Abstract
Epstein-Barr Virus (EBV) encodes multiple microRNAs (miRNAs) from two primary transcripts, BHRF1 and the BARTs. The expression of BHRF1 miRNAs is dependent on the type of viral latency, whereas the BART miRNAs are expressed in cells during all forms of latency. It is not known how these miRNAs are otherwise regulated, though. We have used quantitative, stem-loop, real-time PCR to measure the expression of EBV’s miRNAs and found them to differ nearly 50- and 25-fold among all tested cell lines and among EBV-positive Burkitt’s lymphomas, respectively. In addition, the expression of individual BART miRNAs within a cell can differ by 50-fold or more despite the fact these miRNAs are likely transcribed together as a single primary transcript. These measurements are illuminating: they indicate that few of EBV’s miRNAs are expressed at levels comparable to those of cellular miRNAs in most cell lines and therefore likely function interdependently.
Keywords: Epstein-Barr Virus, microRNAs, BamHI A rightward transcripts, stem-loop real-time PCR
Introduction
Epstein-Barr virus (EBV) is a successful pathogen that infects more than 90% of all adults in the world (Williams and Crawford, 2006). While adapting to its host EBV has evolved properties that make it particularly interesting to study. It maintains its DNA as a licensed, extra-chromosomal replicon. This viral replicon is present in clonally derived cells with a wide distribution of numbers of DNA molecules per cell (Nanbo et al., 2007). Failures in its synthesis and faithful partitioning drive these distributions both to increase and decrease the numbers of DNA molecules per cell during cellular proliferation (ibid.). This property means that the templates encoding viral genes vary widely in otherwise genetically identical cells and raises the general question of how the expression of these genes is controlled. A second recently appreciated property of EBV is that it encodes about 30 mature miRNAs from 20 pre-miRNAs which, given the size of its genome (approximately 165 kbp), represents a thousand-fold enrichment of this class of genes relative to those in its human host (Cai et al., 2006; Grundhoff et al., 2006; Landgraf et al., 2007; Pfeffer et al., 2004). MiRNAs function dose-dependently with two-fold differences in their levels having significant biological consequences (Raveche et al., 2007; Xiao et al., 2007). That EBV encodes such a relatively high number of miRNAs indicates its evolutionary selection for them and raises the question of how the expression of these particular viral genes is controlled.
EBV’s miRNAs are encoded in two primary transcripts, the BHRF1 transcript which also encodes the BHRF1 orf and the BamHI A rightward transcripts (BARTs) which are a set of long, alternatively spliced transcripts, which may encode the BARF0 orf (Fig. 1). The BARTs contain two clusters of miRNAs, which have been termed Cluster 1 and Cluster 2 (Fig. 1). Some of these miRNAs have been characterized by their being cloned; some have been detected by Northern blotting; while others have been predicted by various computational approaches (Cai et al., 2006; Grundhoff et al., 2006; Landgraf et al., 2007; Pfeffer et al., 2004). These analyses have detected different signal intensities of EBV’s miRNAs in Northern blots but have not determined the numbers of molecules in the various cells studied. We therefore have used stem-loop real-time PCR and synthetic standards to quantify the average number of EBV’s miRNAs in 17 cell lines under various conditions. We have assayed miRNAs encoded by the BHRF1 transcript and the BARTs to identify the conditions under which they are co- or differentially regulated. We have also measured the levels of the miRNAs following infection of primary B-cells by EBV and in cell lines induced to support the viral lytic cycle in order to elucidate their regulation throughout the viral life-cycle.
Fig. 1.
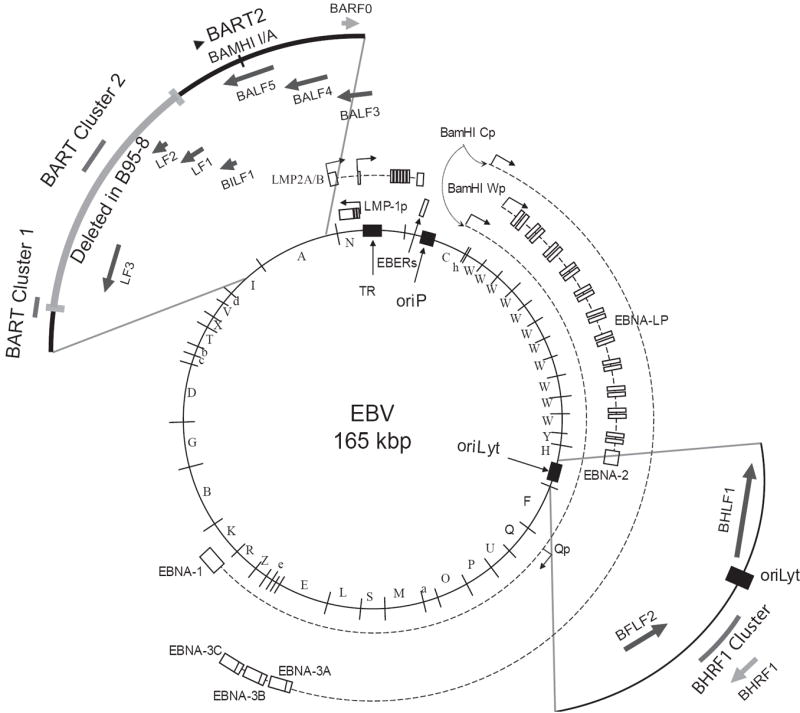
EBV’s miRNAs are processed from two transcripts. The mRNA encoding BHRF1 encodes BHRF1-1 in the 5’ untranslated region (UTR), and BHRF1-2 and BHRF1-3 are encoded in the 3’ UTR of the transcript. The BARTs encode 28 miRNAs found in two clusters. BART Cluster 1 encodes 11 mature miRNAs from 8 pre-miRs in a 1kb region, whereas BART Cluster 2 encodes 15 mature miRNAs from 11 pre-miRs in a 3 kb region. The two clusters are separated by 5 kb. BART2 is encoded near the end of the genome, encodes two mature miRNAs, and is complementary to BALF5, the EBV lytic DNA polymerase. In the B95-8 strain of EBV a 14 kb region of the BARTs encoding six BART Cluster 1 miRNAs and all BART Cluster 2 miRNAs are deleted.
Our measurements indicate that the primary transcripts encoding EBV’s BART miRNAs likely encode an intrinsic mode of regulation such that the rank order of individual miRNAs between cells is similar. These relative levels, for example, are recapitulated when a primary transcript encoding most BART miRNAs is expressed from a retroviral vector in EBV-negative cells. The measurements show also that the absolute levels of the miRNAs vary by up to 50-fold between cell types and are independent of the average number of viral templates per cell. These findings make it likely that the expression of EBV’s miRNAs is regulated intrinsically by their primary transcripts and likely also by the efficiency of transcription of those primary transcripts.
Results
EBV’s BART miRNAs accumulate to different levels among EBV-infected cell lines
It has been shown that the levels of BART miRNAs probably differ in EBV-positive BLs (Cai et al., 2006; Edwards et al., 2008; Lo et al., 2007; Pfeffer et al., 2004; Xia et al., 2008). We wanted to measure both the number of individual miRNAs within one cell line and their numbers between cell lines. These numbers would help determine which mRNAs could be targeted by EBV’s miRNAs and the extent to which given mRNAs could be regulated by the viral miRNAs. We used stem-loop, quantitative, real-time PCR (qPCR) to measure eight BART miRNAs and the BHRF1-3 miRNA along with known quantities of synthetic miRNAs identical to the mature miRNA sequence to generate standard curves for these experiments. The miRNAs analyzed were selected to span the range of levels of expression derived from published Northern blots (Cai et al., 2006; Edwards et al., 2008; Grundhoff et al., 2006; Lo et al., 2007). We assayed six kinds of EBV-infected cells to uncover possible relationships between the structure of the viral genome, its overall pattern of transcription, and the steady-state levels of viral miRNAs. These cell lines included ones with large deletions within the coding regions for BART Cluster 1 and Cluster 2 (B958IID6 and 721) (Figure 1); Type I BLs which express the fewest viral genes among EBV-positive tumors (Akata, BL-5, Dante, KemI, MutuI, and SavI); Type III BLs which have evolved upon propagation in cell culture to express additional viral genes found in type III latency (MutuIII and Raji); variant BLs with deletions in EBNA2 (Daudi, OkuI, and GG68); and carcinoma cell lines (C666-1 and SNU719). The numbers of molecules of each miRNA per 10 pg of total RNA were determined in 17 EBV-positive cell lines (Fig. 2). No miRNA was detected at more than 1 molecule per 10 pg of total RNA in the EBV-negative BL cell line, BJAB (data not shown). We chose to compare the number of miRNA molecules per unit of total RNA mass because different amounts of total RNA were isolated per cell among all the cell lines (Supplementary Table 2). In measuring the amount of a miRNA per 10 pg of RNA, we compared the expression of each miRNA in relation to all RNAs in the cell, which may be a more accurate measurement for the activity of a miRNA than total number per cell. We were able to detect both mature BART15 and BART20-5p, which have been largely undetected by Northern blot (Grundhoff et al., 2006; Lo et al., 2007). BART15 was expressed at levels similar to BART1-5p, where BART20-5p was consistently the least well expressed of all the BART miRNAs assayed.
