Abstract
In this article, a first aim was to develop a minimal modeling approach to noninvasively assess hepatic insulin extraction in 204 healthy subjects studied with a standard meal by coupling the already available meal C-peptide minimal model with a new insulin model. The ingredients of this model are posthepatic IDR, which in turn is described in terms of pancreatic ISR and hepatic insulin extraction HE, and a linear monocompartmental model of insulin kinetics. Even if ISR is provided by the C-peptide minimal model, the simultaneous assessment of HE and insulin kinetics is critical, since compensations may arise between parameters describing these two processes. Therefore, as a second aim of this study, a method was developed to predict standard values of insulin kinetic parameters in an individual on the basis of the individual's anthropometric characteristics. The statistical analysis, based on linear regression of insulin kinetic parameters estimated from IM-IVGTT data performed on the same subjects, demonstrated that insulin kinetic parameters can be accurately predicted from age and body surface area. Once kinetic parameters of the new insulin model were fixed to these values, HE profile and indexes during a meal were reliably estimated in each individual, indicating a significant suppression during the meal since the overall index of HE, equal to 60 ± 1% in the basal state, is reduced to 40 ± 1% during a meal. However, standard parameters provide an approximation of the individual one; thus, the third aim was to define the impact on estimated indexes of using standard instead of individually estimated values. Our results showed that the 25% uncertainty affecting as an average insulin kinetic parameters of an individual, when they are predicted from age and body surface area, translates into a similar relative uncertainty in the individual's hepatic insulin extraction indexes.
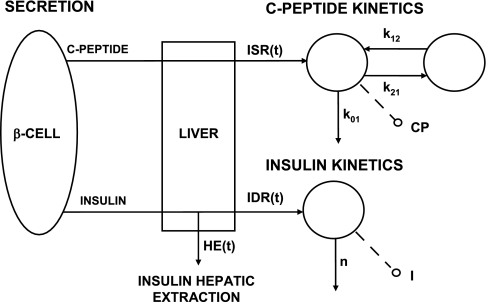
the liver plays a major role in determining circulating insulin levels since a significant fraction (∼50%) of secreted insulin is extracted in every passage through it. Thus, a quantitative assessment of hepatic insulin extraction (HE) in an individual, in the basal state, and/or during a glucose perturbation, especially under normal life conditions, e.g., during a meal, is an essential step in deriving an overall parametric picture of the glucose regulatory system (6). Since the direct measurement of HE requires invasive protocols, with catheters placed in artery and hepatic vein, indirect approaches based on mathematical models are essential to infer hepatic extraction from plasma measurements (7). We recently proposed a modeling method to measure the time course and indexes of hepatic extraction during an insulin-modified intravenous glucose tolerance test (IM-IVGTT) (12) based on the rationale illustrated in Fig. 1; insulin and C-peptide are equimolarly secreted from the β-cells and pass through the liver, where insulin, but not C-peptide, undergoes hepatic extraction (11). The simultaneous modeling of insulin secretion rate (ISR) from C-peptide concentration and insulin delivery rate (IDR) in plasma after its passage through the liver from insulin concentration provides an estimate of hepatic extraction profile and index:
| (1) |
| (2) |
Fig. 1.
Rationale for assessing hepatic extraction from insulin and C-peptide data. I (pmol/l), plasma insulin concentration, accessible to measurement; n (min−1), rate constant of insulin disappearance; CP (pmol/l), C-peptide concentration in the accessible compartment; k01, k21, and k12, transfer rate parameters; IDR (pmol/min−1), posthepatic insulin delivery rate; ISR (pmol/min), insulin secretion rate; HE (%), hepatic insulin extraction.
In Eq. 2, T is the time at which glucose, insulin, and C-peptide concentrations reach their end-test values after the perturbation.
Two models were employed to predict ISR and IDR from plasma measurements, the C-peptide minimal model and the insulin minimal model. These models, identified on plasma concentration data, integrate the ISR and IDR models with a two-compartment model of C-peptide kinetics and a single-compartment model of insulin kinetics, respectively. However, the simultaneous estimation of secretion and kinetics from a single experiment is in general difficult due to undesired compensations between the two processes (13). Thus, C-peptide kinetic parameters were fixed to standard values calculated according to the method proposed (15) from individual anthropometric characteristics, whereas for insulin the use of IM-IVGTT data rendered possible a reliable estimation of both insulin kinetics and IDR in each individual thanks to the decay of insulin concentration observed after the short insulin infusion administered from 20 to 25 min after the glucose bolus.
The purpose here is to extend the minimal model approach to measure HE during a meal test (or an oral glucose load) in humans. The extension is not straightforward for two reasons. First, although a model of C-peptide secretion and kinetics during an oral test is already available (4), a model of IDR and kinetics is not, and second, a priori knowledge of insulin kinetics is required to derive a reliable estimate of IDR from insulin concentration.
