Abstract
Many cellular structures are assembled from networks of actin filaments and the architecture of these networks depends on the mechanism by which the filaments are formed. Several classes of proteins are known to assemble new filaments, including the Arp2/3 complex, which creates branched filament networks, and Spire, which creates unbranched filaments1, 2. We find that JMY, a vertebrate protein first identified as a transcriptional co-activator of p53, combines these two nucleating activities by both activating Arp2/3 and assembling filaments directly using a Spire-like mechanism. Increased levels of JMY expression enhance motility while loss of JMY slows cell migration. When slowly migrating HL-60 cells are differentiated into highly motile neutrophil-like cells, JMY moves from the nucleus to the cytoplasm, and is concentrated at the leading edge. Thus, JMY represents a new class of multifunctional actin assembly factor whose activity is regulated, at least in part, by sequestration in the nucleus.
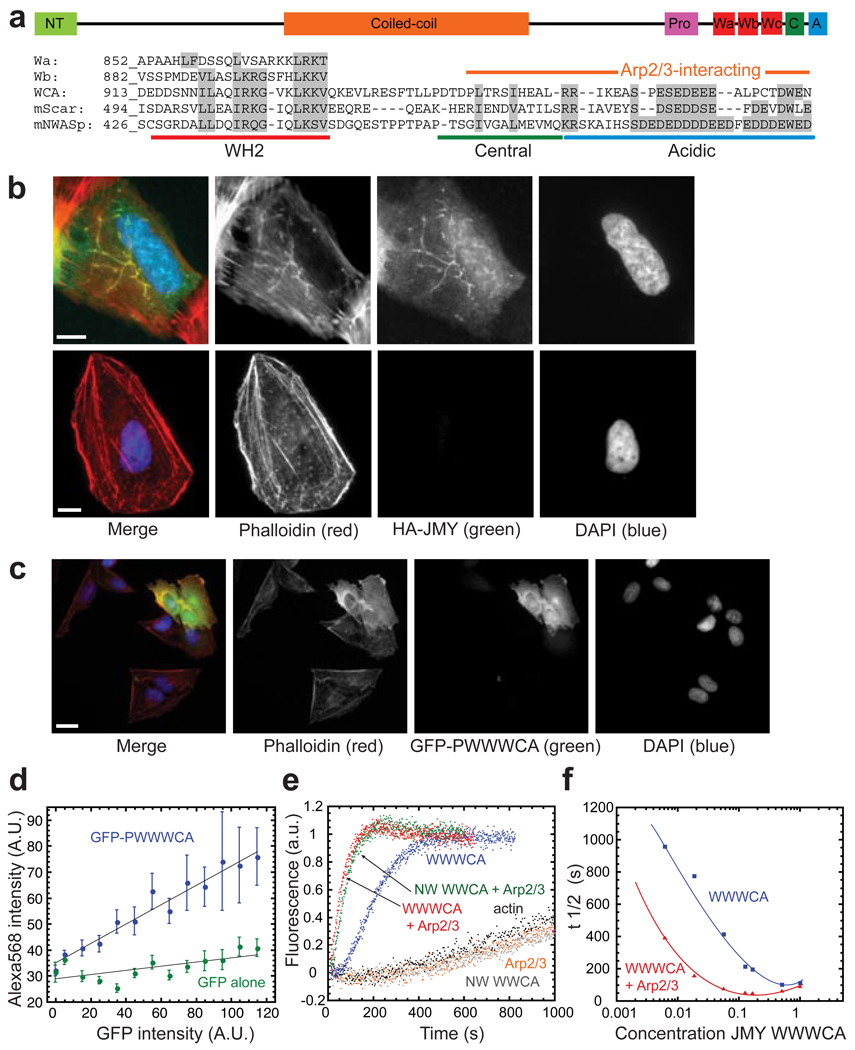
By searching genome databases for sequences related to the WASp Homology 2 (WH2) domain we discovered a potential Arp2/3-activating sequence, WWWCA, in the vertebrate protein JMY (Fig. 1a). This sequence is composed of three tandem repeats of the actin monomer-binding WH2 domain (WWW); an actin- and Arp2/3-binding central domain (C); and an Arp2/3-binding acidic domain (A). These sequence elements, first identified in WASp-family proteins1, 3, collaborate in activating Arp2/3. The identification of these elements in JMY was surprising, since JMY localizes primarily to the nucleus and was originally discovered as a binding partner of p300, a coactivator for many transcription factors, including the tumour-suppressor p534. In fibroblasts JMY accumulates in the nucleus in response to DNA damage, where it enhances p53-dependent transcription of pro-apoptotic genes4, 5.
Figure 1. JMY nucleates actin filaments and activates the Arp2/3 complex.
(a) Domain structure of JMY. The C-terminus of JMY is homologous to activators of Arp2/3. A poly-proline (P) domain28 is followed by three tandem actin monomer-binding WH2 domains (Wa through Wc), an actin and Arp2/3-binding central domain (C), and an Arp2/3-binding acidic domain (A). Alignment shows individual WH2 domains of JMY, and compares the sequences of the WCA regions of JMY, Scar, and N-WASp. JMY WCA is 28% identical to N-WASp WCA (ClustalW), and residues putatively involved in binding actin and Arp2/329, 30 are 100% conserved between all available JMY sequences.
