Abstract
Background
ECE-CYC2 clade genes known in patterning floral dorsoventral asymmetry (zygomorphy) in Antirrhinum majus are conserved in the dorsal identity function including arresting the dorsal stamen. However, it remains uncertain whether the same mechanism underlies abortion of the ventral stamens, an important morphological trait related to evolution and diversification of zygomorphy in Lamiales sensu lato, a major clade of predominantly zygomorphically flowered angiosperms. Opithandra (Gesneriaceae) is of particular interests in addressing this question as it is in the base of Lamiales s.l., an early representative of this type zygomorphy.
Results
We investigated the expression patterns of four ECE-CYC2 clade genes and two putative target cyclinD3 genes in Opithandra using RNA in situ hybridization and RT-PCR. OpdCYC gene expressions were correlated with abortion of both dorsal and ventral stamens in Opithandra, strengthened by the negatively correlated expression of their putative target OpdcyclinD3 genes. The complement of OpdcyclinD3 to OpdCYC expressions further indicated that OpdCYC expressions were related to the dorsal and ventral stamen abortion through negative effects on OpdcyclinD3 genes.
Conclusion
These results suggest that ECE-CYC2 clade TCP genes are not only functionally conserved in the dorsal stamen repression, but also involved in arresting ventral stamens, a genetic mechanism underlying the establishment of zygomorphy with abortion of both the dorsal and ventral stamens evolved in angiosperms, especially within Lamiales s.l.
Background
One important event during the evolution of angiosperms is the emergence of flower bilateral symmetry, i.e. zygomorphy, a key innovation associated with important adaptive radiations [1]. Several zygomorphic clades have independently evolved successfully from actinomorphic ancestors in angiosperms, including Lamiales sensu lato that includes a major genetic model organism snapdragon (Antirrhinum majus) [2,3].
In A.majus, CYCLOIDEA (CYC) and DICHOTOMA (DICH) are essential for the development of dorsoventral asymmetry in flowers due to their dorsal identity function, i.e. controlling the fate of the dorsal floral organs in the second and third whorls [4,5]. CYC promotes cell expansion in the dorsal petals, while it arrests the growth of the dorsal stamen to become a staminode [4,5]. Meanwhile, DICH activity affects the internal asymmetry of the dorsal petals [4,5]. The ability of CYC to arrest the dorsal stamen depends on its negative effect on expression of cell-cycle genes, such as cyclin D3b [3,6]. CYC and DICH encode proteins within the ECE-CYC2 clade (ECE lineage, CYC/TB1 subfamily) in the TCP family of transcription factors with TCP domain related to cell proliferation [3,7-12]. In legumes, distantly related to A. majus, several CYC homologues, such as LjCYC2 in Lotus and PsCYC2 in pea, also have the function in establishing dorsal identity in legume flowers [13,14]. In Arabidopsis thaliana, a model eudicot species with ancestrally actinomorphic flowers, and its close relative Iberis amara, CYC homologues, TCP1 and IaTCP1 genes are characteristic of dorsal identity function, in which IaTCP1 dorsal-specific expression represses the two dorsal petal development in Iberis amara [15,16]. Recent studies in the sunflower family (Asteraceae) show that CYC-like genes, i.e. RAY1, RAY2 in Senecio and GhCYC2 in Gerbera, have played a key role in the establishment and evolution of the capitulate inflorescence [17,18]. Therefore, it is suggested that CYC-like TCP genes have been recruited multiple times for a role of dorsal identity and its modifications in establishing zygomorphy in core eudicots [3,19]. Even though the genetic control for the floral dorsoventral asymmetry has been intensively studied in model systems, it is still a great challenge to explain how modifications of development led to the transformation among different types of zygomorphy and the morphological diversification of zygomorphy in angiosperms, especially in Lamiales s.l., a major clade of predominantly zygomorphically flowered angiosperms.
Zygomorphy is believed to be ancestral in Lamiales s.l. [2,19,20]. Most zygomorphic groups in Lamiales s.l. have a pentamerous perianth with four stamens plus a dorsal staminode and two carpels as in A.majus. However, there is a great variation in morphology and number of corolla lobes and stamens [1]. The dorsal staminode can be completely lost as in Rehmannia and Veronica (Scrophulariaceae sensu lato) [2,21,22] and the two lateral stamens may become aborted instead of one dorsal staminode as in Mohavea (Scrophulariaceae s.l.) and Chirita (Gesneriaceae) [23-25]. In some cases, the two ventral stamens may become staminodes rather than the lateral stamens and the dorsal one, such as in Opithandra and Epithema (Gesneriaceae) [24]. In extreme cases, each flower may have only a single stamen as in Hippuris (Scrophulariaceae s.l.) [20]. In Mohavea, a close relative of A. majus, there is a derived floral morphology with abortion of both the dorsal and lateral stamens unlike the flowers of A.majus with abortion of only the dorsal stamen [23]. The derived floral morphology of Mohavea is correlated with the expression changes of McCYC/McDICH via CYC/DICH, i.e. expansion from the dorsal to both the dorsal and lateral stamens [23]. A similar correlation of expanded expression of CYC-like genes with abortion of both the dorsal and lateral stamens is also observed in Chirita (Gesneriaceae) [25]. However, we are still not clear about the abortion of the ventral stamens that has been involved in the evolutionary shifts of stamen number during the morphological diversification of zygomorphy in Lamiales s.l. [1].
In addition, in the backpetals mutant in A.majus, the ectopic expression of CYC in the lateral and ventral positions results in a dorsalized corolla. However, it seems likely that the androecial development is not affected by the ectopic expression of CYC because the two lateral and two ventral stamens are still fertile [5,26]. In the actinomorphic flower of legume Cadia, no LegCYC1B mRNA is detected in stamens [27]. It is hard to determine whether LegCYC1B or other CYC homologues in legumes have a role in controlling androecial development from data to date [13,14,21]. RAY2 in Senecio and GhCYC2 in Gerbera (Asteraceae) mainly promote the growth of the ligule (the ventral petals) in ray florets and are excluded from the dorsal rudimentary petals [17,18]. In Veronica and Gratiola (Scrophulariaceae s.l.), some of CYC-like genes have dorsal-specific expressions while some have lost this feature, but their expressions do not correlate with ventral stamen arrest [21]. Therefore, expression data correlated with the ventral stamen abortion have not been reported yet for members of the ECE-CYC2 clade. It is uncertain whether abortion of the ventral stamens is related to CYC-like gene activities or to the effect of an unknown analogous counterpart of CYC-like genes, such as members of ECE-CYC3 clade or other factors [3,12,20].