To reach our goals, we analyzed meal data of 204 health subjects who also underwent IM-IVGTT. IM-IVGTT data, analyzed with the minimal model of insulin secretion and kinetics (12), provided the basis for determining standard parameters for insulin clearance to be used to assess posthepatic insulin secretion and thus HE from plasma insulin data during the meal test with a new model. Finally, the impact on HE indexes and profile of using standard rather than individually derived insulin kinetic parameters was evaluated.
DATABASE
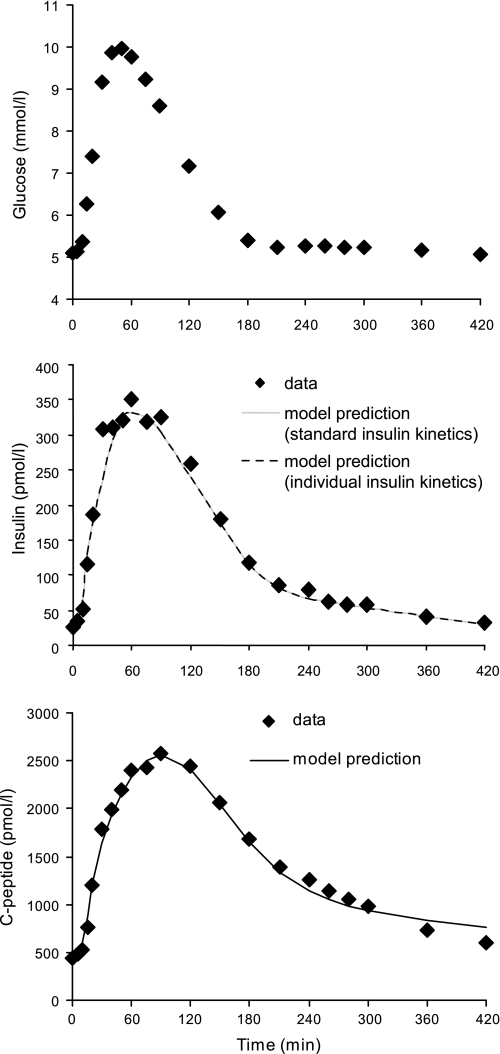
The data originated from a previous study (3) to which we refer for details on subjects, experimental protocols, and analytical methods. Briefly, 204 normal subjects [117 males and 87 females, age 55.5 ± 1.5 yr (means ± SE), body mass index (BMI) 26.6 ± 0.2kg/m2, body surface area (BSA) 1.90 ± 0.01 m2] underwent an oral test consisting of the ingestion within 15 min of a standard mixed meal (10 kcal/kg body wt, 45% carbohydrate, 15% protein, 40% fat). The blood samples were collected at −120, −30, −20, −10, 0, 5, 10, 15, 20, 30, 40, 50, 60, 75, 90, 120, 150, 180, 210, 240, 260, 280, 300, 360, and 420 min for the measurement of glucose, C-peptide, and insulin concentrations, with time 0 considered as the start of the meal. Average glucose, insulin, and C-peptide concentrations are shown in Fig. 2.
Fig. 2.
Mean plasma glucose (top), insulin (middle), and C-peptide (bottom) concentrations in 204 healthy subjects during a meal. Predictions of insulin and C-peptide minimal models are shown as solid or dotted lines.
On a different day, subjects underwent an IM-IVGTT consisting of an intravenous injection of glucose (0.3 g/kg body wt) followed by a square-wave (from 20 to 25 min) insulin infusion (0.02 U/kg body wt). Blood was sampled at 0, 2, 4, 6, 8, 10, 15, 20, 22, 25, 26, 28, 31, 35, 45, 60, 75, 90, 120, 180, and 240 min after the glucose injection, and glucose, C-peptide, and insulin concentrations were measured. Average glucose, insulin, and C-peptide concentrations in a subset of 88 subjects were reported in Ref. 2.
METHODS
Insulin minimal model.
A linear monocompartmental model of insulin kinetics was adopted to link IDR in plasma IDR (pmol/min) to plasma insulin data I (pmol/l):
| (3) |
where n (min−1) is the rate constant of insulin elimination, VI (pmol/l) is the insulin volume of distribution, and Ib is basal insulin concentration. By exploiting Eq. 1, IDR was expressed as the fraction (1-HE) of pancreatic insulin secretion (ISR) that is not secreted by the liver:
| (4) |
ISR was predicted from C-peptide and glucose levels of the oral test, according to the minimal model of C-peptide secretion and kinetics proposed in Ref. 4, and is reported briefly in the appendix 1. For HE, a parametric description was adopted so that its reconstruction was stated as a parameter estimation problem. The most general form is a piecewise linear function with a given number of break points:
| (5) |
where HEi are the unknown parameters representing the values of HE(t) at the break times. Since HE(t) is expected to vary more rapidly in the first portion of the test, intervals were shorter at the beginning and became longer toward the end. A preliminary analysis indicated that six intervals, and thus seven break points, are a good compromise between model flexibility and number of parameters to be estimated from the data. The break points were allocated at 0, 20, 40, 90, 150, 210, and 420. HEb appearing in Eq. 5 is the basal insulin hepatic extraction, which can be expressed as a function of basal levels of ISR and IDR as
| (6) |
where ISRb is to be determined from the minimal model of C-peptide secretion and kinetics (see appendix 1), whereas for IDRb, its expression in terms of insulin model parameters n, VI, and Ib, obtained by solving Eq. 3 in steady state, is used.