(b) Expression of HA-JMY (top panels; visualized by indirect immunofluorescence of HA, green) in U2OS cells induces the formation of filamentous actin structures (visualized with Alexa Fluor 568 phalloidin, red). These elongated actin structures colocalize with JMY and are not seen in untransfected cells (bottom panels). Nuclei were visualized with DAPI (blue). Scale bar, 10 µm.
(c) Expression of GFP-PWWWCA (green) in U2OS cells increases cellular F-actin (Alexa Fluor 568 phalloidin, red).
(d) Quantification of the increase in F-actin induced by GFP-PWWWCA expression. Phalloidin intensity was plotted as a function of GFP-PWWWCA intensity and shows a linear increase in F-actin content with increased expression of GFP-PWWWCA (n=457). In contrast, expressing GFP alone has only a minor effect on the red intensity detected, likely due to a small amount of bleed-through (n=458).
(e) Pyrene-actin polymerization assays show that JMY WWWCA both activates Arp2/3 (R) and nucleates actin in the absence of Arp2/3 (red and blue traces). N-WASp (NW) WWCA activates Arp2/3 (green), but does not nucleate actin on its own (grey).
(f) Intrinsic nucleation and activation of Arp2/3 by WWWCA are dose-dependent. Pyrene-actin polymerization assays were conducted in the absence (blue) or presence (red) of Arp2/3, with increasing concentrations of JMY WWWCA. Time to half-maximal polymerization was plotted as a function of WWWCA concentration. Pyrene-actin polymerization assays were in 1x KMEI and contained 2 µM actin, 167 nM JMY or N-WASp, and 2.5 nM Arp2/3, where noted.
To determine whether JMY also plays a role in assembly of the actin cytoskeleton we tested the effect of JMY expression on actin organization in vivo. Overexpression of JMY in human U2OS cells induces formation of elongated actin filament structures that colocalize with JMY (Fig. 1b), similar to overexpression of WASp-family proteins and the actin nucleation factor Spire 2, 6. Truncation mutants demonstrate that the WH2 cluster is required for this effect but, curiously, the Arp2/3-binding CA domain is not (Supplementary Fig. S1). Expression of the C-terminal region of JMY fused to GFP (GFP-PWWWCA) produces a linear, dose-dependent increase in the cellular concentration of filamentous actin (Fig. 1c) as judged by correlating Alexa-568 phalloidin staining with GFP fluorescence (n=457; Fig. 1d). In contrast, expressing GFP alone has no significant effect on cellular levels of filamentous actin (n=458; Fig. 1d, Supplementary Fig. S1).
To study how JMY affects actin assembly in vitro, we first verified that all three putative WH2 domains (Wa, Wb, and Wc, from N- to C-terminal) bind monomeric actin (Supplementary Fig. S2). JMY activates Arp2/3 in vitro, as determined by pyrene-actin polymerization assays using a C-terminal fragment of JMY (WWWCA) (Fig. 1e; t1/2 (JMY + actin + Arp2/3) = 42.2 ± 2.3 s; versus t1/2 (actin + Arp2/3) = 1193 ± 28 s). JMY WWWCA also induces rapid actin polymerization in the absence of Arp2/3 (Fig. 1e; t1/2 = 188.5 ± 4.4 s). The JMY-dependent increase in polymerization rate is dose-dependent, both with and without Arp2/3 (Fig. 1f). This result is quite surprising and distinguishes JMY from all other proteins known to activate Arp2/3. N-WASp WWCA, for example, activates Arp2/3 to a similar extent as JMY, but does not accelerate polymerization in the absence of Arp2/3 (Fig. 1e).
By itself JMY could accelerate actin assembly by: (1) nucleating new filaments, (2) increasing the rate of filament elongation, or (3) severing existing filaments to create new barbed ends2, 7. JMY does not affect elongation of preformed filaments, arguing that it neither accelerates elongation nor severs, but rather nucleates new filaments (Supplementary Fig. S3a–c). Indeed, we observe greater than 12-fold more filaments in the presence of JMY WWWCA compared to actin alone (Fig. 2a–b). In the presence of Arp2/3, JMY WWWCA produces branched filaments, consistent with nucleation by Arp2/3 (Fig. 2c, left panel), while, by itself, JMY nucleates unbranched filaments (Fig. 2a). In assembly reactions filament length is inversely proportional to the rate of nucleation, and filaments made in the presence of JMY are shorter than those made with actin alone. Arp2/3-nucleated filaments are even shorter (Fig. 2d). JMY produces twice as many branches per micron as Scar1/WAVE1 (Fig. 2e), consistent with JMY inducing a faster rate of Arp2/3 activation than Scar.
Figure 2. Mechanistic dissection of JMY.
