Abstract
Cellular and tissue defects associated with insulin resistance are coincident with transcriptional abnormalities and are improved after insulin sensitization with thiazolidinedione (TZD) PPARγ ligands. We characterized 72 human subjects by relating their clinical phenotypes with functional pathway alterations. We transcriptionally profiled 364 biopsies harvested before and after hyperinsulinemic-euglycemic clamp studies, at baseline and after 3-month TZD treatment. We have identified molecular and functional characteristics of insulin resistant subjects and distinctions between TZD treatment responder and nonresponder subjects. Insulin resistant subjects exhibited alterations in skeletal muscle (e.g., glycolytic flux and intramuscular adipocytes) and adipose tissue (e.g., mitochondrial metabolism and inflammation) that improved relative to TZD-induced insulin sensitization. Pre-TZD treatment expression of MLXIP in muscle and HLA-DRB1 in adipose tissue from insulin resistant subjects was linearly predictive of post-TZD insulin sensitization. We have uniquely characterized coordinated cellular and tissue functional pathways that are characteristic of insulin resistance, TZD-induced insulin sensitization, and potential TZD responsiveness.
Keywords: muscle and adipose tissue, transcriptional mechanisms, diabetes, branched chain amino acid (BCAA), inflammation
Insulin resistance is a pathological state in which insulin action is impaired in target tissues including liver, skeletal muscle, and adipose tissue. Insulin resistance is a defining feature of the metabolic syndrome and the primary defect leading to type 2 diabetes (1, 2). Impaired insulin-stimulated glucose uptake in skeletal muscle and lipid metabolism in adipocytes are central characteristics of insulin-resistance. Other manifestations of the condition include elevated intramuscular fat content (3), dysregulation of adipokine secretion, and chronic low-grade inflammation in adipose tissue (4). Macrophage infiltration in adipose tissue activates inflammatory pathways that induce insulin resistance and modulate the effects of adipose tissue on whole-body metabolism (5). Several studies have shown that decreased mitochondrial protein and oxidative phosphorylation (OXPHOS) in skeletal muscle and adipocytes are also underlying factors of insulin resistance (6, 7).
Thiazolidinediones (TZDs) are insulin-sensitizing drugs used to treat type 2 diabetes. TZDs enhance insulin sensitivity by improving glucose and lipid metabolism, altering adipokine secretion, and reducing adipose tissue inflammation (4, 8). Although TZDs improve insulin sensitivity and the glycemic, lipid, and inflammatory profiles of most patients, approximately 30% of diabetic subjects do not respond to TZD treatment, as gauged by fasting plasma glucose or HbA1c levels (9, 10). TZDs are ligands of peroxisome proliferator-activated receptor gamma (PPARγ) through which they alter the expression of hundreds of genes in skeletal muscle, adipocytes, and macrophages. PPARγ-mediated gene regulation is the predominant mode of TZD-enhanced insulin sensitivity and metabolism. However, the mechanisms by which TZD-induced gene expression changes lead to insulin sensitization or by which TZD-induced insulin sensitization is prevented are poorly understood.
We have conducted a mechanistic analysis of the gene expression profiles of adipose tissue (AT) and skeletal muscle (SM) from 72 subjects ranging from insulin-sensitive to insulin-resistant. The subjects underwent multiple SM and AT biopsies, hyperinsulinemic-euglycemic clamps, 3-month treatment with an insulin-sensitizing TZD, and clinical characterization. We have measured the transcriptional alterations associated with insulin-resistance in AT and SM in the fasting state and SM in the insulin-stimulated (postclamp) state. Insulin resistant subjects responded to TZD treatment with varied improvements in insulin sensitivity. Thus, we characterized transcriptional alterations in AT and SM associated with TZD-induced insulin-sensitization. We ranked the insulin resistant subjects by their degree of TZD response (i.e., increased insulin sensitivity based on rate of glucose disposal during the hyperinsulinemic-euglycemic clamp) to define responder and nonresponder subgroups. Based on our analysis of SM and AT from these subjects, we have defined pathways and key mechanisms of disease that distinguish the responder and nonresponder subgroups.