In addition to the index of hepatic extraction in the basal state (Eq. 6), a global index of hepatic extraction during the oral test was estimated by using Eq. 2.
Standard parameters of insulin kinetics.
Since a reliable estimation of HE in an individual requires a priori knowledge of the individual's insulin kinetic parameters, we exploited the potentiality of the available data set, with IM-IVGTT performed in conjunction with the meal in all of the 204 subjects, to derive standard parameters for insulin kinetics. The application of the minimal model of insulin secretion and kinetics (12) to IM-IVGTT data of our data set provided estimates of insulin kinetic parameters in a population of 204 healthy subjects heterogeneous in terms of age, sex, and body weight. On the basis of these data, linear regression models relating insulin kinetic parameters such as n, VI, and insulin clearance CLI = n·VI (l/min) to individual anthropometric characteristics were developed, following the approach that was used in Ref. 15 to calculate standard kinetic parameters for C-peptide clearance. The subjects were grouped according to sex (male and female) and BMI (lean, i.e., BMI <27, and obese, i.e., BMI ≥27), and the significance of differences of insulin kinetic parameters between the groups and the dependence on covariates (age, weight, and BSA) were tested across all groups (i.e., with a 2 × 2 comparison across all the categories) with multivariate analysis of variance (MANOVA) and general linear regression models with backward elimination (P = 0.2 to enter, P = 0.05 to remove). Only sufficiently independent covariates (i.e., covariates with r < 0.95) were considered in the model. Normality and homogeneity of variance in all groups, a prerequisite of MANOVA, were tested by using the Shapiro-Wilks and the Levene tests, respectively. The parameterization VI, n, and CLI is redundant; thus we associated with VI the parameter between n and CLI, best predicted from anthropometric characteristics, based on the value of the regression coefficient. A P value <0.05 was considered significant. Statistical analysis was performed with Statistica version 6.1 software (StatSoft).
In view of using the regression models to predict insulin kinetic parameters in an individual for whom IM-IVGTT data are not available, we quantified the prediction accuracy for each kinetic parameter, as the square root of the mean squared error of prediction [root mean square error (RMSE)] (5), from the deviations e(i) between standard (i.e., predicted from anthropometric characteristics) and individual (i.e., estimated from IM-IVGTT) value in all subjects:
| (7) |
where Np is the number of parameters of the regression model.
RMSE potentially underestimates prediction errors, since the same data were used to both build the regression models and assess the predictive power. To avoid this bias, a leave-one-out cross-validation strategy was adopted as a second step; the regression model was identified 204 times on all of the possible combinations of 204 subjects and then used to predict the insulin kinetic parameters of the subject not included in the training set. The RMSE index was recalculated with a formula similar to Eq. 7 and denoted as CV-RMSE (5):
| (8) |
Model identification.
Identification of C-peptide and insulin models was performed in each subject by using SAAM II software (1). First, ISR profile was predicted from ISR parameters of the C-peptide model estimated from plasma C-peptide data with C-peptide kinetic parameters fixed to standard population values (15). Second, ISR profile was used in the insulin model, Eq. 4, and HE parameters were estimated from plasma insulin data, with insulin kinetic parameters fixed to the standard values calculated with the regression model described in the previous section.
A second identification was performed with insulin kinetic parameters fixed to individual values estimated in each subject during IM-IVGTT. Absolute weights were chosen, equal to the inverse of the variance of the measurement errors, and assumed to be independent, gaussian, and zero mean with a variance linked to insulin and C-peptide measurements (12).
RESULTS
Standard parameters of insulin kinetics.
VI, CLI parameterization showed a slightly higher correlation (see below) with anthropometrical characteristics compared with VI, n and thus was adopted to describe insulin kinetics. Estimates of VI and CLI in 204 subjects from IM-IVGTT are shown in Table 1 together with basal, i.e., before the meal, insulin secretion and concentration in lean vs. obese and male vs. female subjects. Basal insulin concentration and secretion were significantly different between lean and obese but not between male and female. VI was significantly different in both lean vs. obese and male vs. female, whereas CLI was significantly different only between male and female.
Table 1.
Insulin secretion, kinetics, and hepatic extraction parameters
| ISRb, pmol/min | Ib, pmol/l | VI, l | CLI, l/min | HEb, % | HE, % | |
|---|---|---|---|---|---|---|
| Subjects (no.) | ||||||
| All (204) | 116.95±4.67 | 27.4±1.27 | 9.31±0.18 | 1.7±0.03 | 60±1 | 40±1 |
| Lean (108) | 96.23±3.59 | 23.97±1.37 | 8.67±0.17 | 1.71±0.03 | 59±1 | 39±1 |
| Obese (96) | 140.27±5.2 | 31.26±1.08 | 10.03±0.17 | 1.69±0.03 | 62±1 | 41±1 |
| Female (87) | 112.38±9.29 | 28.48±2.57 | 8.24±0.25 | 1.57±0.04 | 61±2 | 38±2 |
| Male (117) | 120.35±4.28 | 26.60±1.10 | 10.11±0.22 | 1.80±0.04 | 60±1 | 42±1 |
| P value | ||||||
| Lean vs. obese | 1.4·10−6 | 1.4·10−6 | 9.1·10−5 | 0.67 | 0.16 | 0.35 |
| Female vs. male | 0.40 | 0.47 | 6.2·10−8 | 2.7·10−4 | 0.77 | 0.25 |
Values are means ± SE.