(a) JMY nucleates unbranched filaments, and increases the number of filaments over actin alone. Filaments made in the presence (left) or absence (right) of 167 nM JMY WWWCA were fixed with Alexa Fluor 488 phalloidin and Latrunculin B at 6 minutes (t9/10 of JMY WWWCA reaction) prior to dilution and spotting on poly-L-lysine coverslips24,2. Scale bars, 5 µm.
(b) Quantification of filaments per field in images from a demonstrates that JMY nucleates new filaments (JMY, 42.7 ± 4.2 filaments per micron, n=35 fields; actin alone, 3.2 ± 0.5 filaments per micron, n=30 fields).
(c) Filaments prepared as in a, in the presence of JMY plus Arp2/3 (left), Scar plus Arp2/3 (centre), or actin alone (right). The concentration of filaments was kept constant by arresting reactions at their individual t9/10s (see Methods). Filaments nucleated in the presence of JMY and Arp2/3 are branched, consistent with JMY activating Arp2/3, as are filaments made in the presence of Scar and Arp2/3. The shorter, more abundant filaments seen here are due to Arp2/3 nucleating actin more rapidly than intrinsic nucleation by JMY.
(d) Quantification of filament length at t9/10. The rate of nucleation is inversely proportional to the length of filament (rate: JMY+Arp2/3 > JMY > Scar+Arp2/3 ≫ actin alone).
(e) Quantification of filament branching in each condition. n>300 filaments per condition.
(f) Tandem WH2 domains from JMY are sufficient for actin nucleation. Actin polymerization is as fast with WWW as it is with WWWCA. WWW lacks the Arp2/3 binding CA domain, so adding Arp2/3 to WWW does not accelerate polymerization over WWW alone. N-WASp WW does not nucleate actin.
(g) JMY WCA is sufficient to activate Arp2/3, but does not nucleate actin. The rate of actin polymerization in the presence of JMY WCA and Arp2/3 is similar to reactions containing Scar WCA and Arp2/3. In the absence of Arp2/3, JMY WCA has no effect on actin polymerization. Experimental conditions as in Fig. 1.
Some proteins that cap barbed ends in vivo (e.g. capping protein) nucleate filaments that elongate from their pointed end in vitro8. To determine whether JMY caps barbed ends, we made filaments and depolymerized them in the presence of JMY WWCA. JMY does not inhibit disassembly, arguing that it does not cap barbed ends but promotes polymerization by nucleating new filaments that elongate from their barbed ends (Supplementary Fig. S3d, e). Direct nucleation appears to explain the unexpected ability of JMY to induce actin assembly in vivo in the absence of the Arp2/3-binding CA domain (Supplementary Fig. S1).
We hypothesized that, similar to Spire2, JMY uses its tandem WH2 domains to nucleate new filaments. We tested the activity of constructs consisting of either all three WH2 domains (JMY WWW), or the two C-terminal WH2 domains (JMY WbWc). JMY WWW nucleates actin as well as WWWCA, indicating that the CA region plays no role in direct nucleation (Fig. 2f). Moreover, addition of Arp2/3 to JMY WWW does not further increase the rate of polymerization (Fig. 2f). As with Spire, the two C-terminal WH2 domains (JMY WbWc ) are sufficient for nucleation, but all three JMY WH2 domains are required for maximal activity (Supplementary Fig. S3h, i).
In contrast, a fragment composed of a single WH2 domain (Wc) and the CA domains (JMY WCA) activates Arp2/3, but does not nucleate filaments on its own (Fig. 2g). JMY contains a conserved tryptophan residue known to be important for Arp2/3 binding in all WASp family proteins9. Replacing this tryptophan with an alanine in JMY decreases activation of Arp2/3 without affecting intrinsic nucleation activity (Supplementary Fig. S3j). These results argue that nucleation and Arp2/3 activation are separable activities, and that JMY activates Arp2/3 by the same mechanism as WASp-family proteins.
Similarities in the sequences and activities of JMY and Spire suggested that the two proteins nucleate filaments by a common mechanism. We compared the kinetics of nucleation by JMY and Spire over a range of actin concentrations. At all concentrations of actin tested, JMY and Spire display nearly identical kinetics (Supplementary Fig. S3k–l). Also, similar to Spire, high concentrations of JMY sequester actin monomers (Supplementary Fig. S3m–n). Unlike Spire, however, JMY WWCA does not prevent dissociation of monomers from the pointed end of filaments (Supplementary Fig. S3f–g). It is possible that the full-length JMY interacts with filament ends, but this is not required for nucleation.
Spire nucleates actin by stitching monomers together using tandem WH2 domains and a novel actin binding motif (previously called “Linker 3”2) that we designate the Monomer Binding Linker, or MBL. Spire-MBL, a short (~15 AA) sequence connecting the third and fourth WH2 domains, is sufficient to promote weak nucleation2. We compared the sequences of Spire-MBL with the region between the two C-terminal WH2 domains of JMY (Wb and Wc) and N-WASp. Most residues conserved between Spire homologs are also conserved in JMY, but not in N-WASp (Fig. 3a). Another WH2-containing nucleation factor, Cordon Bleu (Cobl), is thought to operate by a different mechanism10 and, consistent with this, we see no conservation between the linker regions of Cobl and Spire (Fig. 3a).