Results
Characterization of Insulin Sensitive Subjects and Insulin Resistant TZD-Responder and Nonresponder Subjects.
We generated tissue gene expression profiles from 72 subjects with a broad range of insulin sensitivity, before and after TZD treatment, to identify transcriptional and biochemical pathway signatures that characterize insulin sensitivity, insulin resistance, and pharmacological insulin sensitization. Each subject underwent a hyperinsulinemic-euglycemic clamp at the start of the study and a second hyperinsulinemic-euglycemic clamp after 3-month treatment with one of three TZDs: rosiglitazone, pioglitazone, or troglitazone. Vastus lateralus SM biopsies were harvested immediately before (basal) and after each clamp (postclamp); abdominal s.c. AT biopsies were harvested immediately before each clamp (Fig. 1A). AT biopsies were harvested only from the pioglitazone and rosiglitazone study groups (44 subjects). Dataset S1 shows selected clinical characteristics of the subjects measured before and after TZD treatment. We gauged the insulin sensitivity of each subject by their rate of glucose disposal (Rd) during the clamp. Individuals were divided into two major groups, based on their rate of glucose disposal during the first hyperinsulinemic-euglycemic clamp (Rd1). Subjects with Rd1 values >8 mg/kg per min were classified as insulin sensitive (normal Rd, NRd), and the remaining subjects were classified as insulin resistant (impaired Rd, IRd). From each AT and SM biopsy, we generated gene expression profiles using genome-spanning microarrays and, with these, conducted biochemical pathway analyses. We compared our results from NRd subjects, IRd subjects, and IRd subject subgroups (see Materials and Methods).
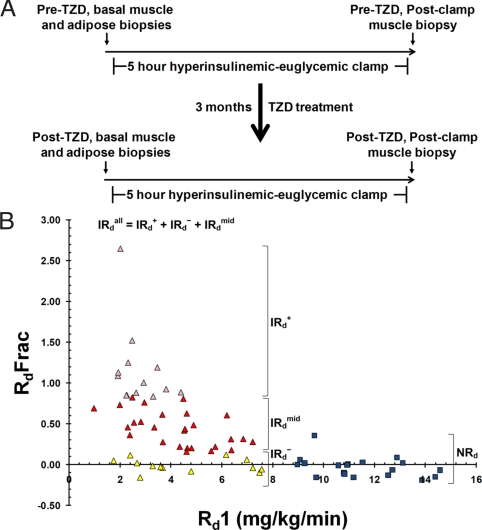
Fig. 1.
Study design and subject insulin sensitivity distribution. (A) Schematic of human insulin-resistance study. (B) Distribution of baseline insulin sensitivity and TZD-mediated insulin sensitization response of study subjects. NRd (insulin sensitive subjects with normal Rd, >8 mg/kg per min, blue square symbols), IRdall (insulin resistant subjects with impaired Rd, <8 mg/kg per min, triangle symbols), IRd+ (subgroup of insulin resistant subjects that are TZD responders, in upper quartile of post-TZD fractional Rd change, pink triangles), IRd− (subgroup of insulin resistant subjects that are TZD nonresponders, in bottom quartile of post-TZD fractional Rd change, yellow triangles), IRdmid (subgroup of insulin resistant subjects in the middle two quartiles of post-TZD fractional Rd change, red triangles). RdFrac − fractional Rd change [(Rd2 − Rd1)/Rd1].