ISRb, basal insulin secretion rate; Ib, basal insulin concentration; VI, insulin volume of distribution; CLI, insulin clearance; HE, hepatic insulin extraction; HEb, basal HE.
Body weight and BSA were highly correlated (r > 0.95), and thus only BSA was included in the model, due to its slightly higher correlation with insulin kinetic parameters. Both VI and CLI were log-normally distributed (P < 0.05) in all groups of subjects, and thus all statistical analysis were performed on natural logarithm of VI [ln(VI)] and CLI [ln(CLI)]. 1) Levene test indicated that ln(VI) and ln(CLI) had the same variance in all groups of subjects (P < 0.05), 2) MANOVA and backward removal general linear regression analysis indicated that the two significant covariates were age and BSA (P < 0.01), and 3) differences of ln(VI) and ln(CLI) between groups of subjects were not significant when BSA and age were taken into account.
The linear regression models linking ln(VI) and ln(CLI) to age and BSA, with their coefficient of multiple correlation r, are
| (9) |
These formulas were applied to the 204 subjects of our database, and the relationship between standard (i.e., predicted from age and BSA by using Eq. 9) and individual (estimated from IM-IVGTT) values (Fig. 3) indicated a good correlation for VI (r = 0.51, P < 0.0001), whereas a lower (r = 0.36, P < 0.0001) but still significant correlation was obtained for CLI. Accuracy of Eq. 9 in predicting insulin kinetic parameters, measured by the RMSE between standard and individual values, was 2.20 and 0.44 for VI and CLI, respectively, i.e., <25% of the average VI and CLI values.
Fig. 3.
Comparison between individual and standard insulin kinetic parameters CLI (l/min) and VI (l). The line represents the linear regression between individual and standard parameters.
Validation by leave-one-out technique confirmed the predictions of the regression model, since correlation coefficients between standard and individual values were r = 0.50 and 0.31 for VI and CLI, respectively, and CV-RMSE = 2.21 and 0.44 for VI and CLI, respectively. The results of leave-one-out cross-validation also indicated a low impact of outliers in the determination of the coefficients of the regression models, since the mean values of the model coefficients in the N identifications were virtually equal to those obtained identifying the model on all subjects (Eq. 9), and the variability among different identifications was low. CV of model coefficients ranged from 1 to 5%, except for the intercept of the CLI model (CV = 40%), but this was expected due to the low impact of this coefficient in the calculation of CLI.
Model of insulin hepatic extraction and kinetics.
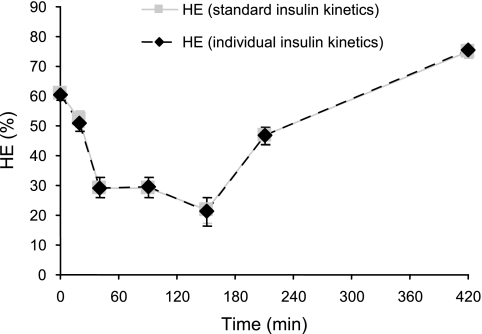
Prediction of the insulin minimal model, identified by using standard insulin kinetics, is shown in Fig. 2, superimposed to insulin data. Similarly, predictions of the C-peptide minimal model are superimposed to C-peptide data. Estimated values of HE (%) at break points were 52 ± 2, 29 ± 2, 29 ± 2, 22 ± 3, 47 ± 2, and 75 ± 1 (at times 20, 40, 90, 150, 210, and 420, respectively), and their precision was good, since the average CVs (%) of the estimates were 12, 23, 29, 32, 9, and 6, respectively. The average HE profile is depicted in Fig. 4; starting from a basal value HEb = 61 ± 1%, HE was suppressed to ∼20–30% for ∼150 min following the glucose load and then slowly returned to steady state. The value of HE reached at the end of the experiment was somewhat higher than HEb measured pretest, which was expected due to differences in insulin and C-peptide concentrations. The total index of hepatic extraction HE indicates that, on the average, 40 ± 1% of secreted insulin is extracted by the liver during a standard meal.
Fig. 4.
HE during a meal (mean ± 90% confidence interval in 204 healthy subjects), estimated with standard (solid gray line) and individual (dotted black line) insulin kinetic parameters.
Despite the differences observed between lean and obese in basal insulin secretion and concentration, hepatic extraction indexes (Table 1) were not significantly different, nor was HE profile (data not shown), between the two groups.
Impact of standard parameters of insulin kinetics on estimation of hepatic extraction.