Figure 3. JMY nucleates actin by the same mechanism as Spire.
(a) The region between JMY Wb and Wc is homologous to the short actin nucleation motif from Spire (monomer-binding linker, MBL). The figure shows an alignment between Spire-MBL and the same region of JMY (mJMY AA 903–917) with homologous residues coloured grey. The MBL sequence is not homologous to the analogous position in N-WASp, WHAMM31, or Cordon Bleu10. The position of the glycine-serine repeats in b is underlined, and the sequences of N-WASp gain of function mutations (NW WJW and WSW) in c are shown.
(b) JMY-MBL is important for actin nucleation. Replacing the MBL in JMY WbWc and Spire CD (the two C-terminal WH2 domains in Spire) with a flexible linker of glycine-serine repeats (gs5) causes a nucleation defect in both JMY and Spire. The analogous N-WASp mutant does not promote nucleation, but inhibits spontaneous polymerization.
(c) Gain of function. Replacing the linker region between the WH2 domains of N-WASp WW with JMY- or Spire-MBL (NW WJW or WSW) converts N-WASp into a weak actin nucleator. This shows that JMY- and Spire-MBL are sufficient for nucleation. Mutated amino acids of WJW and WSW are shown in a. Reactions in b and c contained 4 µM actin. Experimental conditions as in Fig. 1.
(d) t/12s of reactions from b–c. Reactions were repeated > 3 times each. Error bars, s.e.m.
(e) Actin nucleation and activation of Arp2/3 are distinct activities of JMY that overlap spatially. Tandem JMY WH2 domains and the MBL (star in figure) nucleate actin, similar to Spire, and JMY WCA activates Arp2/3, similar to N-WASp and Scar.
Do JMY and Spire nucleate actin by the same mechanism? We replaced the JMY linker with a set of glycine-serine repeats (JMY WbWc(gs5)), and compared it to a similar Spire mutant (Spire CD(gs5)). JMY WbWc(gs5) has a modest defect in nucleation, while replacing the linker in Spire has a more pronounced effect (Fig. 3b, d; Supplementary Fig. S3o). Interestingly, inserting either the JMY- or Spire-MBL between the N-WASp WH2 domains (NW WJW or WSW) converts N-WASp into a nucleator (Fig. 3c,d, Supplementary Fig. S3p), while inserting a flexible linker (NW WW(gs5)) does not (Fig. 3b,d). NW WJW and WSW do not nucleate as well as JMY or Spire. Thus, nucleation by JMY and Spire requires unique properties of both the linker (MBL) and the WH2 domains (Fig. 3e).
To investigate its role in actin assembly in vivo, we determined the localization of JMY in multiple cell types. JMY is primarily nuclear in mouse embryonic fibroblasts, B16-F10 mouse melanoma cells, NIH 3T3 cells, and primary rat neurons (Supplementary Fig. S4 and data not shown). In ruffling B16-F10 cells, we observe a small fraction of JMY colocalized with actin filaments at the leading edge (Supplementary Fig. S4b). Interestingly, in highly motile, primary human neutrophils, JMY is almost entirely excluded from the nucleus, and colocalizes with filaments at the leading edge (Fig. 4d). JMY does not bind actin filaments in vitro, so this localization does not simply reflect direct interaction of JMY with filaments (Supplementary Fig. S4d).
Figure 4. JMY localizes to the leading edge of motile cells.
(a–c) Redistribution of JMY from the nucleus to the leading edge in HL-60 cells. (a) JMY is primarily nuclear in undifferentiated HL-60 cells. (b) Following differentiation into motile cells by culturing in 1.3% DMSO for 5–7 days, JMY colocalizes with filamentous actin in the cytoplasm. (c) Differentiated HL-60 cells were polarized by exposure to 100 nM fMLP, a chemoattractant. JMY is distributed throughout the cytoplasm, where it colocalizes strongly with filamentous actin at the leading edge. Cells were fixed and stained with Alexa Fluor 568 phalloidin (red), anti-JMY (green), and DAPI (blue). Scale bars, 10 µm.
(d) Human primary neutrophils were obtained by finger pinprick26, stimulated with 20 nM fMLP, and fixed and stained as above. JMY (green) colocalizes with filamentous actin (red) at the leading edge. Scale bar, 5 µm.
(e) Western blots of JMY and binding partner p300 in undifferentiated (U) and differentiated (D) HL-60 cells. p300 is expressed in undifferentiated, but not differentiated, HL-60 cells.