Fig. 2.
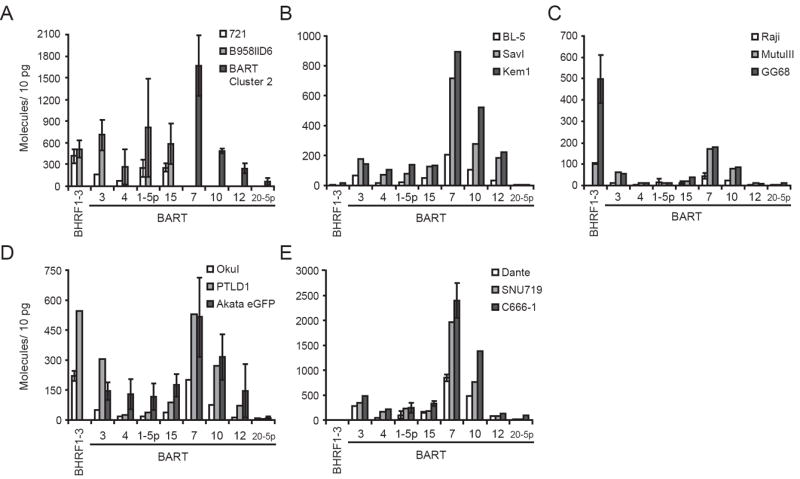
Some EBV miRNAs are expressed in all tested EBV-positive cell lines, but do not accumulate to the same steady-state levels. B95-8 infected LCLs (A), type I BLs (B, D, E), type III BLs (C, D), and recombinant EBV (D), PTLD (D), NPC (E), and gastric carcinoma (E) cell lines were assayed for their steady-state levels of one BHRF1 miRNA and eight BART miRNAs by stem-loop qPCR. We also measured the expression of EBV miRNAs in a BJAB clone, which conditionally expresses EBV’s BART Cluster 2 miRNAs (A). Steady-state levels of each miRNA were calculated by generating standard curves with known quantities of synthetic miRNAs identical to the mature miRNA sequence. Each figure has a different range on the y-axis depending on the levels of the miRNAs in the cell lines assayed. The error bars for BHRF1-3 and BARTs 3, 1-5p, 7, and for all cell lines in (A), Raji and MutuIII (C), OkuI (D), and Dante and C666-1 (E) represent the standard deviation of the average of two independent experiments, each consisting of triplicate qPCR measurements. For Akata eGFP (D), all miRNAs were measured in more than one independent experiment. The samples without error bars are the average of triplicate qPCR measurements from one experiment. BART miRNAs are shown in the order as they appear in the wild-type EBV genome. BARTs 3, 4, 1-5p, and 15 are encoded by BART Cluster 1, and miR-BARTs 7, 10, 12, and 20-5p are encoded by BART Cluster 2.
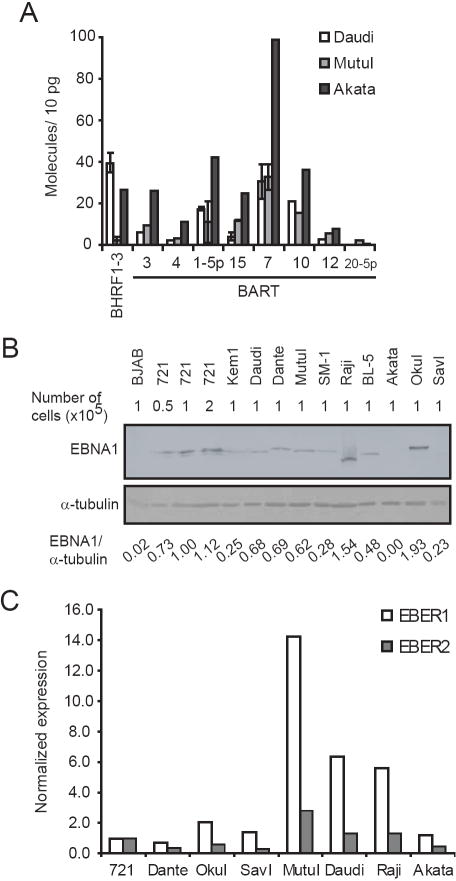
BHRF1-3 accumulated in cells in which EBV exhibited a type III latency (Fig. 2A, C, D) and was also detected in five BL cell lines not known to express BHRF1 miRNAs (Fig. 2B, D, 4A). OkuI cells expressed BHRF1-3 at similar steady-state levels to type III BLs (~200 molecules per 10 pg of total RNA), while BL-5, KemI, Daudi, and Akata all expressed BHRF1-3 at one-fifth or less the levels of type III BLs. The steady-state levels of BART miRNAs differed among EBV-positive cell lines. Expression of each miRNA was normalized to the expression across all cell lines maintaining the full-length EBV genome (Fig. 3). In agreement with what has been previously estimated with Northern blots, EBV BART miRNAs were differentially expressed among cell lines. We found the levels of expression of BART miRNAs to be greatest in the NPC cell line, C666-1, and the gastric carcinoma cell line, SNU719, which were at least two-fold greater than in any of the BL cell lines tested (Fig. 3). On average, the expression of BART miRNAs in C666-1 cells were 50-fold more abundant than in Daudi cells (Fig. 3). Even in comparing BL cell lines we saw a 25-fold greater expression of BART miRNAs in KemI than in Daudi cells, which are the BLs that express the most and least miRNAs, respectively. The Akata eGFP was found to express BART miRNAs 7-fold greater on average than Akata. This difference likely reflects Akata’s evolving to lose its EBV and Akata eGFP being a derivative of Akata which had lost EBV and into which EBV was newly introduced. The original EBV genomes in Akata likely accumulated methylations and possibly other forms of epigenetic regulation not shared by the EBV newly introduced into Akata eGFP.
Fig. 4.
Three BLs that spontaneously lose EBV in culture express low levels of BART miRNAs, but do not have a general defect in transcription. MutuI, Akata, and Daudi cells were assayed for expression of BHRF1-3 and eight BART miRNAs by stem-loop qPCR (A). To confirm expression was not a general defect in transcription, EBNA1 protein expression (B) was measured by western blot, and expression of the EBV non-coding RNAs, EBER1 and EBER2, was measured by qPCR (C). Expression of EBER1 and EBER2 was normalized to the expression of the cellular mRNA, GAPDH.
Fig. 3.
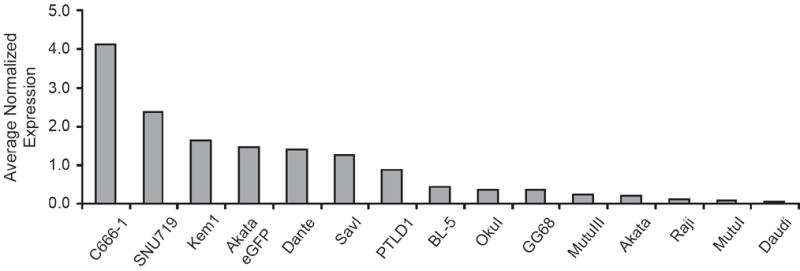
The expression of EBV’s BART miRNAs differ among EBV-positive cell lines. The steady-state levels of each BART miRNA assayed by stem-loop qPCR were normalized to the average levels of that BART miRNA across 15 cell lines maintaining full-length EBV. Shown is the average normalized expression for the eight assayed BART miRNAs. The error bars represent the standard deviation of the average expression for the eight assayed BART miRNAs.