The family Gesneriaceae is sister to the remainder of Lamiales s.l. [28] and has diverse forms of zygomorphy relating to the floral organ differentiation early in the order [1,2,24]. In Gesneriaceae, Opithandra exhibits a peculiar floral morphology, where only the two lateral stamens are fertile and both the dorsal and ventral stamens are aborted in the third whorl (Figure 1). Phylogenetic analyses suggest that the floral morphology of Opithandra is likely derived from a weakly zygomorphic flower with four fertile stamens and a dorsal staminode [24]. Therefore, Opithandra represents an ideal candidate for exploring a potentially novel genetic mechanism underlying the establishment of zygomorphy with ventral stamen arrest in angiosperms, especially in Lamiales s.l.
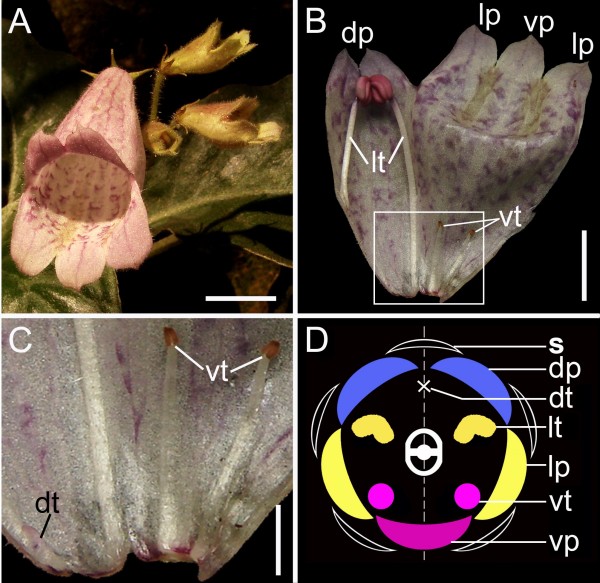
Figure 1.
Flower morphology of Opithandra dinghushanensis. A) Inflorescence with flowers near or at anthesis, showing strongly zygomorphic corolla; B) An opened corolla at anthesis showing two dorsal petals smaller than two lateral and one ventral petals, and androecium with two fertile lateral stamens and two ventral and one dorsal staminodes; C) Magnification of the framed part in (B), showing two infertile lateral stamens with short filaments and small sterile anthers, and a tiny dorsal staminode that is barely visible; D) Floral diagram; Scale bars, 10 mm (A), 7 mm (B) and 3 mm (C). dp, dorsal petal (in blue); dt, dorsal staminode; lp, lateral petal (in yellow); lt, lateral stamen (in yellow); s, sepal; vp, ventral petal (in pink); vt, ventral staminode (in pink).
Here we report that there is a correlation between OpdCYC gene expressions and abortion of both the dorsal and ventral stamens in Opithandra, strengthened by the negatively correlated expression of their putative direct target OpdcyclinD3 genes. The novel patterns of CYC-like gene expressions in Opithandra indicate that ECE-CYC2 clade TCP genes are involved in the ventral stamen repression evolved within Lamiales s. l..
Results
Sequence and phylogenetic analyses of OpdCYC and OpdcyclinD3
We isolated four CYC-like genes from Opithandra dinghushanensis, named OpdCYC1C, OpdCYC1D, OpdCYC2A and OpdCYC2B. The full length open reading frames (ORFs) of OpdCYC1C, OpdCYC1D, OpdCYC2A and OpdCYC2B are 1017 base pair (bp), 1038 bp, 1044 bp and 993 bp, respectively (Additional file 1A). Sequence analyses show that they share 43-48% and 45-51% identity with AmCYC at nucleotide and amino acid levels, respectively. When comparing the TCP and R domains, they share 90-95% identity with AmCYC at the amino acid level, suggesting they are functionally related. Phylogenetic analyses show that OpdCYC genes have a close relationship with AmCYC and AmDICH, and, along with AmCYC and TCP1, belong to the ECE-CYC2 clade in the ECE lineage (CYC/TB1 subfamily) of TCP gene family [12] (Figure 2A) (We have not found a member of the ECE-CYC3 clade yet in Gesneriaceae, probably failed to amplify them because of difficulty in designing specific primers for this clade). OpdCYC genes are closely related to GCYC from Oreocharis among GCYC genes in Gesneriaceae (Figure 2B).
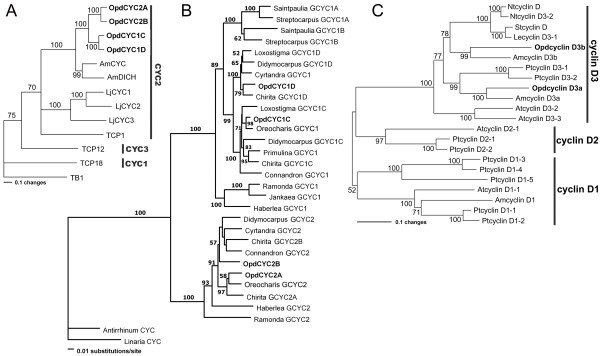
Figure 2.
Neighbor-joining trees of proteins encoded by CYC-like and D-type cyclin genes. A) Neighbor-joining tree of proteins encoded by the ECE lineage genes in CYC/TB1 subfamily, showing that OpdCYC1C/1D and OpdCYC2A/2B form a branch that is sister to AmCYC/AmDICH from Antirrhinum majus, which belong to the CYC2 clade in the ECE lineage. B) Phylogram of GCYC, showing the phylogenetic relations of OpdCYC genes with other GCYC in Gesneriaceae. C) Neighbor-joining tree of proteins encoded by D-type cyclin genes, showing that OpdcyclinD3a and OpdcyclinD3b are clustered with cyclinD3a and cyclinD3b clades, respectively, in the cyclinD3 lineage. For sequence information see Methods. Phylogenetic analyses were conducted using PAUP*4.0b4a, and bootsrap values over 50% (1,000 replicates) are indicated for each branch.
Two D3-type cyclin genes, designated as OpdcyclinD3a and OpdcyclinD3b, were isolated from O. dinghushanensis with full length ORFs of 1563 bp and 1200 bp, respectively (Additional file 1B). The two D3-type cyclin genes contain a cyclin box [29] and the putative (Rb)-binding motif (LxCxE, where x is any amino acid) which are found both in animals [30,31] and plants [6,32,33]. Phylogenetic analyses show that OpdcyclinD3 genes belong to cyclinD3a and cyclinD3b clade, respectively, in the cyclinD3 lineages, in which they have close relations with AmcyclinD3a and AmcyclinD3b from A.majus, respectively (Figure 2C).