Fig. 1B shows the TZD-induced fractional Rd change [(Rd2 − Rd1)/Rd1] for each subject correlated with their starting Rd (Rd1). TZD treatment did not increase the insulin sensitivity (Rd) of most NRd subjects (see also Dataset S1). As expected, most IRd subjects responded to TZD treatment, demonstrating improved insulin sensitivity (Rd2 > Rd1). Insulin-sensitizing efficacy was not significantly different between the three TZDs. Approximately 25% of the IRd subjects did not respond to TZD treatment, demonstrating minimal or no improvement in insulin-sensitivity. To contrast the expression profiles of IRd TZD-responders (those having the best insulin sensitization response) with IRd TZD-nonresponders (those having the worst insulin sensitization response), all of the IRd subjects (referred to below as IRdall) were ranked by their fractional Rd change and divided into quartile subgroups. IRd subjects in the top quartile of fractional change in Rd (>83% increase in Rd) were classified as IRd responders (IRd+). IRd+ subjects not only had the largest fractional Rd change but also had the greatest improvement in plasma insulin levels (Dataset S1). IRd subjects in the bottom quartile of fractional Rd change (<13% increase in Rd) were classified as IRd nonresponders (IRd−). IRd subjects in the middle two quartiles of fractional Rd change (13–83% increase in Rd) were classified as IRd midresponders (IRdmid). Fig. 1B shows the distribution of these IRd subgroups. Thus, the IRdall subject group refers to the sum of the IRd+, IRd−, IRdmid subject groups.
Insulin Resistance Is Associated with Impaired Insulin-Induced Metabolic Gene Expression Changes in Skeletal Muscle, Which Are Largely Normalized After TZD Treatment.
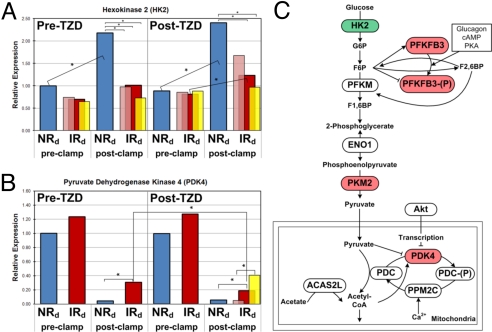
We analyzed SM gene expression profiles from the subject groups and identified ≈150 genes whose pre-TZD, basal and/or postclamp expression levels were significantly different in IRdall subjects (αBonf = 0.05) compared with NRd subjects (Dataset S2). Among these genes were critical regulators of lipid metabolism. For example, expression of lipoprotein lipase (LPL), a critical regulator of SM lipid metabolism (11) and a target gene of PPARγ (12), was 48% lower in IRdall subjects in the pre-TZD fasting state (αBonf = 0.05), was fully rescued by TZD treatment, and was positively correlated with Rd (r = 0.53). All genes with baseline expression values that correlate with Rd are shown in Dataset S3. We also identified 237 insulin-target genes, i.e., genes suppressed or activated in the insulin-stimulated (postclamp) state in NRd subjects. One hundred sixteen insulin-target genes were functionally involved with metabolism (Dataset S2). The insulin-induced response of 42 genes was significantly altered in the IRdall subjects, of which 19 genes were functionally involved with metabolism. IRdall subjects exhibited impaired insulin-induced expression changes in hexokinase 2 (HK2) and pyruvate dehydrogenase kinase 4 (PDK4) compared with NRd subjects. HK2 expression is normally induced by insulin, but this induction was blunted in IRdall subjects (Fig. 2A). Consequently, the postclamp HK2 expression level in IRdall subjects was 50% lower (αBonf = 0.05) than in NRd subjects. Insulin activation of HK2 commits glucose to the intracellular compartment in muscle, facilitating glucose metabolism in the fed state. PDK4 expression is normally suppressed by insulin, but this effect was blunted in IRdall subjects (Fig. 2B). As a result, the postclamp PDK4 expression level in IRdall subjects was 7-fold higher than in NRd subjects. PDK4, in its active state, inhibits pyruvate dehydrogenase complex conversion of pyruvate into acetyl-CoA, effectively disrupting glucose utilization. The schematic in Fig. 2C shows how defective insulin-induced regulation of HK2 and PDK4 in insulin resistant subjects would reduce glycolytic flux. Additional genes involved in the glycolytic pathway (PFKFB3 and PKM2) exhibited defective insulin-induced regulation in IRdall subjects, also shown in Fig. 2C. We observed insulin-induced, transcriptionally coordinated repression of the insulin signaling pathway (Fig. S1). After TZD treatment, IRdall subjects exhibited improved insulin-induced HK2 and PDK4 expression changes and postclamp HK2 and PDK4 expression levels compared with NRd subjects (red and blue bars, respectively, in Fig. 2 A and B). We contrasted these HK2 and PDK4 expression patterns with those from the responder (IRd+) and nonresponder (IRd−) subgroups (pink and yellow bars, respectively, in Fig. 2 A and B). Interestingly, post-TZD improvements in the expression of HK2 and PDK4 were more robust in the IRd+ subject subgroup compared with the IRdall subject group and absent in the IRd− subject subgroup (Fig. 2 A and B). Thus, TZD-improved insulin-regulation of HK2 and PDK4, and presumably glycolytic flux, is related to the degree of TZD-improved insulin sensitization.