As a further validation of the regression model, we evaluated the impact on hepatic extraction indexes and profile of using standard insulin kinetic parameters (predicted by regression of population data), as we did, rather than individual ones (available for these subjects thanks to IM-IVGTT). As an average, our results indicated no significant impact, since the model fits of plasma insulin concentration were virtually the same in the two situations (Fig. 2), as were the model predictions of HE profile (Fig. 4) and HE indexes (HEb: 60 ± 1 vs. 61 ± 1; HE: 40 ± 1 vs. 40 ± 1). Regarding the individual values of HE indexes, there was a good correlation between the two sets of estimates (HEb: r = 0.73, P < 0.0001; HE: R = 0.70, P < 0.0001). The square roots of the mean squared deviations between HE values estimated with insulin kinetic parameters fixed to either standard or individual values were 10 and 16, respectively, indicating that reasonably accurate estimates of HE indexes were obtained in an individual with standard insulin kinetic parameters.
To further support these findings, the impact of errors on insulin kinetic parameters on the HE estimation was evaluated in a typical subject, having C-peptide and insulin concentration as well as age and BSA equal to the population means. Varying insulin kinetic parameters within the 95% confidence interval (estimated from RMSE; Eq. 9) resulted in a range of HE estimates equal to ±0.16, thus supporting the use of standard parameters of insulin kinetics.
DISCUSSION
In this article, a first aim was to develop a minimal modeling approach to noninvasively assess hepatic extraction during a meal by coupling the already available meal C-peptide minimal model (4) with a new insulin model. The ingredients of this model are posthepatic IDR, which in turn is described in terms of pancreatic insulin secretion ISR and HE, and a linear monocompartmental model of insulin kinetics. Even if ISR is provided by the C-peptide minimal model, the simultaneous assessment of HE and insulin kinetics is critical since compensations may arise between parameters describing these two processes (13). Therefore, as a second aim of this study, a method was developed to predict standard values of insulin kinetic parameters in an individual on the basis of the individual's age and BSA. Once kinetic parameters of the new insulin model were fixed to these vales, parameters describing HE during a meal were reliably estimated in each individual, yielding an estimate of HE time course and indexes. However, standard parameters provide an approximation of the individual one; thus, the third aim was to define the impact on estimated HE indexes using standard instead of individually estimated values. All of these goals were reached thanks to a unique data set consisting of both a meal and an IM-IVGTT performed on 204 normal subjects on two different occasions.
Insulin minimal model.
A single-compartment model was adopted to describe insulin kinetics, with IDR as model input, which in turn was modeled as the fraction (1-HE) of ISR that is not extracted by the liver. ISR can be reconstructed by modeling C-peptide data, whereas for HE a mathematical representation as a piecewise linear function was adopted with fixed breakpoints. HE values at the breakpoints were considered as model parameters to be estimated by fitting the model on insulin data. Once the insulin kinetics was fixed to standard values, HE parameters were the only unknown parameters. Seven break points were considered to be a good compromise since the model was able to reproduce the experimental data fairly well (Fig. 2), and the HE parameters were estimated with good precision.
This approach is somewhat different from the one used to estimate HE during an IM-IVGTT (12), which combined ISR and IDR predictions provided by C-peptide and insulin model, respectively. These two models adopted similar functional descriptions of glucose control on ISR and IDR but with different parameter values, since IDR parameters also accounted for HE. A similar approach was not successful with meal data, since an insulin model with an IDR description similar to that adopted for ISR in the oral C-peptide model (4) was not able to provide a reasonable fit of insulin data. In other words, during an oral test, the functional description able to describe ISR was not appropriate to describe IDR, even if parameter values were different. IDR combines ISR and HE; thus, a possible explanation can be found in the more prolonged HE perturbation observed after an oral test (see below) due to differences in glucose patterns during intravenous and oral tests as well differences in secretory mechanisms, e.g., the effect of incretin hormones.
The approach here developed is of more general validity since the piecewise linear description for HE is very flexible, able to adapt to a sustained and prolonged HE perturbation.
Standard parameters of insulin kinetics.
To determine standard parameters for insulin clearance, we analyzed the IM-IVGTT data in our 204 healthy subjects with the minimal model of insulin secretion and kinetics (12). Although this population was highly heterogeneous in terms of age, sex, and body weight, insulin kinetic parameters CLI and VI showed modest interindividual variability, 28 and 27%, respectively, suggesting that they are reproducible from standard population values. The relationships between insulin kinetic parameters in 204 individuals and their anthropometric characteristics were formalized via statistical analysis on the basis of MANOVA and stepwise linear regression. A main result is that insulin kinetic parameters were affected significantly from age and BSA but that the differences between groups of subjects, i.e., male and female, lean and obese, were not significant when effects of age and BSA were taken into account. Therefore, standard values of CLI and VI in a normal subject (either male or female, lean or obese) can be derived by using Eq. 9 as a function of the subject's BSA and age. A fairly good correlation was found between VI and the anthropometrical characteristics (r = 0.55), whereas correlation for CLI was lower (r = 0.35). These values are similar to those associated with the regression models widely used to predict parameters of C-peptide kinetics (15), namely r = 0.50 in men and r = 0.34 in women between C-peptide volume of distribution and BSA and r = 0.28 between C-peptide long half-life and age. Despite these modest correlation values, C-peptide standard kinetic parameters (predicted with the regression models) allowed the estimation of insulin secretion with good approximation (15). Moreover, it was shown in Refs. 8 and 9 that the use of standard instead of individual C-peptide kinetics does not affect, on average, estimates of insulin secretion profile and indexes but only the confidence intervals, which becomes wider but still acceptable. Interestingly, in that study, both the number of subjects (200) and the interindividual variability of parameters defining C-peptide kinetics (20–36%) were similar to those of the present study.