Gene mRNA expression patterns
To assess the potential role of CYC-like genes in floral development, we conducted in situ hybridization complemented by gene-specific RT-PCR on O. dinghushanensis. As petal and stamen primordia began to emerge, OpdCYC1C mRNA was detected in all five petal and stamen primordia (Figure 3A). Weak mRNA signals were also detectable in the lateral edges and vascular tissue of sepals (Figure 3A). After primordial initiation of petals and stamens, OpdCYC1C expression signals were gradually weakened in the two lateral stamens (Figure 3B-D) with weak mRNA detected in the ring meristem of the corolla-tube outside the stamen primordia (Figure 3B-C). Figures 3C and 3D were the successive sections from the same individual flower across the base of stamen primordia (3C) and over their upper parts (3D), respectively, which showed a size reduction from the base to the upper part of the dorsal and ventral stamens. The mRNA signal of OpdCYC1C was weak in lateral stamens while strong in both dorsal and ventral staminodes in which its mRNA signal was stronger in the upper part than at the base (Figure 3C-D). OpdCYC1C mRNA also accumulated less in lateral petals than in dorsal and ventral petals (Figure 3D). As the lateral stamens enlarged laterally, weak OpdCYC1C mRNA shifted to peripheries and gradually became undetectable in the two lateral stamens, while OpdCYC1C transcripts continued to accumulate to a high level in the dorsal and ventral staminodes (Figure 3E-F). Meanwhile, OpdCYC1C mRNA became weak and not easily detectable in petals (Figure 3E-F). Weak mRNA signals of OpdCYC1C were also detected in the gynoecial primordium (Figure 3E). In the middle developmental stages with flower buds about 8 mm long, as stamens began filament elongation and anther differentiation, OpdCYC1C was strongly expressed in the dorsal region (the dorsal petals and staminode) and ventral staminode shown in RT-PCR while its weak mRNA signal was detected in sepals, lateral and ventral petals, and lateral stamens, in which the signal was much weaker in sepals and lateral stamens (Figure 4A, C). In the late stages, OpdCYC1C transcripts declined in the dorsal region and ventral staminodes, and were undetectable in other regions (Figure 4B-C). The OpdCYC1C mRNA was undetectable in stamens as pollen sacs began development in the two lateral anthers while the two ventral anthers became sterile (Figure 3G).
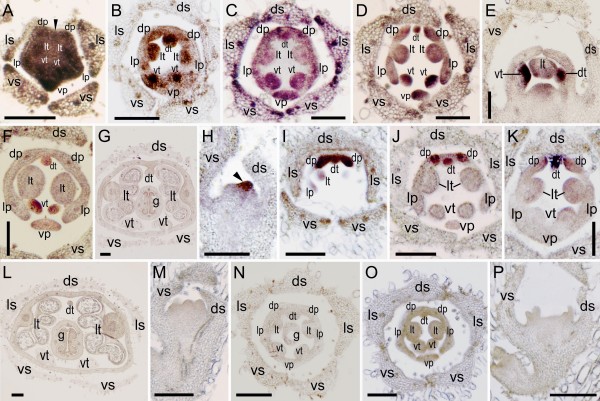
Figure 3.
Tissue-specific expression of OpdCYC1 and OpdCYC2 during floral development in Opithandra dinghushanensis. A-G) RNA in situ hybridizations with antisense probe of OpdCYC1C. A) Its mRNA is first detected in all five petal and stamen primordia with weak signals in lateral edges and vascular tissue of sepals. B-D) Its mRNA then weakened in two lateral stamens but strong in both dorsal and ventral staminodes with weak mRNA in the ring meristem of corolla-tube. C-D) Successive sections from the same individual flower across base (C) and upper parts (D) of stamens. E-F) Its expression shifts to peripheries and becomes undetectable in two enlarged lateral stamens while remains strong in dorsal and ventral staminodes. G) Its mRNA is undetectable in stamens as pollen sacs begin development. H-L) RNA in situ hybridizations with antisense probe of OpdCYC2A. H) Its dense transcript accumulation first restricted to the dorsal side of the floral apex (arrow). I-K) Its strong expression then restricted to two dorsal petals and the dorsal staminode as they are initiated. Note its mRNA later becomes restricted to the dorsal-most parts in two dorsal petals (K). L) Its mRNA is undetectable in stamens as pollen sacs begin development. OpdCYC1D (M) and OpdCYC2B (N) mRNA is not detected in floral tissues. As a negative control, RNA in situ hybridizations with sense probes of OpdCYC1C (O) and OpdCYC2A (P) detect no signal in floral tissues. dp, dorsal petal; ds, dorsal sepal; dt, dorsal staminode; g, gynoecium; lp, lateral petal; ls, lateral sepal; lt, lateral stamen; vp, ventral petal; vs, ventral sepal; vt, ventral staminode. Scale bars, 150 μm.
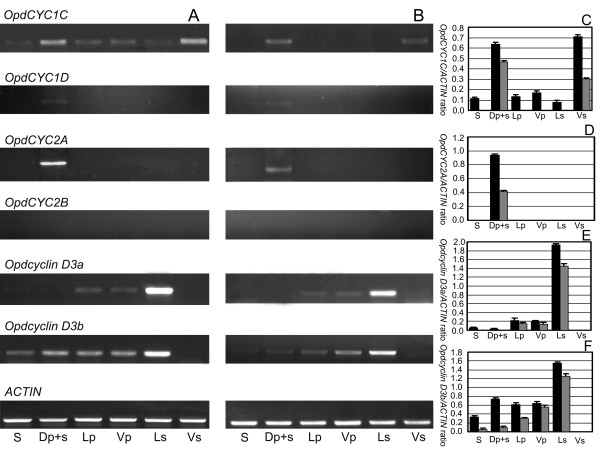
Figure 4.