Fig. 2.
Differential expression of glucose metabolism genes in skeletal muscle. (A and B) Pre- and postclamp HK2 and PDK4 expression data from the subject groups before and after TZD treatment are shown. (A) Insulin-activated expression of HK2 in NRd subjects was completely blocked in IRdall subjects. After TZD-treatment, this defect was improved in IRdall subjects, normalized in IRd+ subgroup subjects and unaffected in IRd− subgroup subjects. (B) Insulin-induced repression of PDK4 expression in NRd subjects was blunted in IRdall subjects. After TZD-treatment, this defect was improved in IRdall subjects, normalized in IRd+ subgroup subjects and unaffected in IRd− subgroup subjects. NRd (blue), IRdall (red), IRd+ subgroup (pink), IRd− subgroup (yellow). (C) Glycolysis schematic highlighting defectively regulated genes by insulin in IRdall subjects compared with NRd subjects before TZD treatment. Green fill indicates significantly blunted insulin-induced expression in IRdall subjects, resulting in low postclamp expression. Red fill indicates significantly blunted insulin-repressed expression in IRdall subjects, resulting in high postclamp expression. *, significant difference between bracketed groups (αbonf = 0.05).
Insulin Resistance Is Associated with Elevated Adipocyte Markers in the Skeletal Muscle, Which Are Increased After TZD Treatment.
Despite the gross exclusion of perimuscular fat from SM biopsies, SM from IRd subjects had higher levels of adipocyte-predominant transcripts (Dataset S2) compared with SM from NRd subjects. Several of these genes are interesting to note, including leptin (LEP), adiponectin (ADIPOQ), and retinol-binding protein 4 (RBP4). Leptin was overexpressed >5-fold in IRdall subjects at baseline and in IRd+ subjects before and after insulin-stimulation and/or TZD treatment. Leptin expression in IRd− subjects was not significantly different from that in NRd subjects in any condition. Both ADIPOQ and RBP4 were overexpressed in IRdall and IRd+ subjects and were significantly further elevated after TZD treatment. ADIPOQ and RBP4 were overexpressed in IRd− subjects only after TZD treatment and this pattern was significantly blunted compared to the over-expression we observed in IRd+ subjects. Leptin, adiponectin, and RBP4 are principally secreted by adipocytes (13) and have paracrine effects on glucose uptake, fatty acid uptake, and fatty acid oxidation in peripheral tissues (14–17). Thus, altered expression of these genes suggests that the insulin-sensitizing effects of TZDs involve increased adipocyte-myocyte cross-talk.