Since regression coefficients quantify only the extent to which standard and individual values are related but do not give any information on the accuracy of the prediction, we calculated the squared RMSE between standard and individual values (Eq. 7), which was close to 25% of parameter values, even when evaluated via a leave-one-out cross-validation analysis, thus supporting the use of standard values for insulin kinetics. However, it is worth nothing that our database included subjects with ages ranging from 18 to 87 yr. Caution should be taken in applying this model to children, because the mechanism of insulin clearance maturation during the childhood and adolescence requires further studies and investigations.
HE.
HE during an oral test was often quantified as originally suggested in (11) by using the C-peptide to insulin molar ratios from the areas under plasma concentration curves; see, e.g., Ref. 10. As already discussed in Ref. 12, this ratio is only an indicator of hepatic extraction, since it also depends on clearance rates of C-peptide and insulin. A modeling approach to measure HE during an oral glucose tolerance test was proposed in Ref. 14, which combines C-peptide and insulin models. However, it assumed a simplistic single-compartment model for C-peptide kinetics and a constant insulin hepatic extraction, even if direct measurement of this flux via artero-venous differences suggested that hepatic extraction varies in time, and is significantly related to blood flow. By applying their model to normal, obese, and diabetic subjects, these authors estimated a hepatic extraction equal as an average to 59, 56, and 66%, which represents an approximation of the values directly measured in the same subjects by hepatic catheterization (41, 46, and 57%).
In the present study, our model analysis provided a time-varying HE profile during a meal. HE was markedly suppressed within 30 min after glucose ingestion, since from the pretest value equal as an average to HEb = 60 ± 1% it reached a nadir equal to ∼20–30%, which was maintained for ∼2 h; then, it slowly returned to steady state. As an average, 40 ± 1% of insulin passing through the liver was extracted during a meal. During IM-IVGTT on the same subjects, HE was somewhat higher (51 ± 1%) and the profile markedly different (see Ref. 12, where the profile obtained in a representative subset of 20 subjects is shown). A possible explanation is that liver saturation mechanisms are activated at insulin concentration levels observed during a glucose test, either oral or intravenous, but overall HE suppression is more pronounced during a meal, since insulin concentration remains elevated (∼200 pmol/l) for ∼150 min, whereas during an IM-IVGTT, insulin perturbation is more rapid and elevated but vanishes within 60 min.
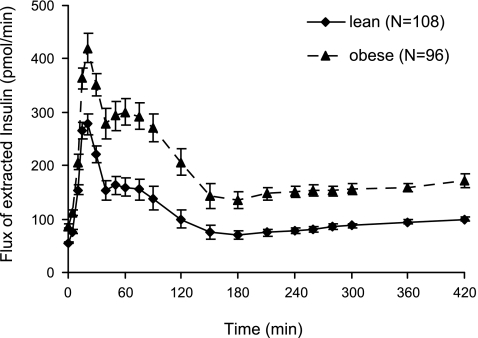
No difference was detected for HE profile (not shown) and indexes between males and females (Table 1). HE profile and indexes were also similar between lean and obese despite the significant differences in insulin secretion and concentration between the two groups. As a consequence, the flux of extracted insulin was significantly higher in obese compared with lean subjects (Fig. 5). Moreover, the inverse relationship between insulin concentration and HE (Fig. 6) is shifted to the right in obese subjects, with saturation of HE occurring at higher insulin levels compared with lean subjects (∼240 vs. 140 pmol/l). Although these findings might suggest a higher liver capacity, in absolute terms, to extract insulin in obese subjects, they require further investigation, also because age may exert a confounding effect (2, 3).
Fig. 5.
Flux of extracted insulin by the liver during a meal (means ± SD) for lean (solid line) and obese subjects (dotted line).
Fig. 6.
Mean HE plotted vs. mean insulin plasma concentration during a meal for lean (solid line) and obese subjects (dotted line). The arrows indicate the direction with respect to time.
HE and standard insulin kinetic parameters.
In the present study, HE was estimated by fixing insulin kinetic parameters to standard values. As previously discussed, standard values provide a very good approximation of insulin kinetic parameters at population level, whereas at single-subject level the average relative accuracy is ∼25%. It is thus important to evaluate the impact on HE estimations of fixing insulin kinetic parameters to standard instead of individual values by comparing HE profile and indexes obtained in the two situations. Our results indicate that, at population level, standard values of insulin kinetic parameters can be used to assess hepatic extraction without loss of accuracy, whereas at the single-subject level, HE accuracy (15% for HEb and 40% for HE) reflects the accuracy of insulin kinetic parameters.