Gene-specific semiquantitative RT-PCR on RNA prepared from dissected Opithandra dinghushanensis flowers. A) sepal (S), dorsal petal+staminode (Dp+s), lateral/ventral petals (Lp/Vp), lateral stamens (Ls) and ventral staminode (Vs) were dissected from flower buds of middle-stage (less than 1 cm long). B) sepal (S), dorsal petal+staminode (Dp+s), lateral/ventral petals (Lp/Vp), lateral stamens (Ls) and ventral staminode (Vs) were dissected from flowers of late-stage (3-4 cm long). C) Relative level of OpdCYC1C mRNA expression in middle-stage (black) vs. late-stage (grey) compared with ACTIN. D) Relative level of OpdCYC2A mRNA expression in middle-stage (black) vs. late-stage (grey) compared with ACTIN. E) Relative level of Opdcyclin D3a mRNA expression in middle-stage (black) vs. late-stage (grey) compared with ACTIN. F) Relative level of Opdcyclin D3b mRNA expression in middle-stage (black) vs. late-stage (grey) compared with ACTIN. ACTIN was used for RT template control. The values (means ± SD) shown are determined from five independent experiments.
In contrast to OpdCYC1C, OpdCYC2A mRNA densely accumulated in the dorsal region of the floral apex as petals and stamens became visible (Figure 3H). Then, the OpdCYC2A expression signal was specifically concentrated in the dorsal petals and the dorsal staminode (Figure 3I-J). As floral organs developed, OpdCYC2A transcripts continued to be highly concentrated at the dorsal staminode and the dorsal-most parts of the two dorsal petals (Figure 3J-K). In the middle stages, the strong mRNA signal of OpdCYC2A was detected in the dorsal region (the dorsal petals and staminode) shown in RT-PCR that declined in late stages with no signal in other regions (Figure 4A-B, D). Its mRNA was undetectable in stamens as pollen sacs began development in the two lateral anthers while the two ventral anthers became sterile (Figure 3L). Even though OpdCYC1D expression was undetectable in floral tissues using in situ hybridization (Figure 3M), its very weak mRNA signals were detected in the dorsal region (the dorsal petals and staminode) from middle to late stages using RT-PCR (Figure 4A-B). OpdCYC2B mRNA was not detectable in floral tissues both using in situ hybridization and RT-PCR (Figure 3N, Figure 4A-B). No signal was detected in floral tissues with sense probes of OpdCYC1C (Figure 3O) and OpdCYC2A (Figure 3P).
To further elucidate the role of OpdCYC genes in floral development of Opithandra dinghushanensis, we carried out semiquantitative RT-PCR studies of two OpdcyclinD3 genes because cyclinD3 genes were previously revealed to be negatively controlled by CYC as shown in the mid-to-late stage flowers in the model organism snapdragon [6]. RT-PCR results showed that OpdcyclinD3a was strongly expressed in lateral stamens from middle to late stages while its weak mRNA signal was also detected in lateral and ventral petals (Figure 4A-B, E). OpdcyclinD3a mRNA was not detected either in the dorsal region (the dorsal petals and staminode) and ventral staminodes (Figure 4A-B, E). OpdcyclinD3b transcripts were widely distributed in floral tissues except ventral staminodes, in which its mRNA signal was strong in lateral stamens from middle to late stages (Figure 4A-B, F). Transcripts of OpdcyclinD3b detected in the dorsal region were likely mainly distributed in the dorsal petals (OpdcyclinD3b mRNA was uneasily detectable in the dorsal staminode using in situ hybridization (data not shown)).
Discussion
The androecium of Opithandra only has two fertile stamens at the lateral positions with three sterile stamens (staminodes) at the dorsal and ventral sides (Figure 1) (also see [24]). The dorsal aborted stamen is tiny and barely detectable at anthesis while the two infertile ventral stamens have short filaments with very small and sterile anthers (Figure 1B-C). Correlative with the differentiation along the dorsoventral axis of the morphologically peculiar androecium, the OpdCYC2A strong expression is restricted to the dorsal staminode while OpdCYC1C transcripts are initially distributed in all five stamen primordia but later are concentrated in the dorsal and ventral staminodes to late stages. In the ECE lineage of CYC/TB1 subfamily, the TCP proteins in ECE-CYC2 clade studied to date function as negative regulators in stamen development, whereas they appear to vary in petal development according to the trait concerned [3-5,9,10,12,17,23,25,34,35]. The abortion of the dorsal stamen in Antirrhinum comes from CYC and DICH activities [4,5]. The CYC-like gene expression expansion from the dorsal to both the dorsal and lateral stamens is correlated with abortion of both the dorsal and lateral stamens in Mohavea and Chirita [23,25]. TB1 gene exhibits a mix feature of ECE-CYC1 and ECE-CYC2 clades and functions to suppress axillary meristem (CYC1) while retard stamen growth (CYC2) in maize [3,12,34]. In Asteraceae, a CYC homologue GhCYC2 from Gerbera functions by disrupting stamen development [17]. Given that CYC-like gene (ECE-CYC2 clade) function is conserved in repressing stamen development, OpdCYC2A expression restricted to the dorsal stamen and OpdCYC1C expression later concentrated in the dorsal and ventral stamens in the third whorl might be related to abortion of the dorsal and ventral stamens in Opithandra. In fact, the successive sections from the same individual flower indicate that correlative with OpdCYC gene strong expression, the early primordial growth have already been retarded in the dorsal and ventral staminodes in comparison with that in the lateral stamens.
Evidence shows that CYC functions to repress stamen development in the third whorl through its negative effect on expression of D3-type cyclin genes, including cyclinD3b, which usually play an important role in locally regulating cell proliferation in floral development [3,6,36]. The negative effects on cell cycle progression have been reported from other TCP genes, such as IaTCP1 from Iberis (Brassicaceae), TCP2 and TCP4 from Arabidopsis, and CIN from Antirrhinum [16,37,38]. We, therefore, investigated the expression pattern of OpdcyclinD3a and D3b to test for further correlation between CYC expression and stamen abortion through cell-cycle regulation, especially the ventral stamens. In strengthening the above suggestion, OpdcyclinD3 genes have expression patterns in floral tissues negatively correlated with those of OpdCYC genes and stamen abortion in Opithandra. Both Opdcyclin D3a and D3b transcripts are not detected, or weakly detected (i.e. dorsal), in the dorsal and ventral staminodes where both OpdCYC2A and OpdCYC1C or OpdCYC1C are strongly and continuously expressed throughout floral development, while their transcripts are much more concentrated in the lateral stamens where there is only a weak expression of OpdCYC1C in early stages. These factors indicate that OpdCYC gene activities may suppress the development of the dorsal and ventral stamens through negatively regulating OpdcyclinD3 genes (Figure 5). Our recent findings of consensus-binding sites of the TCP transcription factor in the 5' upstream regions of OpdcyclinD3 homologues, i.e. ChcyclinD3a and D3b in Chirita heterotricha, further support the direct regulatory relation between CYC-like and cyclinD3 genes in Gesneriaceae (Yang, Xia and Wang, Yin-Zheng, unpublished).
