein occupancy enrichment or depletion levels are quantified using a z-score which represents the probe-by-probe relative signal intensity with respect to the mean, and normalized to the standard-deviation, of signals from replicate hybridizations of whole genomic DNA (Methods).
Figure 1. In vivo protein occupancy display (IPOD).
(A) Schematic for isolation and genome-wide display of protein-bound sites across a bacterial genome. Formaldehyde cross-linking preserves in vivo protein-DNA interactions. Following cell-lysis and sonication, protein footprints are minimized through DNase I treatment. Phenol extraction enriches for protein-DNA complexes at the interface between the aqueous and organic phases. Following interface isolation, cross-links are reversed, the resulting DNA fragments are end-labeled and hybridized to a tiling array. (B) Gel-fractionation shows that DNase I treatment leads to a drop in the mode of fragment-length distribution from ~1000 bp (no DNase I) to ~200 bp (½X DNase I), to below 100 bp for (1X DNase I). The samples were separated on the same gel and extraneous lanes were removed for clarity. (C) Cumulative probability distribution of occupancy (z-score: standard deviations from the mean) for both coding and non-coding regions determined during late exponential phase growth. The z-score values were smoothed by averaging within a moving window of 128 base pairs.
Global protein occupancy profile of the E. coli chromosome
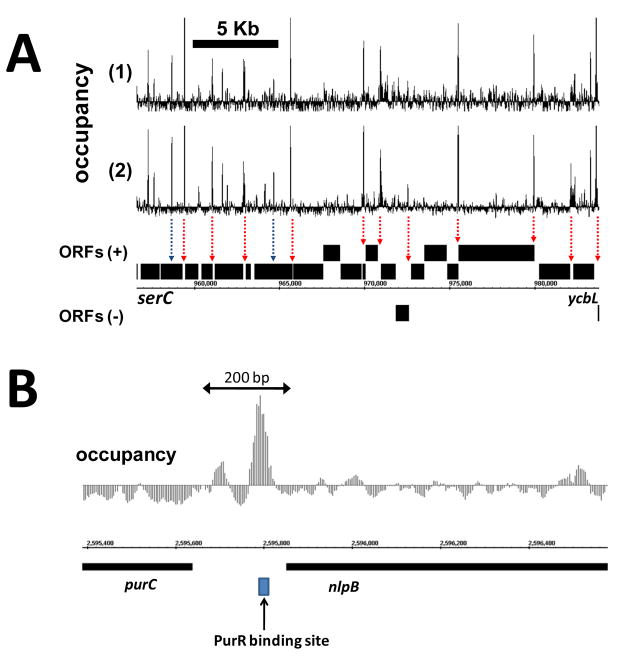
The vast fraction of characterized protein-DNA interactions occur via sequence-specific interactions of transcription factors with DNA within, and in close proximity to, non-coding regulatory regions (Gama-Castro et al., 2008). Consistent with this, we see highly significant occupancy enrichment in non-coding regions as compared to coding regions (Fig. 1C). This difference in occupancy is clearly discernable in a local chromosomal view where high-amplitude peaks are largely confined to the regions between genes (Fig. 2A). Independent biological replicates demonstrate that the position and relative amplitude of these occupancy peaks show a high level of reproducibility (Fig. 2A). Although there is, overall, relative depletion of occupancy within open reading frames (ORFs), occasionally, this is interrupted by a sharp occupancy peak (Fig. 2A, S1). The functional role of these intragenic interactions is not known, but could represent a significant gap in our understanding of bacterial gene expression. At high resolution, occupancies of individual proteins can be readily discerned, displaying footprints on the scale of a typical transcription-factor binding site (Fig. 2B, S2). An automated peak detection algorithm identified ~2063 individual occupied sites in a population of E. coli cells growing in late exponential phase (Fig. S3). The pattern of peaks is reproducible in biological replicates and shows condition-dependent variation (Fig. S4).