Our localization studies suggest that JMY’s presence at the leading edge correlates with motility. To further test this we investigated JMY expression and localization in HL-60 cells, which can exist as non-motile, relatively undifferentiated cells or be induced to differentiate into highly motile cells11. In undifferentiated HL-60 cells, JMY is primarily nuclear, and does not colocalize with filaments (primarily nuclear in 91% of cells, n=246; Fig. 4a, Supplementary Fig. 5). Addition of 1.3% DMSO to the culture medium induces differentiation into highly motile cells that polarize and undergo chemotaxis. During differentiation JMY localization shifts dramatically, becoming almost entirely cytoplasmic. This shift occurs approximately 2 days before cells are competent to polarize in response to chemoattractant (fMLP; Supplementary Fig. S5c). In differentiated cells JMY colocalizes with filamentous actin in the cell cortex (cytoplasmic in 94%, nuclear in 6%, n=212; Fig. 4b), and upon addition of fMLP, JMY localizes strongly to the leading edge where it overlaps with filamentous actin (88% of polarized cells, n=311; Fig. 4c, Supplementary Fig. S5d). Comparing JMY staining to soluble GFP in polarized cells shows that enrichment of JMY at the leading edge is not a volume artefact (Supplementary Fig. S5e–g). These data suggest that translocation of JMY to the cytosol plays a role in building the leading edge. Interestingly, expression of JMY’s nuclear binding partner p300 disappears when HL-60 cells are differentiated, suggesting that the nuclear (p300/p53-dependent transcription) and cytoplasmic (actin nucleation) roles of JMY are regulated by separate pathways (Fig. 4e).
To test JMY’s role in cell motility, we stably expressed GFP-JMY and GFP-JMYΔCA in U2OS cells, grew monolayers, scratched them, and monitored wound healing over time12. Cells expressing GFP-JMY migrate 17% faster than wild-type cells (n=3; p<0.003; Fig. 5a–b, Supplementary Fig. S6). In contrast, cells expressing GFP-JMYΔCA migrate at the same rate as wild-type cells (GFP-JMY, 43.8 ± 1.6 µm/hr; GFP-JMYΔCA, 38.5 ± 1.7 µm/hr; wild-type, 37.5 ± 1.0 µm/hr), suggesting that JMY requires its Arp2/3-activating activity to enhance motility.
Figure 5. JMY contributes to cell motility.
(a–b) Expressing GFP-JMY in U2OS cells significantly increases their motility in wound healing assays. Stable lines of GFP-JMY and GFP-JMYΔCA were wounded by scraping with micropipette tips. Images were acquired at 0, 2, 4, 6, and 12 hours after wounding. (b) Migration rate from 0–6 hours was averaged from a minimum of 4 replicates on each of 3 days. GFP-JMY expression induces cells to migrate 16.6% faster than wild-type cells (n=3, p<0.003). Cells expressing a truncation of JMY lacking the Arp2/3-interacting CA domain migrate at the same rate as wild-type cells. Scale bars, 100 µm.
(c–e) Wound healing assays in U2OS cells indicate that knocking-down JMY by RNAi impairs cell migration. Cells were transfected with JMY or control non-targeting 2 (Dharmacon) siRNA (C) and wounded (red dashed line) as in a. Images were taken at the same position 0, 3, 6, and 24 h after wounding. (d) Wound size at each time point for all conditions. (n=4). (e) Western blots show RNAi efficiency in U2OS cells.
(f) Knocking down JMY expression decreases cellular levels of F-actin. HEK 293 cells were transfected with a vector encoding both GFP and a JMY-specific shRNA25 and fixed and stained with Alexa Fluor 568-phalloidin. Average phalloidin intensity in GFP-negative (non-RNAied) and GFP-positive (RNAied) cells is plotted (n=396, p<0.03, see Methods). Error bars, s.e.m.
(g) JMY localizes to the leading edge of U2OS cells. Cells were grown as above, and fixed and stained for JMY 15 minutes after wounding. JMY is primarily nuclear, but it is also enriched at the leading edge (indirect immunofluorescence, green) where it colocalizes with a subset of actin filaments (Alexa Fluor 568-phalloidin, red). Inset: leading edge, contrast enhanced to show actin filaments and JMY. Scale bar, 10 µm.
(h) Models of in vivo role of JMY. Top, Model 1: JMY nucleates filaments that then serve as substrates for dendritic nucleation by Arp2/3. Bottom, Model 2: JMY evolved to nucleate actin in different cellular contexts. In the nucleus it nucleates unbranched filaments, and in the cytoplasm it both nucleates filaments and activates Arp2/3.
We next knocked-down JMY expression in cultured U2OS and HEK 293 cells by RNAi. Western blotting shows that RNAi was efficient (Fig. 5e, Supplementary Fig. S6). In both cell lines knock-down of JMY expression significantly slows the rate of wound healing (Fig. 5c–d, Supplementary Fig. S6). In the first 6 hours, cells treated with JMY siRNA move 38.0% slower than control U2OS cells (n=4; p<0.05). By 24 hours, control cells fully migrate into the wound, whereas cells treated with JMY siRNA migrate only 86 percent of the distance (Fig. 5a–b; n=4). Control HEK 293 cells migrate 92 percent of the distance by 24 hours, whereas cells treated with JMY siRNA migrate 60 percent (Supplementary Fig. S6; n=7).