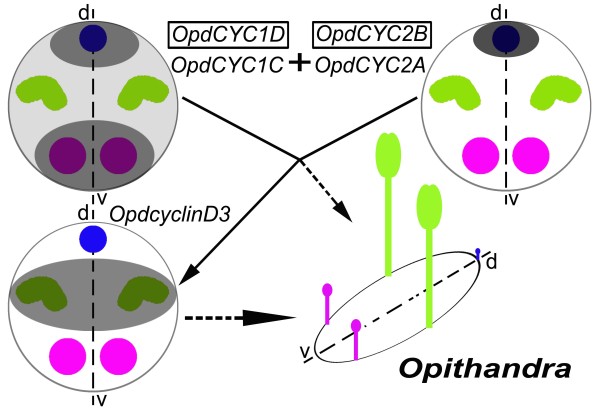
Figure 5.
Diagram showing OpdCYC gene expressions correlated with the floral morphology, especially the infertility of the two ventral stamens in Opithandra, complemented by the expression of their putative negative target OpdcyclinD3 genes. Notes: dorsal staminode is in blue, lateral stamens in green and ventral staminodes in pink; the gene that is not boxed indicates this gene is expressed in a pattern as shown by the shaded parts within the circle (shaded degrees indicate relative levels of gene expression); the gene boxed indicates this gene is not expressed or has very little expression signals in the floral tissues.
It seemingly remains a question whether OpdCYC activity is related to the lateral stamen development or not because OpdCYC1C has a weak expression in the early developing lateral stamens but has no obvious effect on their growth. The evidence that CYC-like genes regulate lateral stamen development comes from both functional analyses in Antirrhinum and expression data in Mohavea and Chirita [3,4,21,23,25]. A gradient of CYC effect along the dorsoventral axis results in abortion of the dorsal stamen and reduced size of lateral stamens in comparison with ventral ones in A. majus, while CYC-like gene strong and continuous expressions in both the dorsal and lateral stamens is correlated with abortion of both of them in Mohavea and Chirita [4,23,25,26]. The TCP1 gene is expressed early in floral buds of Arabidopsis, but the mature flowers of Arabidopsis are actinomorphic because they lack later effects of TCP1 [12,15]. In Iberis amara closely related to Arabidopsis, the IaTCP1 strong dorsal-specific expression represses the two dorsal petal growths to become much smaller than the ventral ones in size, while IaTCP1 has only a weak expressional signal in the natural actinomorphic variants [16]. In the tetrandrous flowers of Oreocharis that is closely related to Opithandra in both morphology and GCYC phylogeny [24] (Figure 2B), there is also a weak expression of CYC homologue ObCYC in the early developing lateral stamens that are reduced in size compared with ventral ones at anthesis as in A. majus [39]. In Bournea, another Opithandra's close relative with actinomorphic flowers in Gesneriaceae, the CYC homologue BlCYC1 is strongly expressed in the dorsal petal and stamen in early development and is downregulated later, which is correlated with the floral development undergoing a morphological transition from initial zygomorphy to actinomorphy at anthesis with five fertile stamens in Bournea [35]. According to Cubas [1], the maintenance of CYC expression after early floral development should be important for generating the morphological asymmetries in the flowers. Preston and Hileman [3] also suggest that early expression of CYC-like genes may be unimportant for establishing mature flower symmetry. The high concentration of OpdcyclinD3a and D3b transcripts in the lateral stamens also indicates that OpdCYC1C has lost negative effects on their expression after early floral development; therefore, the two lateral stamens are fertile at anthesis in Opithandra (Figure 5).
Zygomorphic flowers with three staminodes at the dorsal and lateral positions or at the dorsal and ventral sides have been considered to be derived in the family Gesneriaceae [24,40-42]. In the primitive zygomorphic taxa, such as Haberlea and Oreocharis characteristic of tetrandrous flowers with four didynamous stamens plus a dorsal staminode, and the derived actinomorphic groups (definition see note in [35]), such as Ramonda and Bournea, there is only one single copy of GCYC1 and GCYC2, respectively, found to date (Figure 2B) [35,39,43,44]. However, two copies of GCYC1 are frequently found in the advanced zygomorphic taxa, especially in the zygomorphic genera characterized by diandrous flowers with three staminodes, such as two African genera Streptocarpus and Saintpaulia with GCYC1A and GCYC1B [44,45] and Asian genera Didymocarpus, Chirita and Loxostigma with GCYC1C and GCYC1D [25,46] as well as Opithandra herein (Figure 2B). Recent studies show that two copies of GCYC2 (GCYC2A/2B) are also found in the Asian genera with three staminodes, such as Chirita [25] and Opithandra in this study (Figure 2B). The derived morphology of diandrous flowers might have resulted from subsequent expression differentiation after gene duplication events. In the diandrous flowers of Chirita (also Gesneriaceae) that differs from Opithandra in abortion of both the dorsal and lateral stamens rather than ventral stamens, ChCYC1C is strongly expressed both in the dorsal and lateral stamens while ChCYC1D maintains strong expressions in the dorsal floral regions, and ChCYC2A/2B have no expression signals in floral tissues [25]. No expression of GCYC2 detected in floral tissue is frequently found in Gesneriaceae while GCYC1 is usually conserved in dorsal-specific expression in this family, such as Oreocharis and Bournea [35,39]. Therefore, the peculiar diandrous flowers established in Opithandra might involved not only gained or enhanced expression of OpdCYC1C in ventral staminodes but also the reactivated expression of OpdCYC2A specific to the dorsal staminode accompanied with the downregulation of OpdCYC1D in the dorsal region in the third whorl, a more complicated mechanism than that in another diandrous flowers of Chirita.