Figure 2. Protein occupancy profile of the E. coli genome during late exponential phase growth.
(A) At low spatial resolution, high-amplitude occupancy peaks are largely confined to intergenic (non-coding) regions of the genome (red arrows). However, similar peaks can less frequently be seen within coding regions as well (blue arrows). Two independent biological replicates show highly reproducible occupancy profiles across this region. (B) At high spatial resolution, multiple occupancy peaks are discernable within a single intergenic region. Peaks are localized to the typical footprint of individual transcription factors and often overlap experimentally determined binding sites (PurR, RegulonDB).
Discovery of Extended Protein Occupancy Domains (EPODs)
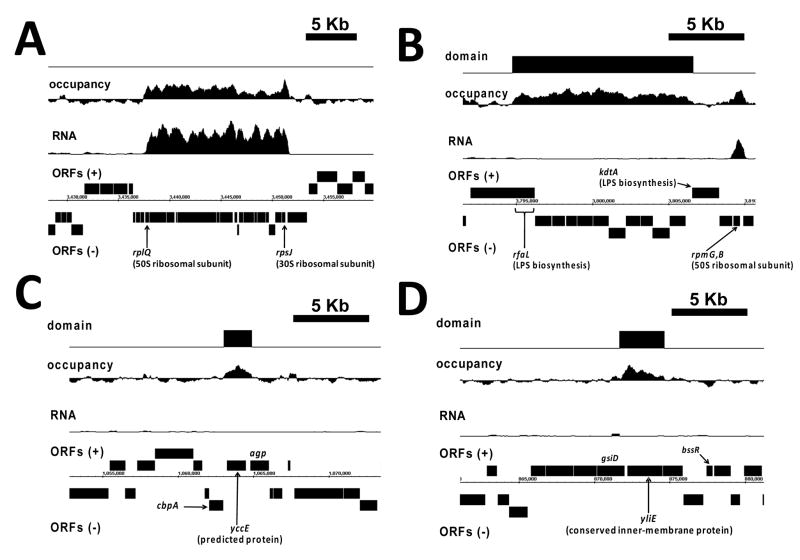
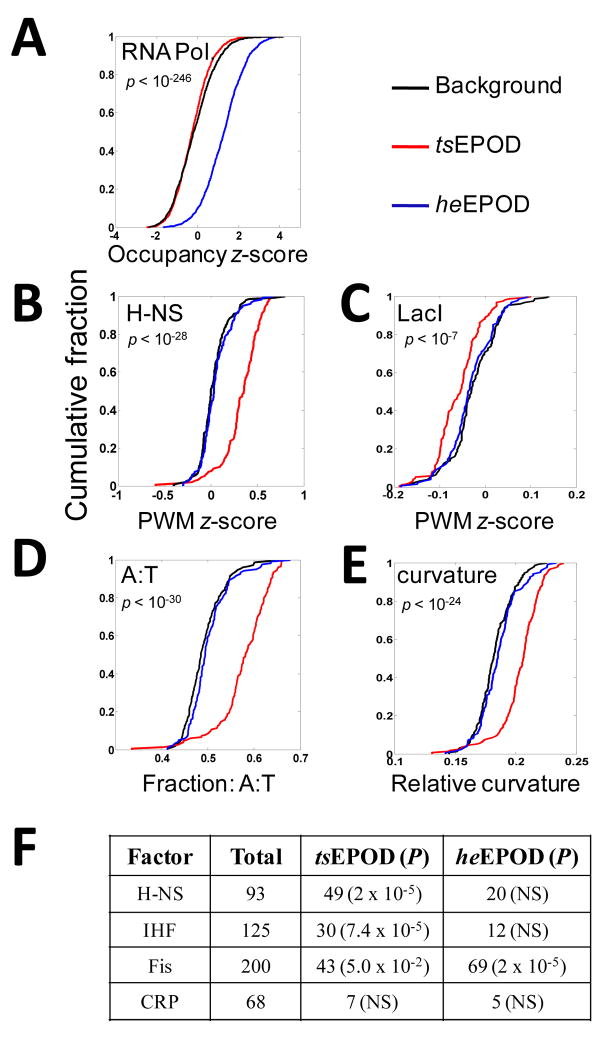
Intriguingly, examination of the entire genome-wide occupancy profile revealed contiguous regions of protein binding, many of which extend beyond a kilobase in length (Fig. 3A–D, S5). We performed a systematic search for these extended protein occupancy domains (EPODs) under early exponential growth using an automated algorithm that identified regions 1024 bp or longer with contiguous median occupancy values above the 75th percentile of all genome-wide values. These domains had a median length of 1.6 kb and extended as long as 14 kb (Fig. S6A). We wondered whether the extreme signal in these domains corresponded to the footprint of RNA polymerase within highly transcribed regions. To test this possibility, we performed transcriptional profiling under identical cellular growth conditions (Methods). As can be seen (Fig. 3A), we found clear cases where the boundaries of an EPOD coincided with those of highly transcribed regions such as those containing ribosomal protein genes (Fig. 3A). However, we found many cases where EPODs existed in a transcriptionally-silent state, across both genes and intergenic regions, and even long operons (Fig. 3B–D, S7). Due to their extreme and bimodal RNA-expression behavior, we performed an automated classification of EPODs by clustering them into two populations using their median expression level across domains (Methods and Supplementary Dataset 1). This resulted in 121 domains in the highly-expressed class (heEPODs) and 151 in the transcriptionally-silent class (tsEPODs). Previously published RNA polymerase ChIP-chip data (Grainger et al., 2005), from cells grown under identical conditions, allowed us to compare RNA polymerase occupancy of tsEPODs and heEPODs relative to a background set generated by randomly sampling genomic sequences from the overall EPOD length distribution (Fig. 4A). As expected, heEPODs showed extremely high levels of RNA polymerase occupancy (P < 10−246). In comparison, tsEPODs showed lower levels of RNA polymerase occupancy (P < 0.02) relative to control.
Figure 3. Extended protein occupancy domains (EPODs).
Protein occupancy and RNA expression profiles are shown for early exponential phase growth, smoothed by averaging within a moving window of 512 base pairs. At the bottom, open reading frames (ORFs) are annotated on both strands. Automatically detected transcriptionally-silent EPODs are shown at the top. (A) A high protein occupancy and high RNA expression domain encompassing a region with genes encoding ribosomal protein subunits. (B) An EPOD automatically detected within a transcriptionally-silent region with genes encoding LPS biosynthesis products. A neighboring region encoding 50S ribosomal protein subunits (rpmG and rpmB) shows an equal level of protein occupancy but shows a high level of RNA expression. (C,D) Transcriptionally-silent EPODs detected within genes of unknown function encoding a predicted protein, yccE (C) and a conserved inner membrane protein, yliE (D).
Figure 4. Distinct occupancy composition and biophysical properties within extended protein occupancy domains.
The cumulative distribution of various measures are shown for transcriptionally-silent EPODs (red), highly-expressed EPODs (blue), and a matched background control (black). The Wilcoxon rank sum test is used to determine statistical significance of observed deviations relative to background. (A) Experimentally-determined relative RNA polymerase occupancy. (B,C) Computationally-scored PWM binding preference for a nucleoid protein (H-NS) and a non-nucleoid transcriptional repressor (LacI) using a genome-wide relative measure (z-score). (D) Cumulative distributions of A:T frequencies within EPODs. (E) Cumulative distribution of predicted relative curvature values within EPODs. (F) Distribution of binding sites for various nucleoid proteins and CRP within tsEPODs and heEPODs.