Defects in wound healing could be caused by: (1) reduced actin assembly, (2) altered transcription, (3) reduced cell division or increased apoptosis, or (4) off-target effects of the siRNA. Consistent with (1), we find that JMY-knockdown cells contain 11.4 ± 0.4% (n=396, p<0.03) less filamentous actin than wild-type cells (Fig. 5f). Loss of either p53 or p300, which are required for all JMY’s known transcriptional effects, leads to increased cell migration13–15, inconsistent with transcriptional effects on motility. In addition, less than 5% of cells close to the wound divide in the first 6 hours of the assay, making it unlikely that cell division contributes to the observed defect. Knockdown of JMY was shown to decrease apoptosis5, ruling out explanation (3). To address possible off-target effects, we used three different siRNAs, from different regions of the human JMY gene, to knock down expression in HEK 293 cells. All three caused migration defects (Supplementary Fig. S6g), while a control siRNA had no effect.
We next tested whether JMY localizes to the leading edge of U2OS cells during migration in response to a wound, as seen in highly motile HL-60 cells. We fixed and stained U2OS cells 15 minutes after wounding, as well as cells in a sub-confluent and non-wounded cultures. Although the majority of JMY is nuclear in these cells, we also observe both endogenous JMY and GFP-JMY colocalized with filamentous actin at the leading edge, (Fig. 5g, Supplementary Fig. S4c), both in cells adjacent to a wound and in ruffling edges of sub-confluent cells (data not shown). Thus, even in cells where JMY is predominantly nuclear, a fraction of the protein can influence actin assembly at the leading edge and promote migration.
JMY combines an unusual set of activities. The combination of transcriptional coactivation and actin nucleation activities suggests that JMY might mediate cellular decisions involving apoptosis and migration. Alternatively, JMY might be a chimera whose transcriptional and cytoskeletal functions are more or less independent. More intriguing is JMY’s combination of Arp2/3-dependent and independent nucleation activities. A cell switching from a resting to a motile state must extensively remodel its cytoskeleton16. The leading edge of most migrating cells is characterized by a highly-branched dendritic arbour of actin filaments nucleated by Arp2/3, and one requirement for generating such a network is the pre-existence of “mother” filaments for Arp2/3 to bind and branch from17,18. Although resting cells contain some actin filaments (cortical actin, stress fibres, etc.) recent work suggests that not all filaments can serve as efficient substrates for Arp2/3-dependent nucleation19, 20. We hypothesize that JMY contributes to cell motility by nucleating filaments to jump-start Arp2/3-dependent nucleation and branching. By first nucleating new mother filaments and then activating Arp2/3 to branch off of these filaments, JMY could promote the rapid formation of a branched actin network. It is also possible that JMY has evolved to promote actin polymerization in different cellular contexts: (1) in the cytoplasm in the presence of Arp2/3, or (2) in the nucleus in the absence of Arp2/3 (see model in Fig. 5h). For example, it would be interesting to test whether inactivation of JMY's function in actin dynamics affects its role as a p300-dependent transcriptional coactivator for p53. Future work is aimed at testing these models.
Methods
Molecular Biology and Biochemistry
Constructs were cloned from full length mouse JMY4, fly Spire2, and rat N-WASp using standard techniques. Primer sequences are available upon request. All constructs were sequenced to ensure no mutations were introduced during cloning. JMY fragments were expressed as GST-fusions in E. coli and purified using a combination of glutathione and cation chromatography. Except for individual WH2 domains (Supplementary Fig. S2), the GST was removed to prevent dimerization of the recombinant protein, which results in a marked increase in the rate of both intrinsic nucleation and activation of Arp2/3 (data not shown). To improve reproducibility of fluorimetry reactions, we mutated the non-conserved cysteine C978 to serine. This mutation does not change the kinetics of activation of Arp2/3 (Supplementary Fig. S6h, i), but is more stable than wild-type peptide. JMY concentrations were calculated using predicted molar extinction coefficients for JMY peptides that contained tryptophan residues (ProtParam), or by quantitative SDS-PAGE with Sypro-Red staining (Invitrogen).
Actin polymerization assays
Actin was purified from Acanthamoeba castellani as described21, labelled with pyrene iodoacetamide as described22, and stored on ice. Arp2/3 was purified from Acanthamoeba as described23 and flash frozen with 10% glycerol. For all assays, Arp2/3 was thawed daily and diluted with 1 mg/mL BSA in Buffer A (0.2 mM ATP, 0.5 mM TCEP, 0.1 mM CaCl2, 0.02% w/v sodium azide, 2 mM Tris, pH 8.0 at 4°C). Actin polymerization assays were performed in 1x KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0). Ca2+-actin was converted into Mg2+-actin by incubation of actin in ME (50 mM MgCl2, 0.2mM EGTA) for two minutes prior to adding 10x KMEI and test components. Pyrene fluorescence was measured with an ISS PCI/K2 fluorimeter. Unless otherwise noted, polymerization reactions contained 2 µM actin (5% pyrene labelled), 2.5 nM Arp2/3 and 167 nM JMY or N-WASp. JMY proteins were diluted with 10 mg/mL BSA in Buffer A to prevent loss of activity. To normalize fluorimetry data, we subtracted the offset from zero then divided by the plateau value of actin alone, so as to not mask the effects of sequestration by JMY. For half-time calculations, reactions were normalized by dividing by their own plateau and solving for time at half-maximal fluorescence (0.5 a.u.).