Genes involved in other aspects of adipocyte biology were differentially expressed in SM from the subject groups. IRdall and IRd+ subjects had higher basal and post-TZD expression of genes that regulate lipid uptake and storage, compared with NRd subjects. Perilipin (PLIN), fatty acid binding protein (FABP4), stearoyl-CoA desaturase (SCD), cell death-inducing DFFA-like effector c (CIDEC) are among these genes. Overexpression of these genes was increased after TZD treatment. Overall, these overexpression patterns were more pronounced in IRd+ subjects and weakest in IRd− subjects suggesting that elevated lipid uptake and storage is related to insulin sensitization. In fact, fractional CIDEC expression change correlated with fractional Rd change (r = 0.57). We observed significant overexpression of genes involved in adipogenesis in SM of IRd subjects, including CCAAT/enhancer binding protein α (CEBPA), sterol regulatory element binding transcription factor (SREBF1), and early growth response 2 (EGR2). CEBPA overexpression and SREBF1 expression each increased significantly after insulin infusion in IRdall subjects. Interestingly, only the IRd+ subjects exhibited significant up-regulation of EGR2 (8.6-fold) after TZD treatment (αBonf = 0.05). Together, these findings suggest that TZD-induced insulin-sensitization is mediated, in part, by stimulation of intramuscular adipocyte differentiation (see Discussion).
Insulin Resistance Is Associated with Increased Inflammatory Markers in Adipose Tissue, Which Are Coordinately Decreased After TZD Treatment.
We identified 588 AT genes that were differentially regulated between NRd and IRd subjects before TZD treatment (Dataset S4). The genes in this set were generally distinct from those we identified in our SM profiling (Compare gene expression profiles in Dataset S5). We used functional analysis (18, 19) to identify enriched function and pathway annotations within this gene set (Dataset S6). We identified statistical enrichment of several inflammation-related ontology categories, including immune system process, extracellular matrix, and MHC class II receptor activity. Several immune system pathways were also significantly enriched, including antigen processing and presentation and leukocyte transendothelial migration. Thirty-four percent of the adipose tissue genes differentially expressed IRdall subjects were associated with inflammation and virtually all of these (192 of 202) were overexpressed. This is shown schematically in Fig. 3A for genes involved in the antigen processing and presentation pathway and leukocyte transendothelial migration pathway (Fig. 3C), two critical pathways regulating tissue inflammation and immune cell infiltration. AT is a heterogeneous mixture of cell types. The enriched functional ontology and pathway annotations, of the differentially expressed genes, associated with the immune system are evidence of increased antigen-presenting cells in AT from the IRdall subjects.
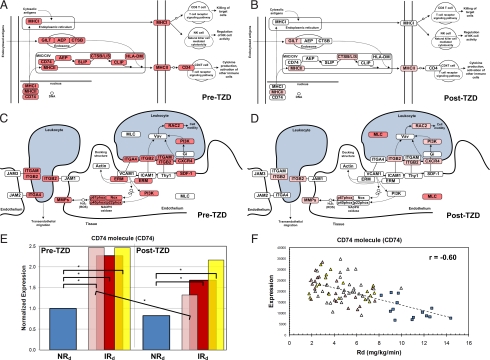
Fig. 3.
Inflammatory marker expression in adipose tissue of insulin resistant subjects. (A and B) Schematics of the antigen processing and presentation pathway highlighting genes that are overexpressed in IRdall subjects before TZD treatment (A) and down-regulated in IRd+ subjects after TZD treatment (B). (C and D) Schematics of the leukocyte transendothelial migration pathway that highlight genes that are overexpressed in IRdall subjects before TZD treatment (C) and down-regulated in IRd+ subjects after TZD treatment (D). White ovals – genes not differentially expressed between NRd and IRdall (A and C) or IRd+ (B and D) subjects. Red ovals – genes significantly overexpressed between NRd and IRdall (A and C) or IRd+ (B and D) subjects. Pink ovals – genes still significantly overexpressed in IRd+ subjects compared with NRd subjects but are significantly down-regulated compared with pre-TZD levels. Schematics are adapted from KEGG. (E) CD74 was significantly overexpressed in all IRd subject groups compared with NRd subjects. After TZD treatment, CD74 expression was significantly down-regulated in IRd+ subgroup subjects (pink) but not in IRdall subjects (red) or IRd− subgroup subjects (yellow). *, significant difference between bracketed groups (αbonf = 0.05). (F) Negative correlation between CD74 expression and Rd (r = −0.59). Graph includes pre- and post-TZD treatment data. IRdall subjects (pink, red, and yellow triangles). IRd+ subgroup subjects (pink), IRd− subgroup subjects (yellow), IRdmid subgroup subjects (red).