In order to gain further insight into the potential role of EPODs, we looked for enrichment of specific functional categories in genes that overlapped them (Table S1). As expected, heEPODs were highly enriched in processes and pathways that are highly expressed, including translation and tRNAs. The most significantly enriched classes within tsEPODs were predicted and hypothetical ORFs, with marginally significant enrichment in prophage and prophage-related genes. On the other hand, tsEPODs, by and large, avoid putatively essential genes (Table S2). The number of tsEPODs, their apparently random, yet widespread genome-wide distribution, and their enrichment within transcriptionally-silent ORFs of unknown function, suggested that they may fulfill an architectural role. In fact, there exists compelling evidence that the E. coli chromosome is organized into domains, subserving both chromosomal compaction and topological domain isolation (Postow et al., 2004). Evidence for such in vivo organization comes from both genetic and biochemical studies (Garcia-Russell et al., 2007; Postow et al., 2004), including visualization of rosette-like structures by microscopy (Delius and Worcel, 1974b; Hinnebusch and Bendich, 1997; Pettijohn, 1996; Postow et al., 2004). However, the formation, composition, maintenance, and dynamics of these domains remain open questions (Bendich, 2001; Postow et al., 2004; Travers and Muskhelishvili, 2007). Investigators have argued that such domains may be organized through the binding and cooperation of abundant proteins collectively referred to as nucleoid proteins (Azam and Ishihama, 1999). These proteins have characteristics that suit them well for this task. These include high abundance, low sequence-specificity, tendency to cause DNA curvature, and propensity to bind curved DNA. In addition, some of these factors (e.g. H-NS) are known to form at least homodimeric interactions (Stella et al., 2005), a capacity that as argued previously (Dame et al., 2000; Skoko et al., 2006) may allow distant chromosomal sites to be brought together to form topologically isolated domains. Low-resolution ChIP-chip studies against known nucleoid proteins (Grainger et al., 2006) revealed both a bias toward interaction in non-coding regions and a correlation with Fis and H-NS binding, suggesting cooperative interaction of nucleoid proteins in maintaining genomic architecture.
We sought evidence for the involvement of nucleoid proteins in the formation of tsEPODs. The availability of probabilistic sequence-specificity models, in the form of position weight matrices, PWM (Gama-Castro et al., 2008), allowed us to determine the relative occupancy potential of these regions through computational analysis of a subset of these factors: H-NS, IHF, and Fis (Methods). We found that, indeed, as a population, tsEPODs have significantly higher PWM scores for all of these nucleoid proteins (e.g. for H-NS P < 10−28). The same was not true for heEPODs, as their PWM score distribution did not deviate significantly from background (Fig. 4B, Fig. S8). On the contrary, the PWM score distribution for LacI (a non-nucleoid transcription factor) showed the opposite trend, with significantly lower values (P < 10−7) within tsEPODs (Fig. 4C). Consistent with the preference of nucleoid proteins for A/T rich DNA (Cho et al., 2008; Grainger et al., 2006) we also saw a highly skewed A:T frequency bias: 59% within tsEPODs, as compared to 49% for the background and 50% for heEPODs (P< 10−30, Fig. 4D). We also found tsEPODs to display extreme biophysical characteristics (Pedersen et al., 2000) such as high curvature (P < 10−24) and stacking energy (P< 10−34), again consistent with the hypothesis that these regions constitute chromosomal organizing centers (Fig. 4E, Fig. S9). Consistent with our computational analyses above, we saw significant enrichment for the high-affinity binding of nucleoid proteins in our tsEPODs relative to background (Fig. 4F) within individual ChIP-chip profiles for H-NS, IHF, and Fis (Grainger et al., 2006). Intriguingly, we also saw a highly significant enrichment for the binding of Fis within heEPODs (Fig. 4F). This is consistent with the locus-specific role of Fis in the regulation of highly-expressed genes, including ribosomal RNAs (Aiyar et al., 2002; Cho et al., 2008; Grainger et al., 2006).