Phylogenetic analyses show that the CYC-like genes isolated from Opithandra and other Gesneriaceae belong to ECE-CYC2 clade as CYC and TCP1 from Antirrhinum and Arabidopsis (Figure 2B) [25,35,39,45]. As outlined above, ECE-CYC2 clade genes are characteristic of dorsal identity function which sometimes expands to lateral stamens [4,5,21,23,25,35,39]. It would be especially interesting to know whether or how CYC-like gene activities are related to abortion of the ventral stamens [3,12,20]. Even though not tested functionally, this positive correlation between CYC-like gene expression and ventral stamen abortion and the complement of cyclinD3 to CYC-like gene expressions suggests a genetic mechanism underlying the establishment of zygomorphy with abortion of both the dorsal and ventral stamen evolved within Lamiales s.l.. However, it has been shown for Veronica and Gratiola (also Lamiales s.l.) that the CYC-like gene expression does not positively correlate with the ventral stamen abortion (Preston et al., 2009). These facts inconsonant in the expression data of ECE-CYC2 clade TCP genes imply that the ventral stamen abortion might have evolved by convergent genetic mechanisms in different lineages of Lamiales s.l.. It merits further research in function and upstream regulatory pathway to determine how the expression divergence is caused among paralogues of OpdCYC in Opithandra. In addition, since the diverse variations of zygomorphy in Lamiales s.l. might have involved independent shifts in stamen number [3,21], further investigation of expression pattern and functional analyses of CYC-like genes with identification of their upstream cis- or trans-regulators as well as research in finding other factors possibly coopted to this regulatory pathway in more zygomorphic groups would shed new lights on the mechanisms that underlie the vast morphological diversity of zygomorphy in Lamiales s.l..
Conclusion
As the first to document the expression domain of ECE-CYC clade genes in the ventral stamens, we here report that the expressions of OpdCYC genes are correlated with abortion of both dorsal and ventral stamens in Opithandra, strengthened by the negatively correlated expression of their putative direct target OpdcyclinD3 genes. The complement of OpdcyclinD3 to OpdCYC gene expressions further indicates that OpdCYC expressions are related to the dorsal and ventral stamen abortion through the negative effect on OpdcyclinD3 genes. The novel patterns of CYC-like gene expressions in Opithandra, along with previous reports, suggest that ECE-CYC2 clade TCP genes are not only functionally conserved in the dorsal stamen repression, but also involved in arresting ventral stamens, a genetic mechanism underlying the establishment of zygomorphy with abortion of both the dorsal and ventral stamens evolved within Lamiales s.l.. It would be important to further find whether ECE-CYC2 clade TCP genes are recruited repeatedly for arresting ventral stamens and (or) whether there is any other genetic pathways underlying the ventral stamen abortion, independently or interacting with ECE-CYC2 clade TCP genes, in different lineages of Lamiales s.l..
Methods
Plant materials
All materials used in this study, including gene cloning, in situ hybridization and RT-PCR, were collected from the wild fields, i.e. Dinghu Mountains, Guangdong province, China, where plants of Opithandra dinghushanensis W. T. Wang are mainly distributed.
Gene cloning and sequence analyses
CYC-like genes were isolated from O.dinghushanensis using degenerate oligonucleotide primers in 3' and 5'-RACE according to described methods [47] and the manufacture's protocol (INVITROGEN), respectively. Two D3-type cyclin genes were also isolated from O. dinghushanensis using the above methods. Total RNA was extracted from the floral buds of O. dinghushanensis using the Plant RNA Purification Reagent (INVITROGEN) according to the manufacture's protocol. First-strand cDNAs were synthesized from total RNA with the Supertranscript™ III RNase H- Reverse Transcriptase (INVITROGEN). To examine the intron/exon structures we isolated and sequenced the corresponding genomic DNA of OpdCYC and OpdcyclinD3 genes from leaves. The Oligonucleotide sequences for primers are included in the Additional Material - Additional file 2.
According to the known sequence information, phylogenetic analyses of CYC-like and D-type cyclin genes were conducted to identify the position of CYC-like and D3-type cyclin genes isolated herein in their gene families, respectively. AmCYC and AmDICH are from A. majus [4,5], and LjCYC1/2/3, TCP1/12/18 and TB1 are from Lotus japonicus [13], Arabidopsis [12] and maize [34]. GCYC1A/1B are from Saintpaulia ionantha and Streptocarpus primulifolius [43-45]. GCYC1C/1D (designated as GCYC1 respectively in some taxa) are from Chirita heterotricha [25], Jankaea heldeichii [43], Conandron ramondioides, Haberlea ferdinandi-coburgii, Primulina tabacum, Ramonda myconi [44], Cyrtandra apiculata, Didymocarpus citrinus and Loxostigma sp. [46]. GCYC2 (2A/2B) are from Chirita heterotricha [25], Conandron ramondioides, Haberlea ferdinandi-coburgii, Ramonda myconi [44], Cyrtandra apiculata [46] and Didymocarpus citrinus (Yin-Zheng Wang, unpublished). Amino acid sequences of D-type cyclin genes are from A. majus (Amcyclin D1/D3a/D3b) [6], Populus trichocarpa (Ptcyclin D1/D2/D3) [48], Nicotiana tabacum (Ntcyclin D/D3) [49], Solanum tuberosum (Stcyclin D) (accession nos.EU325650), Lycopersicon esculentum (Lecyclin D3-1) [50] and Arabidopsis (Atcyclin D1/D2/D3) [32]. Phylogenetic analyses with the neighbor joining method and p-distance were carried out using PAUP*4.0b4a [51] and bootstrap was estimated with 1,000 resampling replicates. The DNA sequences of genes reported in this paper, i.e. OpdCYC1C, OpdCYC1D, OpdCYC2A, OpdCYC2B, OpdcyclinD3a and OpdcyclinD3b have been deposited in the GenBank database (accession nos. FJ710518, FJ710519, FJ710520, FJ710521, FJ710522 and FJ644637).
RNA in situ hybridization
Floral tissue for in situ hybridization was fixed, sectioned and hybridized to digoxygenin-labeled probes of OpdCYC1C, OpdCYC1D, OpdCYC2A and OpdCYC2B with reference to described methods [52]. Four gene-specific fragments of OpdCYC1C, OpdCYC1D, OpdCYC2A and OpdCYC2B in the coding region were amplified, respectively, and then were purified and cloned into pGEM®-T Easy vectors. Digoxygenin-labeled probes of OpdCYC1C, OpdCYC1D, OpdCYC2A and OpdCYC2B were prepared from linearized templates amplified using primer Yt7 and Ysp6 from pGEM®-T plasmids [53]. The oligonucleotide sequences for primers are included in the Additional Material - Additional file 2.