Although the absolute levels of EBV’s BART miRNAs differ among EBV-infected cell lines, the ranked order of the levels of individual BART miRNAs are similar among these cell lines
Measuring the eight assayed BART miRNAs revealed that though the genomic locations of these miRNAs are clustered, their steady-state levels differ from each other in different cell lines. For instance, C666-1 cells expressed BART7 at 6-fold greater levels than BART12, and 50-fold greater than BART20-5p, all three being encoded in BART Cluster 2. BART7’s expression was at least two-fold greater than any assayed miRNA encoded by BART Cluster 1 (Fig. 2E). BART20-5p consistently had the lowest steady-state levels, while BART7 and BART10 were among the highest in all assayed EBV-positive cell lines (Fig. 2).
These measurements demonstrated that EBV-positive cells apparently express their different miRNAs relative to each other similarly. To test this possibility, each of the eight assayed BART miRNAs was ranked based on its steady-state levels in each cell line, and then compared to the ranked expression of the same eight BART miRNAs in all other assayed cell lines. The ranked order of expression of the eight assayed BART miRNAs was statistically significant for 90 of 105 (86%) pairwise comparisons (Spearman rank correlation, p<0.05, Supplementary Fig. 1). The rank order of this expression is independent of cell type; epithelial cell lines and B cell lines both exhibit a similar rank order. This analysis shows that the ranked orders of the steady-state levels of BART miRNAs in most EBV-positive cell lines are similar.
This conclusion is confirmed in the studies of the 721 and B958IID6 cells which are both immortalized by the B95-8 strain of EBV. This strain contains a 14 kbp deletion in the BARTs that removes the coding sequence for part of BART Cluster 1 and all of the Cluster 2 miRNAs (Figure 1). In these cells, the coding sequence for BARTs 3, 4, 1-5p and 15 are all retained. As with the full-length virus, BART4 was expressed at the lowest levels, whereas expression of BARTs 3, 1-5p, and 15 were similar to each other (Fig. 2A). Thus the ranked order of expression of EBV’s BART miRNAs is maintained even when a large portion of the DNA encoding the primary transcript for the BARTs is deleted. That the relative levels of individual viral miRNAs is similar between cell lines is also supported by our expressing BART Cluster 2, which is deleted in the B95-8 strain of EBV, in the EBV-negative cell, BJAB. The order of the steady-state levels of these miRNAs encoded by Cluster 2 in BJAB cells is similar to that in most EBV-positive cell lines (Fig. 2A).
Three BLs that spontaneously lose EBV express the BART miRNAs less efficiently than do the BLs that maintain EBV stably
Three EBV-positive BL cell lines expressed surprisingly low levels of the individual BART miRNAs. These cell lines, Akata, MutuI, and Daudi all spontaneously lose EBV in cell culture (Kitagawa et al., 2000; Nanbo et al., 2002; Shimizu et al., 1994). The levels of miRNAs expressed by these cells are so low that measuring their relative levels becomes difficult (Fig. 4A). In contrast, the type III derivative of MutuI, MutuIII, which maintains EBV in culture, had on average 2.5-fold more of each BART miRNA than did MutuI cells (Fig. 3). We tested if these low levels of miRNAs result from some cells having already lost their EBV genomes. By FISH 90%, 98%, and 84% of Akata, MutuI, and Daudi cells were EBV-positive, respectively, at the time of RNA isolation. Thus the observed low steady-state levels of BART miRNAs in these cell lines did not result from the cells having lost EBV. We asked whether the inefficient expression of BART miRNAs in these cell lines was specific to the miRNAs, or reflected a general defect in transcription. Expression of EBNA1 protein was measured in the Daudi, MutuI, and Akata cell lines and compared to expression in EBV-positive cell lines that do not lose EBV spontaneously (Fig. 4B). Both Daudi and MutuI cells expressed EBNA1 at levels similar to other BL cell lines. We could not detect EBNA1 in the Akata cells with our antisera although it has been shown to be expressed normally in earlier studies (Komano et al., 1998). We also measured the expression of the EBV non-coding EBERs, EBER1 and EBER2 by qPCR (Fig. 4C). All three cell lines expressed both EBER1 and EBER2 at levels comparable to cell lines that efficiently expressed BART miRNAs. The inefficient expression of BART miRNAs in Akata, Daudi, and MutuI cells therefore is not caused by a general defect in the expression of other viral genes. This finding raises the possibility that BART miRNAs provide a selective advantage to EBV-positive lymphomas that retain EBV but not to those that lose the virus. Raji cells, which have not been shown to lose EBV in culture, also have low levels of EBV’s BART miRNAs (Fig. 2C, 3). The origin of viral DNA synthesis used primarily in Raji cells is located within the BART locus which may limit expression of this locus (Norio et al., 2000; Wang and Sugden, 2008).
The steady-state levels of BART miRNAs are not determined by the number of viral templates per cell
The wide variation in the average normalized expression of viral miRNAs in different B cell lines led us to ask if these levels correlated with the average number of viral genomes per cell. Eight EBV-positive BL cell lines that stably maintain the virus were assayed for their number of viral genomes per cell by FISH and compared to the average normalized, steady-state levels of the eight assayed BART miRNAs. The steady-state levels of the BART miRNAs did not correlate with the average number of viral templates (Fig. 5, Spearman rank correlation, p>0.05). This was also true when we compared the number of viral templates to the expression of the BART miRNAs on a per cell, rather than per 10 pg scale (data not shown, Spearman rank correlation, p>0.05). Raji, which has the greatest number of viral genomes, had the lowest steady-state levels of miRNAs among the assayed cell lines. Dante and BL-5 cell lines had the same number of viral templates per cell, but Dante expressed on average three-fold greater levels of each assayed BART miRNA than did BL-5 cells. Both GG68 and OkuI cells had a third the number of the genomes as did BL-5, but on average expressed similar levels of each assayed BART miRNA per cell. Finally, SavI, GG68, and OkuI cells had similar distributions of the number of viral templates per cell, but SavI expressed on average nearly three-fold more of the eight assayed BART miRNAs than did GG68 and OkuI cells.
Fig. 5.
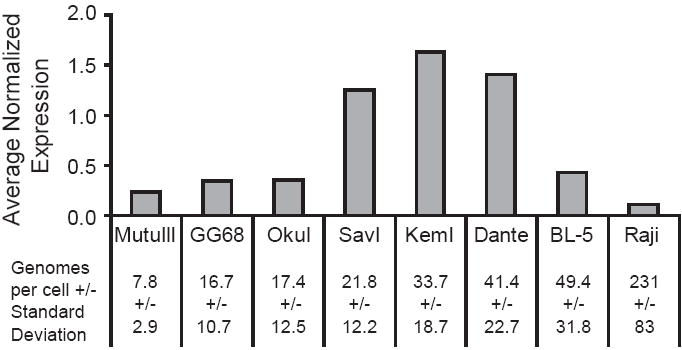
Steady-state levels of BART miRNAs are not determined by the number of viral templates per cell. Eight EBV-positive BL cell lines were assayed for the number of genomes per cell by FISH, and for the levels of BART miRNAs by stem-loop qPCR. Cell lines are ordered left to right in increasing number of viral templates. The average normalized expression for eight assayed BART miRNAs is shown as in Fig. 3. The error bars represent the standard deviation of the average normalized expression.
We were surprised to find that Raji contained on average 230 viral genomes per cell. This is nearly 5-fold higher than previously reported by qPCR (Huang et al., 2003). We confirmed that the cells were Raji with PCR by amplifying the region spanning the EBNA3C deletion identified previously in Raji EBV (Hatfull et al., 1988) and data not shown).