HE and insulin clearance.
In line with previous analysis of IVGTT data (12), a linear single-compartment model was assumed to describe insulin kinetics. The linearity assumption appears reasonable since insulin concentration remains within the linearity range for the largest part of the experiment. On the other hand, the reduction of HE predicted during meal is apparently in contrast with this assumption because HE is a relevant component of insulin clearance. However, as discussed extensively in Ref. 12, the reduction from basal of HE during intravenous as well as oral tests accounts for only a low percentage (<20%) decrease of insulin clearance, which is hard to detect from insulin data. We can thus speculate that insulin clearance is likely to vary in time during a meal due to variations in hepatic extraction, but these variations are modest and difficult to detect from plasma insulin data. Only a constant value of plasma clearance rate can be used, which represents the overall clearance value during meal.
To check the impact of these assumptions on assessment of HE, we applied an alternative model that links insulin clearance to hepatic extraction. The model, presented for IM-IVGTT in Ref. 12 and reported here in appendix 2, describes insulin clearance as the sum of two processes: hepatic clearance, varying in time with a pattern proportional to hepatic extraction profile; and peripheral clearance, assumed to be constant. As for IM-IVGTT data, numerical identification required to fix a priori the ratio between the two processes at basal state, i.e., hepatic clearance, accounts for 60% of the total clearance. Moreover, for meal data, VI and basal insulin clearance were fixed equal to the corresponding values estimated on IM-IVGTT data analyzed with the time-varying clearance model. Thus, the new model was identified on the 17 subjects out of 20 analyzed in Ref. 12 that presented plausible estimates of insulin clearance. It provided a fit virtually superimposable to that of the minimal model (data not shown) and average values of insulin clearance and HE indexes very similar and well correlated to those estimated with the minimal model (CLI = 1.62 ± 0.1 vs. 1.63 ± 0.1 l/min, r = 0.73, P = 0.001; HEb = 63 ± 3 vs. 66 ± 2%, r = 0.77, P = 0.001; HE = 50 ± 3 vs. 45 ± 3%, r = 0.78, P < 0.001). There were some differences in HE profiles estimated with the two models at some specific time points, but the overall correlation between HE profiles was good (r = 0.83, P = 0.001). Summing up, the use of a more complex model that links insulin clearance to HE profile provided similar estimates of average insulin clearance and hepatic insulin extraction. However, numerical identification of this model is required to fix a priori the ratio between the hepatic and peripheral insulin clearance at basal state, and this is a critical issue since the ratio is likely to vary among subjects/conditions. These reasons thus support the assumption that a linear insulin kinetics allows an adequate description of HE processes.
In conclusion, hepatic extraction and IDR during an oral test can be assessed by combining the insulin and C-peptide minimal models. The linear regression models here proposed predict standard values of insulin kinetic parameters from anthropometrical characteristics, which can be safely used to derive reliable estimates of hepatic extraction in experimental protocol-like meal-like oral glucose tolerance test and standard IVGTT, where the simultaneous estimation of insulin kinetic and delivery/hepatic extraction is critical.
GRANTS
This study was partially supported by National Institutes of Health Grants RR-00585 and DK-29953 and by Ministero dell'Università e della Ricerca Scientifica, Rome, Italy.
APPENDIX 1: MINIMAL MODEL DESCRIPTION OF INSULIN SECRETION RATE
The minimal model of C-peptide secretion and kinetics (4) models the three components of pancreatic secretion: basal (ISRb), static (ISRs) controlled by glucose concentration, and dynamic (ISRd) controlled by glucose rate of increase, as normalized to the C-peptide volume of distribution VC, namely isrs, isrd, and isrb (pmol·min−1·l−1):
| (A1) |
isrs is assumed equal to the provision of releasable insulin to β-cells controlled by glucose concentration G (mmol/l) in a linear dynamic fashion, i.e., in response to a glucose step increase above threshold level h (mmol/l), provision, and thus, isrs tends with a rate constant α (min−1) and thus with a delay, T = 1/α (min−1), toward a steady-state value that is linearly related to the glucose step through a parameter Φs (10−9 min−1).
| (A2) |
isrd represents the secretion of insulin from the promptly releasable pool and is proportional to the rate of increase of glucose through parameter Φd (10−9):
| (A3) |
Since ISR model is identified on C-peptide concentration data, it is coupled with a two-compartment model of C-peptide kinetics:
| (A4) |
where CP1 and CP2 (pmol/l) are C-peptide concentrations in the accessible and in the peripheral compartment, respectively, CPb (pmol/l) is the basal plasma C-peptide concentration, and k21, k12, and k01 (min−1) are the constant kinetic parameters. Steady-state solution of Eq. A4 provides the expression of isrb as
| (A5) |
APPENDIX 2: INSULIN MODEL WITH INSULIN CLEARANCE LINKED TO HE(T)
Following the approach adopted for IVGTT and presented in the appendix of Ref. 12 to include in the model a link between insulin clearance and hepatic extraction, insulin clearance was described as the sum of two processes: hepatic clearance, varying in time with a pattern proportional to hepatic extraction; and peripheral clearance, assumed to be constant. Thus, the equation describing insulin kinetics is
| (A6) |
where nH HE(t) (min) represents hepatic clearance, modulated by HE, and nP (min) the peripheral insulin clearance, assumed to be constant during the experiment.