Gene-specific semiquantitative RT-PCR
Flowers of different stages were collected as follows: Flower buds of middle-stage (less than 1 cm long) and flowers of late-stage (3-4 cm long) were collected separately. Sepals were removed from the outer whorl. The petals with corresponding corolla-tube were dissected into dorsal (including the attached dorsal staminode), lateral, and ventral regions. Lateral stamens and ventral staminodes were dissected from the corolla-tube and collected each for RT-PCR. All materials were frozen in liquid nitrogen immediately after collection for ribonucleic acid (RNA) isolation. The extraction of total RNAs, purification of poly (A) mRNAs, and synthesis of the first-strand cDNAs were performed according to the methods described above. The template quantity was regulated to be uniform using the ACTIN gene [54]. PCR was performed by using gene-specific primers of OpdCYC1C, OpdCYC1D, OpdCYC2A, OpdCYC2B, OpdcyclinD3a and OpdcyclinD3b. To make sure that each pair of primers was suitable, we first used them to amplify genomic DNA of O. dinghushanensis. The PCR products were then cloned. At least 20 clones of each PCR product were sequenced, and all the primers used could amplify the specific copies of OpdCYC and OpdcyclinD3 genes. The following thermocycling conditions were employed: initial denaturation at 96°C for 3 min, 30 cycles of 96°C for 30 s, 55-60°C (depending on the Tm value of primer pairs) for 30s, and 72°C for 1 min, and a final extension at 72°C for 10 min. The amplified products were separated on a 1.5% agarose gel, and the density of ethidium bromide-stained bands was determined using a Bioimaging System (Gene Tools Program, Syngene, UK). We repeated the RT-PCR experiments five times independently with a new RNA extraction each time. In addition, all RT-PCR products were cloned into pGEM-T Easy-vector, and at least 20 clones from each product were sequenced to test the gene specificity of RT-PCR. The OpdCYC/ACTIN and Opdcyclin/ACTIN ratios represented the relative level of OpdCYC and OpdcyclinD3 mRNA expression. Data are presented as the mean ± SD of independent RT-PCR experiments, and one-way analysis of variance was used to analyze the expression difference of these transcripts in floral tissue from O. dinghushanensis. A P value less than 0.05 was taken to indicate statistical significance. The oligonucleotide sequences for primers are included in the Additional Material - Additional file 2.
Authors' contributions
CFS and QBL isolated OpdCYC and OpdcyclinD3 genes and performed the laboratory work of in situ hybridization and gene-specific RT-PCR. RHL carried out the phylogenetic analyses of OpdCYC and OpdcyclinD3 genes. YZW conceived of and designed the studies, and CFS and QBL participated in the design. YZW drafted the manuscript and all the authors participated in the editing of the manuscript. All the authors read and approved the final manuscript.
Supplementary Material
Sequence alignment of OpdCYC and OpdcyclinD3 with other related proteins. The data provided the sequence alignment of putative proteins encoded by OpdCYC1C, OpdCYC1D, OpdCYC2A, OpdCYC2B with AmCYC from Antirrhinum majus and Opdcyclin D3a and OpdcyclinD3b with AmcyclinD3a and AmcyclinD3b from A.majus.
Oligonucleotide sequences for primers used in this study. The data provided the oligonucleotide sequences for primers used in molecular cloning, RNA in situ hybridization and gene-specific semiquantitative RT-PCR.
Acknowledgments
Acknowledgements
We thank Dr. James Smith for his critical comments and language improvement on the manuscript. This work was supported by CAS Grant KSCX2-YW-R-135 and National Natural Science Foundation of China Grant, no.30770147.
Contributor Information
Chun-Feng Song, Email: cfsong@ibcas.ac.cn.
Qi-Bing Lin, Email: linqb77@ibcas.ac.cn.
Rong-Hua Liang, Email: liangronger@126.com.
Yin-Zheng Wang, Email: wangyz@ibcas.ac.cn.
References
- Cubas P. Floral zygomorphy, the recurring evolution of a successful trait. BioEssays. 2004;26:1175–1184. doi: 10.1002/bies.20119. [DOI] [PubMed] [Google Scholar]
- Endress PK. Antirrhinum and Asteridae - evolutionary changes of floral symmetry. Symposium Series of the Society of Experimental Biology. 1998;51:133–140. [PubMed] [Google Scholar]
- Preston JC, Hileman LC. Developmental genetics of floral symmetry evolution. Trends in Plant Science. 2009;14:147–154. doi: 10.1016/j.tplants.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum . Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E. Control of organ asymmetry in flowers of Antirrhinum . Cell. 1999;99:367–376. doi: 10.1016/S0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA, Coen E, Doonan JH. The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiology. 2000;122:1137–1148. doi: 10.1104/pp.122.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis-elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant Journal. 2002;30:337–348. doi: 10.1046/j.1365-313X.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant Journal. 1999;18:215–222. doi: 10.1046/j.1365-313X.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Howarth DG, Donoghue MJ. Phylogenetic analysis of the "ECE" (CYC/TB1) clade reveals duplications predating the core eudicots. Proceedings of the National Academy of Sciences, USA. 2006;103:9101–9106. doi: 10.1073/pnas.0602827103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, Chen J, Zheng L, Guo X, Luo J, Sato S, Tabata S, Ma W, Cao X, Hu X, Sun C, Luo D. Control of petal shape and floral zygomorphy in Lotus japonicus . Proceedings of the National Academy of Sciences, USA. 2006;103:4970–4975. doi: 10.1073/pnas.0600681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Luo YH, Li X, Wang LP, Xu SL, Yang J, Weng L, Sato S, Tabata S, Ambrose M, Rameau C, Feng XZ, Hu XH, Luo D. Genetic control of floral zygomorphy in pea (Pisum sativum L.) Proceedings of the National Academy of Sciences, USA. 2008;105:10414–10419. doi: 10.1073/pnas.0803291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Coen E, Zapater JM. Ancient asymmetries in the evolution of flowers. Current Biology. 2001;11:1050–1052. doi: 10.1016/S0960-9822(01)00295-0. [DOI] [PubMed] [Google Scholar]
- Busch A, Zachgo S. Control of corolla monosymmetry in the Brassicaceae Iberis amara . Proceedings of the National Academy of Sciences, USA. 2007;104:16714–16719. doi: 10.1073/pnas.0705338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proceedings of the National Academy of Sciences, USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Cui M-L, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E. Regulatory genes control a key morphological and ecological trait transferred between Species. Science. 2008;322:1116–1119. doi: 10.1126/science.1164371. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, Ree RH, Baum DA. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends in Plant Science. 1998;3:311–317. doi: 10.1016/S1360-1385(98)01278-3. [DOI] [Google Scholar]