The expression of BART and BHRF1 miRNAs is not increased substantially during productive infection
The absence of a correlation between the number of viral templates in BLs during latent infection and their expression of viral BART miRNAs led us to ask if the large increase in the numbers of viral DNA molecules in cells supporting the lytic cycle would affect expression of the miRNAs. Two EBV-positive cell lines, Akata and B958 were induced to support their lytic cycle, and the levels of eight BART and all 3 BHRF1 miRNAs were measured for 93 hours post induction (hpi). The levels of BART miRNAs remained unchanged in cells induced to support the lytic cycle, though the number of available viral templates increased nearly 50-fold in both cell lines (Fig. 6C, D, E, F). BART7, which is deleted in B958, was not detected above background upon induction of lytic replication in this cell line (data not shown). The levels of the BHRF1 miRNAs in the B958 cells remained unchanged, while their levels increased between 4- and 20-fold in the induced Akata cells (Fig. 6A, B). The levels of expression of the three BHRF1 miRNAs in the induced Akata cells were much lower, however, than those found in BLs that exhibit type III latency (Fig. 2A, 6B). The BHRF1 miRNAs are thus regulated differently in the B958 and Akata cells either because of the different viral strains or their different host cells.
Fig. 6.
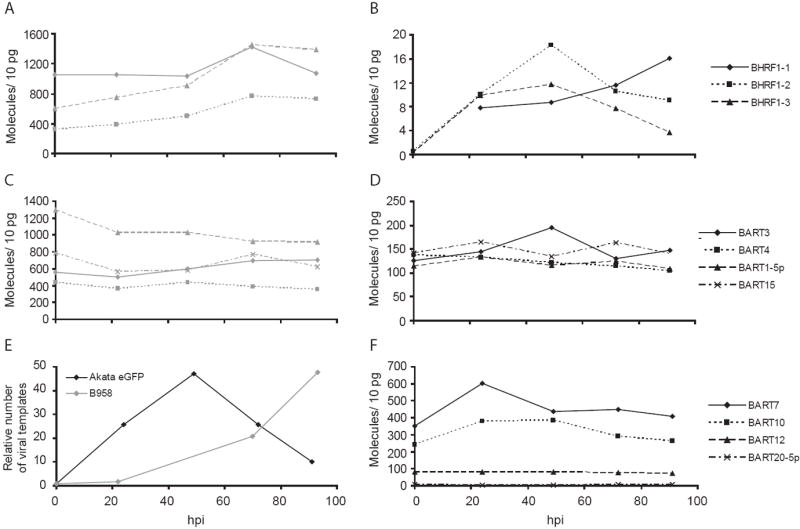
EBV BART miRNAs do not correlate with the expression of viral templates in lytic Akata eGFP or B958IID6 cells. However, BHRF1 miRNAs in Akata eGFP do increase in expression upon lytic induction. Akata eGFP and B958ID6 cells were induced to support lytic replication. B958IID6 cells were harvested at 22, 47, 70, and 93 hours post induction (hpi) and Akata eGFP cells were harvested at 24, 49, 72, and 91 hpi. Expression of the BHRF1 (A) and BART (C) miRNAs in B958IID6 cells were measured by stem-loop qPCR. Likewise, expression of BHRF1 (B), BART Cluster 1 (D), and BART Cluster 2 (F) miRNAs were measured in induced Akata eGFP cells. BHRF1-1 was not detected above background in Akata eGFP cells until 24 hpi. The relative number of viral templates present in induced B958IID6 and Akata eGFP cells is shown (E). B958IID6 measurements are shown in gray, and Akata eGFP measurements are shown in black.
The expression of EBV BART miRNAs increases over the first 8 days post infection
We measured when during infection of primary B cells EBV miRNAs are expressed. Primary B cells were infected with either the B95-8 or the Akata strain of EBV at an MOI of 1, cells were harvested at 1, 2, 4, and 8 days post infection (dpi), and assayed for their expression of miRNAs (Fig. 7). These miRNAs were detectably expressed by 2 dpi, and increased in expression throughout the first 8 dpi (Fig. 7A, B, C). In an independent experiment, where we used a miRNA microarray with probes to EBV’s miRNAs to detect expression of the viral miRNAs, expression of EBV’s miRNAs could be detected 10-fold above background 20 hours post infection (data not shown). Not surprisingly, BART7 was expressed at the highest level in Akata EBV-infected cells (Fig. 7D). BART7 was not detected above background in cells infected with B95-8 EBV (data not shown). We observed that the BART miRNAs in Akata EBV-infected cells were expressed in a similar ranked order as are established cell lines that maintain EBV (Fig. 7D). The mature BART and BHRF1 miRNAs were expressed as early as 2 dpi, and thus could affect regulation of viral and host genes at the beginning of the viral life-cycle.
Fig. 7.
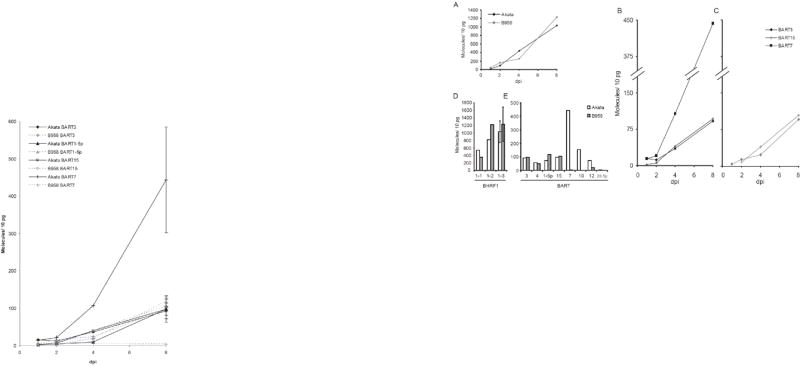
EBV miRNAs increase in expression during the first 8 days post infection (dpi) of primary B cells, and are detected as early as 2 dpi. Expression of BHRF1-3 (A) and BART miRNAs in primary B cells infected with Akata (B, black lines) or B95-8 (C, gray lines) EBV was measured during the first 8 dpi. BART15 was not detected in cells infected with B95-8 EBV until 2 dpi (C). At 8 dpi, expression of BHRF1 miRNAs (D) and eight BART miRNAs (E) was measured.
Raji EBV does not express mature BHRF1-3 because of a mutation in its precursor-miRNA
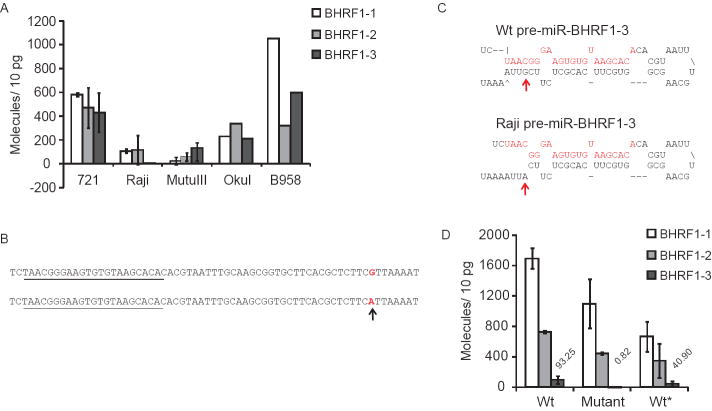
Raji cells, which exhibit type III latency, did not express detectable levels of BHRF1-3. We assayed the three BHRF1 miRNAs by stem-loop qPCR in various EBV-positive cell lines that were found to express BHRF1-3 to determine if Raji’s apparent failure to express this viral miRNA was exceptional. While all cell lines assayed expressed BHRF1-1 and BHRF1-2, only Raji failed to express BHRF1-3 detectably (Fig. 8A).
Fig. 8.
Raji EBV does not express detectable BHRF1-3. Expression of BHRF1 miRNAs in Raji, 721, MutuIII, OkuI and B958IID6 (B958) cell lines was measured by stem-loop qPCR1 (A). Error bars represent the standard deviation of the average expression from at least two independent measurements. Sequence alignment of the wildtype and Raji revealed a single nucleotide mutation in the genomic region encoding pre-miR-BHRF1-3 (B). The mature miRNA sequence is underlined, and the mutation is indicated by an arrow. (C) The stem-loop structures of the wildtype and Raji pre-miR-BHRF1-3 were predicted by Mfold (C). The mature BHRF1-3 sequence is highlighted in red, and the mutation is indicated by an arrow. The steady-state levels of the BHRF1 miRNAs were measured in 293 cells transfected with vectors encoding the BHRF1 sequence from B95-8 EBV (Wt), Raji EBV (Mutant), or Raji EBV in which the mutation in the pre-miR-BHRF1-3 was reverted to the Wt sequence (Wt*) (D). Transfection efficiencies of each vector were determined to be 6.5~9% by calculating percentage of GFP positive cells 48 hours post transfection. Expression of BHRF1 miRNAs was assayed by stem-loop qPCR from total RNA isolates. The number of molecules per 10 pg for BHRF1-3 in the three transfections are listed above the BHRF1-3 bars. Background levels for BHRF1-3 were found to be less than one copy per cell in EBV-negative BJAB cells. Error bars represent the standard deviation of duplicate experiments.