Discussion
In total, our observations argue in favor of a model in which the binding of tsEPODs by nucleoid proteins establishes them as chromosomal organizing centers. We argue that the underlying biophysical properties of these regions may largely dictate this role. IHF is known to have a preference for curved DNA, causing it to bend sharply upon binding; the nucleoid proteins HU and H-NS bind strongly to curved DNA as well (Swinger and Rice, 2004). Fis, H-NS, and IHF restrain supercoils (Pettijohn, 1996) and both H-NS (Dame et al., 2000) and Fis (Skoko et al., 2006) show oligomerization and DNA compaction in vitro. We propose that nucleation starts with nucleoid proteins preferentially binding these curved regions of DNA. Because several of the nucleoid proteins prefer to bind curved DNA, these initial protein-DNA interactions make the region more favorable for further binding events. In this way, a wave of nucleoid proteins may spread across these regions, reinforced through the maintenance of curvature and intra-domain protein-protein interactions. Homo- and heterodimeric protein-protein interactions, for example as shown for H-NS (Stella et al., 2005) can then bring these domains in contact with each other, forming the classic rosette structures visualized by EM (Delius and Worcel, 1974a; Postow et al., 2004).
Our observations do not suggest that every tsEPOD is essential to chromosomal organization at all times. Rather, a subset of tsEPODs could be involved in the formation of higher-order structure in any one cell, or across different environmental conditions. For relevant discussions see (Deng et al., 2005; Postow et al., 2004; Valens et al., 2004). The lack of any discernable fitness deficit for a reduced genome E. coli strain, MDS42 (Kolisnychenko et al., 2002), which is missing 24% of the ORFs contained in tsEPODs, supports this dynamic and redundant picture. In fact, IPOD analysis of this reduced genome showed that the occupancy pattern of the remaining EPODs is largely preserved, with 44% of EPOD sequences in MDS42 exactly overlapping those defined in MG1655 (Fig. S10). Although there are a minority of loci with substantially different occupancy patterns, most of the residual discrepancy is due to differences in the exact definition of EPOD boundaries and not their locations. These observations provide additional support for our proposed model. Namely that specific chromosomal regions, by the virtue of their sequence composition, act as extended protein occupancy domains, which in turn may allow them to participate in organizing large-scale chromosomal topology. However, we also raise the possibility that the establishment of these transcriptionally-silenced protein occupancy domains may subserve other functions. For example, others have argued for the role of nucleoid proteins such as H-NS in the silencing of horizontally-transferred DNA (Dorman, 2007).
A closer inspection of some EPODs suggests that our automated classification of them into the two groups of highly expressed and transcriptionally silent may not capture the full range of their diversity. Indeed, one of the longest tsEPODs is defined over a cluster of genes encoding enzymes in the pathway of lipopolysaccharide (LPS) biosynthesis (Fig. 3B). Analysis of strand-specific RNA-abundance of this locus (Fig. S11) clearly shows that although this region is classified as transcriptionally ‘silent’, there is low-level expression which is mostly confined to the first three genes in the operon (rfaQ, rfaG, and rfaP). These observations suggest that extended protein occupancy may be present at loci with low-level expression, and that it may be caused by processes that are distinct from those operating at absolutely silent loci.
We have developed IPOD, a global, in vivo approach for monitoring the protein occupancy of an entire bacterial genome at the resolution of individual binding sites. Aqueous/organic phase separation has been previously used to enrich on the basis of nucleosome density in S. cerevisiase (Nagy et al., 2003), and Grainger et al demonstrated that cross-linked RNA-polymerase bound sequences are preferentially partitioned to the organic phase in E. coli (Grainger et al., 2006). Here we have shown that localization of small nucleoprotein complexes at the aqueous/organic interface is a simple yet powerful strategy for profiling protein occupancy across an entire prokaryotic genome. Although the identity of the protein bound at each site is not known, increasingly accurate sequence-specificity models of protein-DNA interactions should allow probabilistic assignments to known DNA-binding proteins. In fact, since IPOD analysis allows measurements of correlated occupancy of many sites across different conditions, it should aid in the refinement of existing sequence-specificity models and the discovery of new ones.