- Endress PK. Symmetry in flowers: diversity and evolution. International Journal of Plant Sciences (Suppl) 1999;160:3–23. doi: 10.1086/314211. [DOI] [PubMed] [Google Scholar]
- Preston JC, Kost MA, Hileman C. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytologist. 2009;182:751–762. doi: 10.1111/j.1469-8137.2009.02794.x. [DOI] [PubMed] [Google Scholar]
- Xia Z, Wang Y-Z, Smith JF. Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s.l.) inferred from five gene regions. American Journal of Botany. 2009;96:519–530. doi: 10.3732/ajb.0800195. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Kramer EM, Baum DA. Differential regulation of symmetry genes and the evolution of floral morphologies. Proceedings of the National Academy of Sciences, USA. 2003;100:12814–12819. doi: 10.1073/pnas.1835725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Wang YZ. Plants of Gesneriaceae in China. Zhengzhou, China, Henan Science & Technology Publishing House; 2004. [Google Scholar]
- Gao Q, Tao JH, Yan D, Wang YZ. Expression differentiation of floral symmetry CYC-like genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae) Development Genes and Evolution. 2008;218:341–351. doi: 10.1007/s00427-008-0227-y. [DOI] [PubMed] [Google Scholar]
- Kalisz S, Ree RH, Sargent RD. Linking floral symmetry genes to breeding system evolution. Trends in Plant Science. 2006;11:568–573. doi: 10.1016/j.tplants.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Citerne HL, Pennington RT, Cronk QCB. An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proceedings of the National Academy of Sciences, USA. 2006;103:12017–12020. doi: 10.1073/pnas.0600986103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortley AH, Rudall PJ, Harris DJ, Scotland RW. How much data are needed to resolve a difficult phylogeny? case study in Lamiales. Systematic Biology. 2005;54:697–709. doi: 10.1080/10635150500221028. [DOI] [PubMed] [Google Scholar]
- Nugent JH, Alfa CE, Young T, Hyams JS. Conserved structural motifs in cyclins identified by sequence analysis. Journal of Cell Science. 1991;99:669–674. doi: 10.1242/jcs.99.3.669. [DOI] [PubMed] [Google Scholar]
- Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-F. [DOI] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes & Development. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Shah ZH, Murray JA. A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M, Murray JA. The role and regulation of D-type cyclins in the plant cell cycle. Plant Molecular Biology. 2000;43:621–633. doi: 10.1023/A:1006482115915. [DOI] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-R, Wang Y-Z, Smith JF, Chen RJ. Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae) New Phytologist. 2008;178:532–543. doi: 10.1111/j.1469-8137.2008.02384.x. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development. 2005;132:359–370. doi: 10.1242/dev.01584. [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Du ZY, Wang YZ. Significance of RT-PCR expression patterns of CYC-like genes in Oreocharis benthamii (Gesneriaceae) Journal of Systematics and Evolution. 2008;46:23–31. [Google Scholar]
- Burtt BL. Studies in the Gesneriaceae of the old world XXXI: some aspects of functional evolution. Notes from the Royal Botanic Garden Edinburgh. 1970;15:1–10. [Google Scholar]
- Wang WT, Pan KY, Li ZY. Key to the Gesneriaceae of China. Edinburgh Journal of Botany. 1992;49:5–74. [Google Scholar]
- Weber A. Gesneriaceae. In: Kubitzki K, Kadereit JW, editor. The families and genera of vascular plants, Dicodyledons Lamiales (except Acanthaceae including Avicenniaceae) Vol. 7. Berlin, Germany, Springer; 2004. pp. 63–158. [Google Scholar]
- Möller M, Clokie M, Cubas P, Cronk QCB. Integrating molecular phylogenies and developmental genetics: a Gesneriaceae case study. In: Hollingsworth PM, Bateman RJ, Gornal RJ, editor. Molecular systematics and plant evolution. London, UK, Taylor & Francis; 1999. pp. 375–402. [Google Scholar]
- Citerne H, Möller M, Cronk QCB. Diversity of cycloidea-like genes in Gesneriaceae in relation to floral symmetry. Ann als of Botany. 2000;86:167–176. doi: 10.1006/anbo.2000.1178. [DOI] [Google Scholar]
- Wang L, Gao Q, Wang YZ, Lin QB. Isolation and sequence analysis of two CYC-like genes, SiCYC1A and SiCYC1B, from zygomorphic and actinomorphic cultivars of Saintpaulia ionantha (Gesneriaceae) Acta Phytotaxonomica Sinica. 2006;44:353–361. doi: 10.1360/aps050071. [DOI] [Google Scholar]
- Wang CN, Möller M, Cronk QCB. Phylogenetic position of Titanotrichum oldhamii (Gesneriaceae) inferred from four different gene regions. Systematic Botany. 2004;29:407–418. doi: 10.1600/036364404774195593. [DOI] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: a laboratory manual. Third. Vol. 2. New York, USA, Cold Spring Harbor Laboratory Press; 2001. pp. 8.61–8.65. [Google Scholar]
- Menges M, Pavesi G, Morandini P, Bogre L, Murray JA. Genomic organization and evolutionary conservation of plant D-type cyclins. Plant Physiology. 2007;145:1558–1576. doi: 10.1104/pp.107.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell DA, Combettes B, Chaubet-gigot N, Gigot C, Murray JA. Distinct cyclin D genes show mitotic accumulation or constant levels of transcripts in tobacco bright yellow-2 cells. Plant Physiology. 1999;119:343–352. doi: 10.1104/pp.119.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes J, Walsh D, Raymond P, Chevalier C. Molecular characterization of the expression of distinct classes of cyclins during the early development of tomato fruit. Planta. 2000;211:430–439. doi: 10.1007/s004250000306. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods) Version 40b10. Sinauer, Associates, Sunderland, MA; 2002. [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-R. [DOI] [PubMed] [Google Scholar]
- Divjak M, Glare EM, Walters EH. Improvement of non-radioactive in situ hybridization in human airway tissues: use of PCR-generated templates for synthesis of probes and an antibody sandwich technique for detection of hybridization. Journal of Histochemistry and Cytochemistry. 2002;50:541–548. doi: 10.1177/002215540205000411. [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U. Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Development Genes and Evolution. 2001;211:281–290. doi: 10.1007/s004270100153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of OpdCYC and OpdcyclinD3 with other related proteins. The data provided the sequence alignment of putative proteins encoded by OpdCYC1C, OpdCYC1D, OpdCYC2A, OpdCYC2B with AmCYC from Antirrhinum majus and Opdcyclin D3a and OpdcyclinD3b with AmcyclinD3a and AmcyclinD3b from A.majus.
Oligonucleotide sequences for primers used in this study. The data provided the oligonucleotide sequences for primers used in molecular cloning, RNA in situ hybridization and gene-specific semiquantitative RT-PCR.