We analyzed this defect by isolating a 2.5 kb fragment of EBV DNA containing the BHRF1 mRNA coding sequence from both 721 and Raji cells, cloned, and sequenced these DNAs. There was a single nucleotide mutation in Raji EBV that mapped to the predicted stem sequence of the BHRF1-3 precursor-miRNA (pre-miR), introducing an A:C mismatch in place of a G:C base pair (Fig. 8B). This mutation was confirmed by independently amplifying and sequencing the Raji EBV a second time. Mfold (Mathews et al., 2007; Zuker, 2003) predicted that the A:C mismatch in Raji EBV would disrupt base-pairing near the base of the stem-loop structure of pre-miR-BHRF1-3, shortening the length of the pre-miR stem, and making it likely to inhibit its maturation (Fig. 8C). We tested whether this mutation affected the processing of BHRF1-3 by reverting the mutation in pre-miR-BHRF1-3 of Raji EBV to that of the B95-8 sequence by site-directed mutagenesis. Vectors encoding the three BHRF1 miRNAs from Raji EBV DNA (Mutant), from B95-8 EBV DNA (Wt), and the reverted sequence (Wt*) were introduced into 293 cells. While the mature BHRF1-3 was expressed from the B95-8 derived vector but not detectably from the Raji-derived vector, it was expressed from the reverted Raji-derived vector (Fig. 8D). BHRF1-3 was measured at an approximately 50-fold higher level than the limit of detection in cells transfected with the reverted sequence as compared to the Raji-derived vector, indicating that the mutation detected in Raji cells underlies this strain of EBV’s defect in expressing this miRNA. Thus for strains of EBV with functional BHRF1 miRNA genes, the three genes are expressed coordinately.
Discussion
EBV’s miRNAs have been detected by Nothern blotting or cloning (Cai et al., 2006; Grundhoff et al., 2006; Landgraf et al., 2007; Pfeffer et al., 2004). Among these, BART7, 10, and 12 have been most frequently detected and BART15 and 20-5p rarely or not at all (Cai et al., 2006; Edwards et al., 2008; Grundhoff et al., 2006; Kim do et al., 2007; Lo et al., 2007). We have used stem-loop real-time PCR along with synthetic miRNAs to quantify these five viral miRNAs and six additional ones in a biologically diverse set of cell lines to determine their levels of accumulation within and between these cell lines.
These measurements indicate the two sets of viral miRNAs expressed from individual primary transcripts, the BHRF1 miRNAs and the BART miRNAs, are each expressed coordinately. Generally all three BHRF1 miRNAs are expressed or none is; all BART Cluster 1 and Cluster 2 miRNAs are expressed or none is. An apparent exception to this generalization was the BHRF1-3 miRNA in Raji cells which failed to be expressed while the other two BHRF1 miRNAs were. This exception was explained by finding a mutation in the coding region for the pre-miRNA of BHRF1-3 which would likely prevent its processing (Fig. 8). Another exception was in cell lines immortalized with the B95-8 strain of EBV, which is explained by the absence of sequence that encodes all of BART Cluster 2 and part of BART Cluster 1 (Fig. 2A).
Secondly, BHRF1 miRNAs are thought to arise from the Cp promoter in type III latency (Cai et al., 2006). Transcripts from this promoter encode the EBNA proteins and the miRNAs in these transcripts would be located in known introns (Austin et al., 1988). Based on recent evidence, such transcripts could co-express the EBNAs, and the BHRF1 miRNAs (Kim and Kim, 2007). However, we were able to detect BHRF1-3 above background levels in four cell lines characterized as type I BLs (Fig. 2). It is not clear where transcripts encoding BHRF1-3 originate in these cell, but they could arise from the BHRF1 transcript which has been shown to be transcribed during latency in some EBV-positive BL cell lines (Austin et al., 1988).
A third finding of these analyses is that levels of the individual BART miRNAs differ by 50-fold or more within a cell line even though they are processed from one primary transcript. This wide variation is preserved between cell lines: BART7, for example, accumulates to the highest levels and BART20-5p to the lowest levels for most cell lines studied (Fig. 2). The steady-state levels of these miRNAs reflect their rates of synthesis and degradation. Given that they are all processed from one primary transcript the contribution to their steady-state levels by their synthetic rates would be determined at least in part by their processing. The differential expression of EBV’s BART miRNAs may be explained by the tertiary structure of their primary-miRNA (pri-miR). Steven Chaulk and colleagues have found the individual miRNAs in the human miR-17-92 cluster are differentially expressed, with those expressed more efficiently having their stem-loop structures located on the outside of the pri-miR structure and being more accessible for processing by the microprocessor (S. Chaulk and J. N. M. Glover, submitted for publication). In this model, of the BART miRNAs we assayed, BART7 would be the most accessible to processing by microprocessor, while BART4 would be less accessible. It is also possible that the stability of individual pre-miRs, which has been used to predict pre-miRs in silico, may contribute to the steady state levels of EBV’s BART miRNAs (Brameier and Wiuf, 2007; Pfeffer et al., 2004; Sewer et al., 2005).
A fourth finding of these analyses is that the absolute levels of EBV’s miRNAs vary by 50-fold or more between cell lines (Fig. 3). This finding is important functionally because the levels of miRNAs in cells will determine their capacity to target specific mRNAs: the concentration of a given miRNA will affect its concentration in the RISC complex and the frequency with which that complex encounters a given mRNA. For example, the translation of c-Myb is less efficiently inhibited by miR-150 in B-cells in mice heterozygous for this miRNA than in those cells in wild-type animals carrying two copies of the gene encoding this miRNA (Xiao et al., 2007). Three studied cell lines were outliers with exceptionally low levels of miRNAs (Fig. 4A). These three, Daudi, MutuI, and Akata have all been found to lose EBV DNA spontaneously showing that EBV affords these cell too little selective advantage to be retained in cells in culture. These cell lines do not express other viral genes at disproportionately low levels raising the possibility that the BART miRNAs encode functions most BLs require and that Daudi, MutuI, and Akata cells have evolved to no longer depend on functions of these miRNAs (Fig. 4B, C).
The levels of EBV’s miRNAs not only differ widely between cells but also are not correlated with the number of possible DNA templates encoding them (Fig. 5). This finding could be explained by only a subset of viral DNA molecules in infected cells functioning as templates for transcription. One consequence of this hypothesis is that it would explain how the individual cells within a clone which have a wide distribution of viral DNA molecules per cell regulate their expression of viral genes: only a subset of viral DNAs are transcribed.
It is clear that when EBV’s lytic cycle is induced, there is little or no increased expression of its miRNAs (Fig. 6). Apparently EBV does not regulate its productive cycle by altering the levels of its miRNAs. Clearly one likely exception to this conclusion is BART2, which we have not examined but which is proposed to regulate EBV’s polymerase gene (Barth et al., 2008).
We have found that even for those of EBV’s miRNAs which are highly expressed, most are present at 100’s of molecules per cell while a few rise maximally to 1,000-2,000 molecules per cell in carcinoma cell lines (Fig. 2). In contrast, individual cellular and viral miRNAs which have been associated with functions are often present at 10-100,000 molecules per cell. For example, miR-155 accumulates to levels of 2-10,000 per cell in diffuse large B-cell lymphomas where it may contribute to B-cell tumorigenicity (Costinean et al., 2006; Eis et al., 2005). Three of HSV-1’s miRNAs are found at only 100’s of molecules per cell in lytic infection, but accumulate to 100,000s per cell in neurons where they are thought to contribute to latency (Umbach et al., 2008). Even the abundantly expressed BART7 and BART10 are present at lower levels than human miR-16, which has been measured to range from 500 to 50000 molecules per 10 pg in primary BLs and BL cell lines (Landgraf et al., 2007). One possible explanation for EBV’s relatively low numbers of individual miRNAs is that they act together to target single mRNAs important for EBV’s life-cycle and likely important for its functions of tumor maintenance, too. This explanation is consistent with experiments showing that multiple miRNAs can act additively or synergistically to regulate the same targeted mRNA (Grimson et al., 2007).