To visualize actin filaments we polymerized 2 µM actin under the same conditions used for fluorimetry, then arrested reactions at the time indicated with Alexa Fluor 488 phalloidin and Latrunculin B. This technique preserves the ratio of monomeric to filamentous actin at the moment of quenching24. To keep the concentration of F-actin constant we arrested reactions at their individual t9/10s:1 m 40 s for JMY+Arp2/3; 6 m 20 s for Scar+Arp2/3; 37 m 0 s for actin alone. Filaments were diluted to low nanomolar concentrations and spotted on poly-L-lysine (Sigma) coverslips, using wide-bore pipette tips to minimize shearing.
Cell culture
U2OS (ATCC) and HEK 293 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 2 mM L-glutamine, non-essential amino acids, and penicillin-streptomycin (UCSF Cell Culture facility). For transfection, cells were seeded onto glass coverslips and transfected with HA-JMY4 using GeneJuice (Merck), or GFP-JMY and GFP-RNAi constructs using Lipofectamine LTX (Invitrogen), according to manufacturer’s protocol. HL-60 cells were cultured as described25, in RPMI-1640 with 25 mM HEPES, 2.0 g/L NaHCO3, 10% FBS, 1% antibiotic-antimycotic (Fisher), and were passaged every 3–4 days to a density of 0.2 ×106 cells/mL. Differentiation was in complete medium containing 1.3% DMSO (Hybrimax, Sigma). All cells were grown at 37° with 5% CO2. Primary human neutrophils were obtained by finger pinprick as described26.
Immunofluorescence of HL-60 cells was performed as described25. Briefly, flamed coverslips were treated with 200 µg/mL bovine plasma fibronectin (Sigma) in PBS, washed with PBS, and blocked with 1.8% low endotoxin BSA (Sigma) in modified Hanks buffered saline solution (mHBSS: 150 mM NaCl, 4 mM KCl, 1 mM MgCl2, 10 mM glucose, 20 mM HEPES, pH 7.4 at 22° C) for 5 minutes prior to adhering cells. Cells were pelleted and resuspended in BSA/mHBSS, and adhered to coverslips for 30–60 minutes at 37° C, then washed to remove unbound cells. Stimulation or mock stimulation was in BSA/mHBSS with 100 nM (HL-60s) or 20 nM (human neutrophils) fMLP, or DMSO (carrier), for 5 minutes at 22° C. Cells were fixed in 3.2% formaldehyde in cytoskeletal buffer (138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 320 mM sucrose, 10 mM HEPES, pH 7.2 at 22° C). For immunofluorescence of U2OS cells, cells were plated on flamed, fibronectin-coated coverslips, and fixed for 30 minutes in 3.2% formaldehyde in PBS. Cells were then permeabilized with 0.1% triton in PBS with 1.4 U/mL Alexa Fluor 568 phalloidin (Invitrogen) to stabilize and visualize filaments. For JMY immunolocalization, rabbit polyclonal JMY antibody 12895 or Anti-HA antibody HA11 (Babco) was used at a 1:500 dilution. Alexa Fluor 488-labeled goat-anti-rabbit secondary (Invitrogen) was used at a 1:500 dilution. DAPI (Sigma) was used at 0.5 µg/mL. Samples were mounted with fluorescent mounting medium (DakoCytomation).
Epifluorescence and wide field images were acquired on a Nikon TE300 inverted microscope equipped with a Hamamatsu C4742-98 cooled CCD camera, with Simple PCI software (Compix), using 100x and 60x 1.4 NA Plan Apo objectives (Nikon), or with a 10x 0.6 NA Phase objective, using MicroManager software27. For quantification of F-actin levels in cells expressing GFP-PWWWCA or GFP, micrographs were acquired with an IX Micro automated microscope, using identical illumination conditions. Cells were identified and outlined using ImageJ software, background was subtracted, and the average intensity in the red and green channels were measured. We used ImageJ (National Institutes of Health) and Adobe Photoshop (Adobe) for image analysis and contrast adjustment.
For wound healing experiments, cells were plated on marked coverslips at the same density, scratch wounded with a micropipette tip, washed to removed detached cells, and then given fresh medium and kept at 37°C during image acquisition. Stable lines of U2OS cells stably expressing GFP-JMY and variants were selected in 500 µg/mL G418 for at least 3 weeks, then enriched by FACS (UCSF Flow Cytometry Core). siRNA was used at a final concentration of 25nM (hJMY siRNA from Santa Cruz and control non-targeting 2 from Dharmacon) and transfected into cells with oligofectamine or Dharmafect 1 (both according to manufacturer’s protocols), and cells were grown for 72 hours prior to experiments. To discriminate between RNAied and non-RNAied cells, pL-UGIH plasmid (ATCC) was modified to contain a human JMY-specific hairpin based on the sequence of JMY-specific siRNA-3 by the method of Weiner et al. (2006)25. HEK 293 cells transiently transfected with this construct were grown for 7 days prior to fixation or immunoblotting.