Overexpression of inflammatory markers was universally decreased in IRd+ subjects after TZD treatment. Interestingly, most genes overexpressed in IRdall subjects and subsequently down-regulated after TZD treatment were macrophage-specific (Dataset S4). Several facilitate macrophage chemotaxis, including matrix remodeling genes (MMP7, MMP9) and osteopontin (SPP1). Post-TZD repression of overexpressed genes in IRd+ subjects is shown schematically in Fig. 3 for antigen processing and presentation (compare Fig. 3 A and B) and leukocyte transendothelial migration pathways (compare Fig. 3 C and D). A significantly smaller percentage of inflammatory markers were normalized in IRd− subjects compared with IRd+ subjects (χ2 test, P < 0.05) (shown schematically in Fig. S2). Genes still overexpressed in IRd− but not IRd+ subjects after TZD treatment are shown in Dataset S4. Overexpression of CD74, a MHC class II invariant chain gene, was significantly repressed 46% in IRd+ subjects but not in IRd− subjects after TZD treatment (Fig. 3E). In fact, CD74 expression was inversely correlated with Rd (r = −0.60) (Fig. 3F).
Gene Expression Signatures that Correlate with Insulin Resistance and TZD-Induced Insulin Sensitization.
To further elucidate relationships between expression and insulin sensitivity in AT, we determined the correlations between genome-wide gene expression and Rd values (Dataset S7 shows all correlations with r > 0.45). As expected, a set of 319 genes with robust negative correlations between Rd and expression (r < −0.5) was enriched for genes functionally annotated with immune response (αBonf = 0.05) (Dataset S6), including >60% of the genes shown in Fig. 3 A and C. We used statistical regression test (see Methods) to identify AT expression patterns that significantly correlate with Rd. Expression of CD14 molecule (CD14) (r = −0.71), cathepsin S (CTSS) (r = −0.51), neutrophil cytosolic factor 4 (NCF4) (r = −0.65), MHC class II DM beta (HLA-DMB) (r = −0.70), and 28 other genes was inversely correlated with insulin sensitivity (Rd) (Dataset S7). CD14, CTSS, NCF4, and HLA-DMB were also differentially expressed between NRd and IRdall subjects before TZD treatment (Dataset S4), represented in Fig. 3 A and C. These results suggest that insulin resistance is functionally related to elevated AT expression of CD74, CD14, CTSS, NCF4, and HLA-DMB and indicate that these genes are transcriptional markers of insulin resistance. Negative correlations between insulin sensitivity and expression levels of these and the other genes associated with leukocyte transendothelial migration show the association of AT macrophages and insulin resistance. TZD-induced insulin-sensitization (observed in IRd+ subjects but not IRd− subjects) correlated with normalization of these coordinated pathway systems, perhaps as a result of attenuated immune cell residency and recruitment.