The new insulin model consists of Eq. A6 coupled with Eq. 5 to describe HE and Eq. 1 to describe IDR. It was identified simultaneously with the C-peptide model, Eqs. A1–A5, which provides a description for ISR appearing in Eq. 1.
To achieve numerical identification, a constraint was introduced on the relative contribution of hepatic and peripheral clearance in the basal state; namely, it was assumed that, in all subjects, hepatic and peripheral clearance account for 60 and 40%, respectively, of the basal clearance:
| (A7) |
From Eqs. A6 to A7 and Eq. 1, the formula to calculate Heb can be derived:
| (A8) |
REFERENCES
- 1.Barrett PH, Bell BM, Cobelli C, Golde H, Schumitzky A, Vicini P, Foster DM. SAAM II: Simulation, Analysis, and Modeling Software for tracer and pharmacokinetic studies. Metabolism 47: 484–492, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Basu R, Breda E, Oberg AL, Powell CC, Dalla Man C, Basu A, Vittone JL, Klee GG, Arora P, Jensen MD, Toffolo G, Cobelli C, Rizza RA. Mechanisms of the age-associated deterioration in glucose tolerance: contribution of alterations in insulin secretion, action, and clearance. Diabetes 52: 1738–1748, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Basu R, Dalla Man C, Campioni M, Basu A, Klee G, Toffolo G, Cobelli C, Rizza RA. Effects of age and sex on postprandial glucose metabolism: differences in glucose turnover, insulin secretion, insulin action, and hepatic insulin extraction. Diabetes 55: 2001–2014, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Breda E, Cavaghan MK, Toffolo G, Polonsky KS, Cobelli C. Oral glucose tolerance test minimal model indexes of beta-cell function and insulin sensitivity. Diabetes 50: 150–158, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Brown PJ. Measurement, Regression, and Calibration Oxford, UK: Clarendon, 1993 [Google Scholar]
- 6.Cobelli C, Toffolo GM, Dalla Man C, Campioni M, Denti P, Caumo A, Butler PC, Rizza RA. Assessment of β-cell function in humans, simultaneously with insulin sensitivity and hepatic extraction, from intravenous and oral glucose test. Am J Physiol Endocrinol Metab 293: E1–E15, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Ferrannini E, Cobelli C. The kinetics of insulin in man. II. Role of the liver. Diabetes Metab Rev 3: 365–397, 1987 [DOI] [PubMed] [Google Scholar]
- 8.Magni P, Bellazzi R, Sparacino G, Cobelli C. Bayesian identification of a population compartmental model of C-peptide kinetics. Ann Biomed Eng 28: 812–823, 2000 [DOI] [PubMed] [Google Scholar]
- 9.Magni P, Sparacino G, Bellazzi R, Toffolo GM, Cobelli C. Insulin minimal model indexes and secretion: proper handling of uncertainty by a Bayesian approach. Ann Biomed Eng 32: 1027–1037, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Meier JJ, Holst JJ, Schmidt WE, Nauck MA. Reduction of hepatic insulin clearance after oral glucose ingestion is not mediated by glucagon-like peptide 1 or gastric inhibitory polypeptide in humans. Am J Physiol Endocrinol Metab 293: E849–E856, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Polonsky KS, Rubenstein AH. C-peptide as a measure of the secretion and hepatic extraction of insulin. Pitfalls and limitations. Diabetes 33: 486–494, 1984 [DOI] [PubMed] [Google Scholar]
- 12.Toffolo G, Campioni M, Basu R, Rizza RA, Cobelli C. A minimal model of insulin secretion and kinetics to assess hepatic insulin extraction. Am J Physiol Endocrinol Metab 290: E169–E176, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Toffolo G, De Grandi F, Cobelli C. Estimation of beta cell sensitivity from IVGTT C-peptide data. Knowledge of the kinetics avoids errors in modeling the secretion. Diabetes 44: 845–854, 1995 [DOI] [PubMed] [Google Scholar]
- 14.Tura A, Ludvik B, Nolan JJ, Pacini G, Thomaseth K. Insulin and C-peptide secretion and kinetics in humans: direct and model-based measurements during OGTT. Am J Physiol Endocrinol Metab 281: E966–E974, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Van Cauter E, Mestrez F, Sturie J, Polonsky KS. Estimation of insulin secretion rates from C-peptide levels. Comparison of individual and standard kinetic parameters for C-peptide clearance. Diabetes 41: 368–377, 1992 [DOI] [PubMed] [Google Scholar]