Standard methods were used for Western blotting, using 1:500 1289 or 1:1000 L-16 (Santa Cruz Biotechnology) JMY primary antibodies and 1:5000 HRP-anti-rabbit secondary (Jackson ImmunoResearch). p300-CT primary antibody (Millipore) was used at 1:500, goat-anti human GAPDH (Santa Cruz) was used at 1:10,000, and mouse anti-human actin (Sigma) was used at 1:20,000. HRP secondaries (Dako) were used at 1:10,000, and ECL reagent (SuperSignal West Pico, Pierce) was used according to the manufacturer’s instructions. All error values are standard error of the mean, s.e.m. We used two-tailed unpaired t-tests, assuming unequal variance, to calculate p-values (Microsoft Excel).
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH, the American Heart Association Predoctoral Fellowship (JBZ), MRC-Funding (ASC), and the Burroughs-Wellcome Fund Career Award in the Biomedical Sciences Fellowship (MEQ). We thank A. Kelly and R. Manlove for sequence analysis; O. Akin for help with Matlab scripts and advice; C. Campbell and H. Bourne for critical reading of the manuscript; O. Weiner, S. Wilson, P. Temkin, E. Oh, C. Vizcarra, S. Cai, M. D’Ambrosio, K. Campellone, and members of the Mullins lab for reagents and helpful discussions.
Footnotes
Author contributions
J.B.Z, A.S.C, and M.E.Q conducted the experiments and analyzed the results. J.B.Z, A.S.C, M.E.Q, N.B.T, and R.D.M. conceived the experiments and wrote the manuscript.
References
- 1.Welch M, Mullins R. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 2.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 3.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 4.Shikama N, et al. A novel cofactor for p300 that regulates the p53 response. Mol. Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 5.Coutts A, Boulahbel H, Graham A, La Thangue N. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 2006;8:84–90. doi: 10.1038/sj.embor.7400855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Symons M, et al. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 7.Pollard T, Borisy G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J, Pollard T. Effect of capping protein on the kinetics of actin polymerization. Biochemistry. 1985;24:793–799. doi: 10.1021/bi00324a039. [DOI] [PubMed] [Google Scholar]
- 9.Pan F, Egile C, Lipkin T, Li R. ARPC1/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators. J Biol Chem. 2004;279:54629–54636. doi: 10.1074/jbc.M402357200. [DOI] [PubMed] [Google Scholar]
- 10.Ahuja R, et al. Cordon-Bleu Is an Actin Nucleation Factor and Controls Neuronal Morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins S, Ruscetti F, Gallagher R, Gallo R. Normal functional characteristics of cultured human promyelocytic leukemia cells (HL-60) after induction of differentiation by dimethylsulfoxide. J Exp Med. 1979;149:969–974. doi: 10.1084/jem.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalski JR, et al. Cortactin regulates cell migration through activation of N-WASP. J Cell Sci. 2005;118:79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- 13.Sablina A, Chumakov P, Kopnin B. Tumor suppressor p53 and its homologue p73alpha affect cell migration. J. Biol. Chem. 2003;278:27362–27371. doi: 10.1074/jbc.M300547200. [DOI] [PubMed] [Google Scholar]
- 14.Gadea G, de Toledo M, Anguille C, Roux P. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J. Cell Biol. 2007;178:23–30. doi: 10.1083/jcb.200701120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krubasik D, et al. Absence of p300 induces cellular phenotypic changes characteristic of epithelial to mesenchyme transition. Br J Cancer. 2006;94:1326–1332. doi: 10.1038/sj.bjc.6603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southwick FS, Dabiri GA, Paschetto M, Zigmond SH. Polymorphonuclear leukocyte adherence induces actin polymerization by a transduction pathway which differs from that used by chemoattractants. J Cell Biol. 1989;109:1561–1569. doi: 10.1083/jcb.109.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullins R, Heuser J, Pollard T. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 19.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 20.Shao D, Forge A, Munro P, Bailly M. Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil Cytoskeleton. 2006;63:395–414. doi: 10.1002/cm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon DJ, Eisenberg E, Korn ED. Characterization of cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J Biol Chem. 1976;251:4778–4786. [PubMed] [Google Scholar]
- 22.Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 23.Dayel MJ, Holleran EA, Mullins RD. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc Natl Acad Sci U S A. 2001;98:14871–14876. doi: 10.1073/pnas.261419298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiner O, et al. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cassimeris L, McNeill H, Zigmond S. Chemoattractant-stimulated polymorphonuclear leukocytes contain two populations of actin filaments that differ in their spatial distributions and relative stabilities. J. Cell Biol. 1990;110:1067–1075. doi: 10.1083/jcb.110.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuurman N, Amodaj N, Vale RD. Micro-Manager: Open Source software for light microscope imaging. Microscopy Today. 2007;15:42–43. [Google Scholar]
- 28.Petrella E, Machesky L, Kaiser D, Pollard T. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35:16535–16543. doi: 10.1021/bi961498d. [DOI] [PubMed] [Google Scholar]
- 29.Kelly A, Kranitz H, Dötsch V, Mullins R. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J. Biol. Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreishman-Deitrick M, et al. NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry. 2005;44:15247–15256. doi: 10.1021/bi051065n. [DOI] [PubMed] [Google Scholar]
- 31.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.