Materials and Methods
Plasmid Construction
The BHRF1 fragment of EBV was amplified via PCR from 721 (Wt) or Raji (Mutant) genomic DNA, (Forward primer 5’-ATG TAC GCG TTC ACT AGC CAC TAA GCC-3’ and reverse primer 5’ GCT ACT CGA GGT TGA AAT ATC TAG CAC GGC 3’) and then digested with MluI and XhoI. This product was ligated into the multiple cloning site (MCS) of the retroviral plasmid MCS-IRES-eGFP, which was also digested with MluI and XhoI. QuikChange (Stratagene) was used to revert specifically the mutation in pre-miR-BHRF1-3 derived from Raji to the wild-type sequence (Wt*) using the forward primer 5’ GCG GTG CTT CAC GCT CTT CGT TAA AAT AAC ACA AGG 3’ and reverse primer 5’ CCT TGT GTT ATT TTA ACG AAG AGC GTG AAG CAC CGC 3’. QuikChange was again used to mutate the first start codon (ATG) of the BHRF1 protein coding sequence to ATC in all three vectors using as a forward primer 5’ CCA GAT CTT GTA GAG CAA GAT CGC CTA TTC AAC AAG GG 3’ and as a reverse primer 5’ CCC TTG TTG AAT AGG CGA TCT TGC TCT ACA AGA TCT GG 3’. All PCR fragments were verified by restriction enzyme digestion and sequencing.
To clone the BART Cluster 1 vector, genomic DNA from Akata cells was PCR amplified with forward primer 5’ TTG CTG AGC TAT CCT AGA GGT CCT ACC G 3’ and reverse primer 5’ CCT TCG AAC TGT GGT TAC ATG GTG CAT TTG C 3’ specific to the BARTs. The amplified product contained nucleotides 138997 to 140144 of EBV (GenBank accession no. AJ507799), and contains all currently known BART Cluster 1 miRNAs annotated in miRBase (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008). The amplified product was cut with BlpI and BstBI restriction enzymes, and ligated into the MCS of the parent retroviral plasmid tet-inducible-MCS-IRES-eGFP vector which was also cut with BlpI and BstBI.
BART Cluster 2 vector was also cloned using genomic DNA from Akata cells. Akata DNA was PCR amplified with forward primer 5’ AAT TCG AAT AGG TCA CCT AAC GTG GAA GCC 3’ and reverse primer 5’- GTA CGC GTT GTC CTT CTG TCA CAA CC -3’. The amplified product includes nucleotides 145770 to 149022 of EBV (GenBank accession no. AJ507799), and contains all currently known BART Cluster 2 miRNAs as annotated in miRBase (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008). The amplified product was cut with BstBI and MluI restriction enzymes, and ligated into the MCS of the parent retroviral plasmid tet-inducible-MCS-IRES-eGFP vector which was also cut with BstBI and MluI.
Cell Lines, Culture Conditions, and RNA Isolation
The cell lines used in this study, their EBV status and type of latency are listed in Table 1. The B cell lines, including BLs, PTLDs, and EBV-positive lymphoblastoid cell lines (LCLs) were cultured in RPMI 1640 (Invitrogen) supplemented with L-glutamine, 10% fetal bovine serum (FBS), and antibiotics (200 U/mL penicillin and and 200 μg/mL streptomycin). Akata eGFP were additionally supplemented with 500 μg/ml G418 for maintenance of the recombinant EBV in these cells. The cell lines 293 and 293T, the NPC cell line, C666-1, and the gastric carcinoma cell line, SNU719 cells were cultured in Dulbeccos’ modified Eagle’s medium (Invitrogen) supplemented with 10% FBS and antibiotics (D10F). C666-1 cells were additionally grown on fibronectin. All cells were incubated in 5% CO2 at 37°C. RNA was isolated using TRIzol reagent (Invitrogen) as previously described (Chomczynski and Sacchi, 1987). Total RNA was precipitated with linear acrylamide (Ambion) as previously described (Gaillard and Strauss, 1990).
Table 1.
Cell type, EBV status, and latency type of cell lines used in this study
| Cell Line | Cell Type | EBV status | Latency Type | Reference |
|---|---|---|---|---|
| BJAB | BL | - | - | (Menezes et al., 1975) |
| MutuI | BL | + | I | (Gregory, Rowe, and Rickinson, 1990) |
| Daudi | BL | + | I | (Kitagawa et al., 2000) |
| Akata | BL | + | I | (Takada et al., 1991) |
| OkuI | BL | + | I | (Kelly, Bell, and Rickinson, 2002) |
| SavI | BL | + | I | (Kelly, Bell, and Rickinson, 2002) |
| KemI | BL | + | I | (Kelly, Bell, and Rickinson, 2002) |
| Dante | BL | + | I | (Leao et al., 2007) |
| BL-5 | BL derived from HIV-positive donor | + | I | (Lee et al., 1999) |
| Akata eGFP | Recombinant BL | + | I | (Kanda et al., 2004) |
| SNU719 | Gastric carcinoma | + | II | (Han et al., 2004) |
| C666-1 | NPC | + | II | (Cheung et al., 1999) |
| 721 | LCL | + | III | (Kavathas, Bach, and DeMars, 1980) |
| B958IID6a | LCL | + | III | (Miller et al., 1975) |
| GG68 | BL | + | III | (Weigel and Miller, 1983) |
| Raji | BL | + | III | (Pulvertaft, 1965) |
| MutuIII | BL | + | III | (Gregory, Rowe, and Rickinson, 1990) |
| PTLD1 | PTLD | + | III | Gift from C. Rooney |
Clonal derivative of B95-8
Induction of Viral Lytic Replication in EBV-infected cell lines
Akata eGFP and the marmoset cell clone B958IID6 were induced to support lytic replication of EBV. Akata eGFP was treated with 10 μg IgG per 2×10ˆ6 cells at a concentration of 1×10ˆ6 cells per mL for one hour, and then diluted to 5×10ˆ5 cells per mL on day 0, and allowed to grow in the presence of IgG for 4 days. B958IID6 cells were treated with 20 μg/mL TPA and 35 mM sodium butyrate for four days. At each timepoint, cells were counted, and a portion of the cells was harvested for total RNA isolation and genomic DNA isolation. The remaining cells were cultured between 7×10ˆ5 and 1×10ˆ6 cells per mL for the duration of the induction.
Infection of Primary B cells
Primary B cells were isolated from whole blood and infected with recombinant B95-8 EBV and Akata EBV as previously described at an multiplicity of infection (MOI) of 1 (Lee and Sugden, 2007).
Transformation of BART Cluster 1 and 2 into inducible BJAB cell clones and induction of expression of miRNAs
Retroviral particles containing BART Cluster 1 or BART Cluster 2 vectors were generated in 293T cells as previously described (Lee and Sugden, 2008). After 24 hours, transfected 293T cells were irradiated at 3000 rads, and then BJAB cells engineered to express constitutively a tet-KREB fusion protein were transduced by co-cultivation with the 293T cells. Clones were selected for GFP expression, and then grown in R10F + 1 μg/mL puromycin.
To induce the expression of BART Cluster 1 or 2 miRNAs in BJAB tet-KREB, the cells were cultured in R10F and treated with 10 ng/mL of doxycycline for 96 hours. Expresssion was confirmed prior to harvesting cells by the expression of GFP.
Transfection
293 cells were plated in 100-mm dishes and grown to ~70 % confluence. For each dish, 10 μg of vector was diluted in 500 μl of Opti-MEM (Invitrogen) and then combined with pre-diluted Lipofectamine-Opti-MEM (Invitrogen) mixture (30 μl of Lipofectamine diluted in 500 μl of Opti-MEM, incubated 5 minutes at room temperature). DNA-Lipofectamine complexes were incubated for 25 minutes at room temperature. Cells were washed once with PBS and plated with 5 mL DMEM with 1 ml transfection mixture and incubated for 4 hours at 37°C at 5% CO2. Following the incubation, the medium was replaced with 10 mL of fresh D10F. The transfection efficiency was determined 48 hours later by counting GFP-positive cells and the RNA was isolated.