The ability to simultaneously monitor both protein occupancy and transcriptional output, at high spatial and temporal resolution, promises to allow true systems-level modeling of transcriptional network dynamics and chromosomal organization. At large spatial scales, these data have revealed the existence of transcriptionally-silent protein occupancy domains. Our diverse observations implicate these regions as the long proposed domain organizing centers of the E. coli chromosome.
Supplementary Material
Acknowledgments
We thank the members of the Tavazoie laboratory for helpful comments on the manuscript. TV was supported by a NASA pre-doctoral fellowship. AKH was assisted by fellowship #08-1090-CCR-EO from the New Jersey State Commission on Cancer Research. ST was supported by grants from the NSF (CAREER), DARPA, NHGRI, NIGMS (P50 GM071508), and the NIH Director’s Pioneer Award (1DP10D003787-01). The oligonucleotide array data are deposited at NCBI Gene Expression Omnibus with accession number GSE16414.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiyar SE, McLeod SM, Ross W, Hirvonen CA, Thomas MS, Johnson RC, Gourse RL. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J Mol Biol. 2002;316:501–516. doi: 10.1006/jmbi.2001.5390. [DOI] [PubMed] [Google Scholar]
- Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- Bendich AJ. The form of chromosomal DNA molecules in bacterial cells. Biochimie. 2001;83:177–186. doi: 10.1016/s0300-9084(00)01209-8. [DOI] [PubMed] [Google Scholar]
- Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide Analysis of Fis Binding in Escherichia coli Indicates a Causative Role for A-/AT-tracts . Genome Res. 2008 doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H, Worcel A. Electron microscopic studies on the folded chromosome of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1974a;38:53–58. doi: 10.1101/sqb.1974.038.01.008. [DOI] [PubMed] [Google Scholar]
- Delius H, Worcel A. Letter: Electron microscopic visualization of the folded chromosome of Escherichia coli. J Mol Biol. 1974b;82:107–109. doi: 10.1016/0022-2836(74)90577-4. [DOI] [PubMed] [Google Scholar]
- Deng S, Stein RA, Higgins NP. Organization of supercoil domains and their reorganization by transcription. Mol Microbiol. 2005;57:1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Russell N, Orchard SS, Segall AM. Probing nucleoid structure in bacteria using phage lambda integrase-mediated chromosome rearrangements. Methods Enzymol. 2007;421:209–226. doi: 10.1016/S0076-6879(06)21017-6. [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Bendich AJ. The bacterial nucleoid visualized by fluorescence microscopy of cells lysed within agarose: comparison of Escherichia coli and spirochetes of the genus Borrelia. J Bacteriol. 1997;179:2228–2237. doi: 10.1128/jb.179.7.2228-2237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolisnychenko V, Plunkett G, 3rd, Herring CD, Feher T, Posfai J, Blattner FR, Posfai G. Engineering a reduced Escherichia coli genome. Genome Res. 2002;12:640–647. doi: 10.1101/gr.217202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci U S A. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AG, Jensen LJ, Brunak S, Staerfeldt HH, Ussery DW. A DNA structural atlas for Escherichia coli. J Mol Biol. 2000;299:907–930. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- Pettijohn DE. In: The NucleoidEscherichia coli and Salmonella. Neidhardt F, editor. Washington DC: ASM; 1996. pp. 158–166. [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Skoko D, Yoo D, Bai H, Schnurr B, Yan J, McLeod SM, Marko JF, Johnson RC. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J Mol Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. Embo J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Church GM. Quantitative whole-genome analysis of DNA-protein interactions by in vivo methylase protection in E. coli. Nat Biotechnol. 1998;16:566–571. doi: 10.1038/nbt0698-566. [DOI] [PubMed] [Google Scholar]
- Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. Embo J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.