Western Blotting
Cells were harvested and resuspended in a 1:1 volume of 1x NET [50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA] and 2x Sample Buffer [100 mM Tris pH 6.8, 10% SDS, 10% glycerol, 50 μl/mL b-mercaptoethanol, 0.04% bromophenol blue]. Lysates were run on a 12% SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and blocked overnight at 4°C in BLOTTO [5% non-fat milk (w/v), 0.075% Tween-20, 1xPBS]. Blots were probed with primary antibody, followed by secondary antibody conjugated to alkaline phosphatase (donkey anti-mouse at 1:1000, Jackson Laboratories). Bands were visualized using a solution of 5-bromo-4-chloro-3’-indolyphosphate p-toluidine salt and nitroblue tetrazolium chloride. Western blots were scanned and quantified using ImageQuant 5.2 software. Primary antibodies include: IH4 mouse monoclonal anti-EBNA-1 antibody at 1:50 and mouse monoclonal anti-α tubulin antibody (Sigma-Aldrich) at 1:10,000.
Stem-loop real-time PCR
EBV BHRF1 and BART miRNAs were specifically reverse transcribed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Stem-loop primers to the assayed miRNAs were designed in a similar manner to those designed for reverse transcription of human miRNAs (Chen et al., 2005). For each miRNA assayed, 200 ng of total RNA was reverse transcribed as described by the manufacturer. For measurements in primary B cells infected with Akata or B95-8 EBV, 10 ng of total RNA was used.
Each 20 μl PCR reaction contained 1.5 μM forward primer, 0.7 μM reverse primer, 0.2 μM probe, and 1x TaqMan Universal Master Mix, no UNG AmpErase (Applied Biosystems) as previously reported (Chen et al., 2005). Probes were labeled with 5’ FAMRA and 3’ TAMRA. The sequences of primers and probes are listed for each miRNA assayed (Supplementary Table 1). Primers and probes for EBV miRNAs were ordered from IDT (IDT DNA). A U38B kit was used to detect the snoRNA U38B (Applied Biosystems). The reactions were incubated in a 384-well plate at 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 54°C for 1 minute. The absolute copy number of each miRNA in test samples was determined with reverse transcription and amplification of synthetic miRNAs identical to the mature miRNA sequence as annotated in miRBase in March, 2008(Griffiths-Jones, 2004; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008). Synthetic miRNAs were ordered from IDT (IDT DNA).
Measuring the precision of stem-loop real-time PCR
The method traditionally used to detect miRNAs, Northern blot, is difficult to render both sensitive and quantitative. We used two distinct methods to detect and determine the levels of EBV’s miRNAs sensitively in various EBV-positive cell lines. Stem-loop quantitative real-time PCR (qPCR) measures levels of single miRNAs and can be rendered quantitative with standard curves, permitting comparisons. This method has the added benefit that it requires no more than one-hundredth the RNA used for Northern blotting.
To gauge the precision of qPCR, we determined the standard deviation for three different types of measurements. First, we measured the precision of triplicates from the same stem-loop reverse transcription reaction for the snoRNA, U38B. U38B was recommended as a constant cellular standard for stem-loop real-time PCR by Applied Biosystems. The average standard deviation of the average number of U38B molecules was 3%. We did not, however, find the levels of U38B to be the same in all cell lines. Rather, we found the levels to be the greatest in the marmoset cell line, B958IID6, which contained approximately 550 U38B molecules per 10 pg of RNA. In human cell lines, the levels of U38B ranged from 355 to 145 molecules per 10 pg of RNA in KemI and 721 cells respectively, and the average U38B steady-state levels were 246 +/- 64 molecules per 10 pg of RNA in all assayed EBV-positive cell lines. Second, we measured the precision of triplicate qPCR measurements from the same reverse transcription reaction for the nine assayed EBV miRNAs, and found the average standard deviation to be 9% and range between 0.3% and 32% of the average molecules per 10 pg of RNA. Finally, for seven cell lines we measured the levels of four EBV miRNAs twice from two independent RNA isolates. The average standard deviation of the replicate experiments for EBV miRNAs was found to be 45% of the average molecules per 10 pg of RNA. These results indicate that two-fold differences measured between the quantities of miRNAs are within the error of those measurements.
Real-Time PCR of EBERs
1 μg of total RNA was reverse transcribed with Superscript II RT (Invitrogen) according to manufacturer’s instructions using EBER-specific reverse primers at a final concentration of 0.5 μM and GAPDH with oligo d(T) at a final concentration of 5 μM. Reverse transcribed cDNA was amplified and detected by qPCR under the following conditions: 1x Amplitaq Gold PCR Master Mix (Applied Biosystems), 0.5 μM each primer, 0.2 μM probe, 1x ROX reference dye (Invitrogen) and water to 20 μl. PCR cycling conditions were 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds, and 60°C for 60 seconds. The following oligonucleotide sequences were used for RT-qPCR: GAPDH, forward primer: 5’-TCA ACG ACC ACT TTG TCA AGC T-3’, reverse primer: 5’-CCA TGA GGT CCA CCA CCC T-3’, probe: 5’-TTC CTG GTA TGA CAA CGA ATT TGG CTA CAG C-3’, EBER1, forward primer: 5’-TTT GCT AGG GAG GAG ACG TGT GT-3’, reverse primer: 5’-AAG CAG AGT CTG GGA AGA CAA CCA-3’, probe: 5’-TAC AAG TCC CGG GTG GTG AGG A-3’, EBER2, forward primer: 5’-TTG CCC TAG TGG TTT CGG ACA CA-3’, reverse primer: 5’-AAT AGC GGA CAA GCC GAA TAC CCT-3’, and probe: 5’-TTC CCG CCT AGA GCA TTT GCA AGT CA-3’. Probes were labeled with 5’ FAMRA and 3’ TAMRA.
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) was performed on EBV-positive and negative B cell lines as described previously (Nanbo et al., 2007).
Isolation of Genomic DNA and quantification of EBV genomes
Genomic DNA from cells induced to support the lytic replication of EBV was isolated and quantified by qPCR as previously described (Perrigoue et al., 2005). Briefly, 10 ng of genomic DNA was amplified and detected with primer-probe sets to the EBV gene, BALF5 (Forward: 5’-CGG AAG CCC TCT GGA CTT C-3’, Reverse: 5’-CCC TGT TTA TCC GAT GGA ATG-3’, and Probe: 5’-TGT ACA CGC ACG AGA AAT GCG CC-3’), or β-actin (Forward: 5’-TCA CCC ACA CTG TGC CCA TCT ACG A-3’, Reverse: 5’-TCA GGT AGT CAG TCA GGT CCC G-3’, and Probe:5’-ATG CCC TCC CCC ATG CCA TCC TGC GT-3’) as a loading control. Probes were labeled with 5’ FAMRA and 3’ TAMRA. Genomes were quantified using a known quantity of the HindIII D fragment of EBV’s genomic sequence.
Statistical Methods
Statistical tests were performed using MStat 5.01 provided by Norman Drinkwater (Drinkwater, 2008).
Supplementary Material
The rank order of expression of eight BART miRNAs among EBV-positive cell lines is similar. The expression of BART miRNAs was measured by stem-loop qPCR, as shown in Fig. 2. The expression of each miRNA was ranked from highest to lowest expression. The rank orders of all cell lines were compared by a Spearman Rank Correlation. 90 of 105 (86%) of the comparisons were correlated with p<0.05.
Standard curves for the cellular snoRNA U38B (A), EBV’s BHRF1-1 (B), BHRF1-2 (C), and BHRF1-3 (D) miRNAs were generated with synthetic copies of the RNAs. For each synthetic miRNA, stem-loop real-time PCR was performed, and a Ct value was calculated. The standard curves were used to generate the data in Figure 2. Error bars in (A) and (D) represent the standard deviation of measurements from independent standard curves.
Standard curves for BART3 (A), BART4 (B), BART1-5p (C), BART15 (D), BART7 (E), BART10 (F), BART12 (G), and BART20-5p (H) were generated with synthetic copies of the miRNAs. For each synthetic miRNA, stem-loop real-time PCR was performed, and a Ct value was calculated. The standard curves were used to generate the data in Figure 2. Error bars in each panel represent the standard deviation of measurements from independent standard curves.
Low copy standard curves for U38B (A), BHRF1-3 (B), BART3 (C), BART1-5p (D), BART15 (E), and BART7 (F) were generated with synthetic copies of the RNAs. The standard curves were used to generate the data in Figure 7, in which 10 ng of total RNA was used for stem-loop real-time PCR. Error bars in (A) represent the standard deviation of measurements from independent standard curves.
Acknowledgments
This work was supported by grants from the NIH, CA070723, CA022443, and T32CA009135. Bill Sugden is an American Cancer Society Research Professor.
We thank Cliona Rooney, William J. Harrington, Jr., Shannon Kenney, Alan Rickinson, and Kenzo Takada for cell lines. We also thank Elaine Alarid, Paul Lambert, JiSook Lee, and David Vereide for their insightful editorial help. Finally, we thank Steven Chaulk and J.N. Mark Glover for sharing their unpublished findings.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin PJ, Flemington E, Yandava CN, Strominger JL, Speck SH. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc Natl Acad Sci U S A. 1988;85:3678–3682. doi: 10.1073/pnas.85.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Wiuf C. Ab initio identification of human microRNAs based on structure motifs. BMC Bioinformatics. 2007;8:478. doi: 10.1186/1471-2105-8-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, Wong N, Whitney BM, Lee JC. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J Cancer. 1999;83:121–126. doi: 10.1002/(sici)1097-0215(19990924)83:1<121::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkwater N. Mstat. 5.1. McArdle Laboratory for Cancer Research; Madison: 2008. [Google Scholar]
- Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. 2008;82:9094–9106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C, Strauss F. Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res. 1990;18:378. doi: 10.1093/nar/18.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CD, Rowe M, Rickinson AB. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J Gen Virol. 1990;71:1481–1495. doi: 10.1099/0022-1317-71-7-1481. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SU, Kim HT, Seong DH, Kim YS, Park YS, Bang YJ, Yang HK, Kim SJ. Loss of the Smad3 expression increases susceptibility to tumorigenicity in human gastric cancer. Oncogene. 2004;23:1333–1341. doi: 10.1038/sj.onc.1207259. [DOI] [PubMed] [Google Scholar]
- Hatfull G, Bankier AT, Barrell BG, Farrell PJ. Sequence analysis of Raji Epstein-Barr virus DNA. Virology. 1988;164:334–340. doi: 10.1016/0042-6822(88)90546-6. [DOI] [PubMed] [Google Scholar]
- Huang J, Chen H, Hutt-Fletcher L, Ambinder RF, Hayward SD. Lytic viral replication as a contributor to the detection of Epstein-Barr virus in breast cancer. J Virol. 2003;77:13267–13274. doi: 10.1128/JVI.77.24.13267-13274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Yajima M, Ahsan N, Tanaka M, Takada K. Production of high-titer Epstein-Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J Virol. 2004;78:7004–7015. doi: 10.1128/JVI.78.13.7004-7015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavathas P, Bach FH, DeMars R. Gamma ray-induced loss of expression of HLA and glyoxalase I alleles in lymphoblastoid cells. Proc Natl Acad Sci U S A. 1980;77:4251–4255. doi: 10.1073/pnas.77.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G, Bell A, Rickinson A. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med. 2002;8:1098–1104. doi: 10.1038/nm758. [DOI] [PubMed] [Google Scholar]
- Kim do N, Chae HS, Oh ST, Kang JH, Park CH, Park WS, Takada K, Lee JM, Lee WK, Lee SK. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J Virol. 2007;81:1033–1036. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa N, Goto M, Kurozumi K, Maruo S, Fukayama M, Naoe T, Yasukawa M, Hino K, Suzuki T, Todo S, Takada K. Epstein-Barr virus-encoded poly(A)(-) RNA supports Burkitt’s lymphoma growth through interleukin-10 induction. EMBO J. 2000;19:6742–6750. doi: 10.1093/emboj/19.24.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano J, Sugiura M, Takada K. Epstein-Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt’s lymphoma cell line Akata. J Virol. 1998;72:9150–9156. doi: 10.1128/jvi.72.11.9150-9156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao M, Anderton E, Wade M, Meekings K, Allday MJ. Epstein-barr virus-induced resistance to drugs that activate the mitotic spindle assembly checkpoint in Burkitt’s lymphoma cells. J Virol. 2007;81:248–260. doi: 10.1128/JVI.01096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27:2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- Lee J, Sugden B. A membrane leucine heptad contributes to trafficking, signaling, and transformation by latent membrane protein 1. J Virol. 2007;81:9121–9130. doi: 10.1128/JVI.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RK, Cai JP, Deyev V, Gill PS, Cabral L, Wood C, Agarwal RP, Xia W, Boise LH, Podack E, Harrington WJ., Jr Azidothymidine and interferon-alpha induce apoptosis in herpesvirus-associated lymphomas. Cancer Res. 1999;59:5514–5520. [PubMed] [Google Scholar]
- Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Turner DH, Zuker M. RNA secondary structure prediction. Curr Protoc Nucleic Acid Chem. 2007;Chapter 11(Unit 11 2) doi: 10.1002/0471142700.nc1102s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes J, Leibold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- Miller G, Robinson J, Heston L, Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection and interference. IARC Sci Publ. 1975;11(Pt 1):395–408. [PubMed] [Google Scholar]
- Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002;21:954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–4262. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL, Yates JL. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J Virol. 2000;74:8563–8574. doi: 10.1128/jvi.74.18.8563-8574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrigoue JG, den Boon JA, Friedl A, Newton MA, Ahlquist P, Sugden B. Lack of association between EBV and breast carcinoma. Cancer Epidemiol Biomarkers Prev. 2005;14:809–814. doi: 10.1158/1055-9965.EPI-04-0763. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Pulvertaft JV. A Study of Malignant Tumours in Nigeria by Short-Term Tissue Culture. J Clin Pathol. 1965;18:261–273. doi: 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, Toro JR, Zenger VE, Metcalf RA, Marti GE. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Horinouchi K, Ono Y, Aya T, Osato T, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Sugden B. Identifying a property of origins of DNA synthesis required to support plasmids stably in human cells. Proc Natl Acad Sci U S A. 2008;105:9639–9644. doi: 10.1073/pnas.0801378105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel R, Miller G. Major EB virus-specific cytoplasmic transcripts in a cellular clone of the HR-1 Burkitt lymphoma line during latency and after induction of viral replicative cycle by phorbol esters. Virology. 1983;125:287–298. doi: 10.1016/0042-6822(83)90202-7. [DOI] [PubMed] [Google Scholar]
- Williams H, Crawford DH. Epstein-Barr virus: the impact of scientific advances on clinical practice. Blood. 2006;107:862–869. doi: 10.1182/blood-2005-07-2702. [DOI] [PubMed] [Google Scholar]
- Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ., Jr EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The rank order of expression of eight BART miRNAs among EBV-positive cell lines is similar. The expression of BART miRNAs was measured by stem-loop qPCR, as shown in Fig. 2. The expression of each miRNA was ranked from highest to lowest expression. The rank orders of all cell lines were compared by a Spearman Rank Correlation. 90 of 105 (86%) of the comparisons were correlated with p<0.05.
Standard curves for the cellular snoRNA U38B (A), EBV’s BHRF1-1 (B), BHRF1-2 (C), and BHRF1-3 (D) miRNAs were generated with synthetic copies of the RNAs. For each synthetic miRNA, stem-loop real-time PCR was performed, and a Ct value was calculated. The standard curves were used to generate the data in Figure 2. Error bars in (A) and (D) represent the standard deviation of measurements from independent standard curves.
Standard curves for BART3 (A), BART4 (B), BART1-5p (C), BART15 (D), BART7 (E), BART10 (F), BART12 (G), and BART20-5p (H) were generated with synthetic copies of the miRNAs. For each synthetic miRNA, stem-loop real-time PCR was performed, and a Ct value was calculated. The standard curves were used to generate the data in Figure 2. Error bars in each panel represent the standard deviation of measurements from independent standard curves.
Low copy standard curves for U38B (A), BHRF1-3 (B), BART3 (C), BART1-5p (D), BART15 (E), and BART7 (F) were generated with synthetic copies of the RNAs. The standard curves were used to generate the data in Figure 7, in which 10 ng of total RNA was used for stem-loop real-time PCR. Error bars in (A) represent the standard deviation of measurements from independent standard curves.