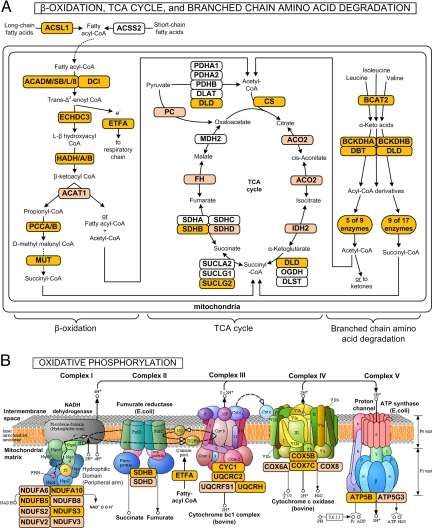
We observed robust positive correlations (r > 0.5) between Rd and the pre- and post-TZD expression of 315 genes. This positively correlated gene set was statistically enriched for genes annotated with mitochondria functional pathways (Dataset S6). Expression of the majority of genes involved in mitochondrial β-oxidation of fatty acids (even-, odd-, mono-, and polyunsaturated and saturated fatty acids) and TCA cycle was correlated with insulin sensitivity (Fig. 4A), including hydroxyacyl-CoA dehydrogenase (HADH, r = 0.73), dodecenoyl-CoA delta isomerase (DCI, r = 0.64), and succinate-CoA ligase (SUCLG2, r = 0.68). Expression of genes encoding subunits from all five complexes in the oxidative phosphorylation pathway was correlated with insulin sensitivity (Fig. 4B), including NADH dehydrogenase (NDUFB5, r = 0.62), electron-transfer-flavoprotein alpha (ETFA, r = 0.69), and cytochrome c reductase (UQCRC2, r = 0.66). Expression of genes involved in branched chain amino acid (BCAA) metabolism (several also involved in TCA cycle) is also correlated with insulin sensitivity (Fig. 4A). Importantly, this set includes all of the genes that regulate the initial, rate-limiting steps of BCAA metabolism (BCAT2, DBT, DLD, BCKDHA, and BCKDHB). Overall, these results indicate that impaired fatty acid and BCAA metabolism and oxidative phosphorylation are proportional to the degree of insulin resistance.
Fig. 4.
Mitochondrial metabolic function in adipose tissue is correlated with insulin sensitivity. Schematics of mitochondrial metabolic pathways highlighting genes whose adipose tissue expression (before and after TZD treatment) was positively correlated with Rd. (A) β-oxidation of fatty acids, TCA cycle, and branched chain amino acid degradation. (B) Oxidative phosphorylation. Dark orange ovals – genes with expression vs. Rd correlations of r > 0.5. Light orange ovals – genes with expression vs. Rd correlations of r = 0.45–0.50.
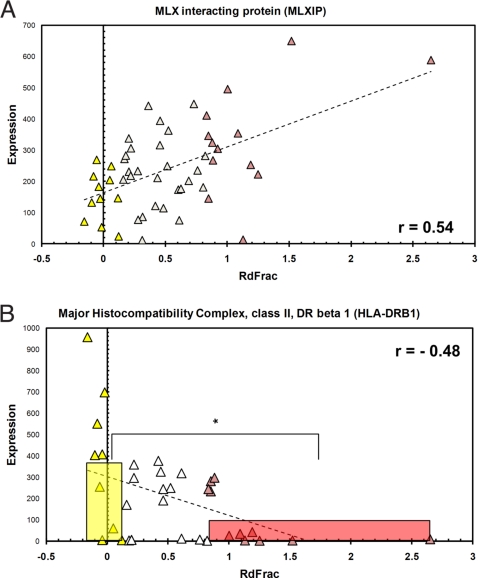
Several published studies have attempted to identify plasma components and other clinical parameters that predict the insulin-sensitizing effect of TZDs on individual patients (9, 20). We have identified gene expression predictors of TZD responsiveness in SM and AT from IRdall subjects, i.e., pre-TZD gene expression that correlates with fractional Rd change [(Rd2 − Rd1)/Rd1] (Dataset S3). MLX interacting protein (MLXIP) in SM and major histocompatibility complex class II DR beta 1 (HLA-DRB1) in AT had gene expression signatures that predict insulin sensitization by TZDs. MLXIP is a transcription factor that activates glycolytic genes (including HK2) in SM (21). Pre-TZD, postclamp SM expression of MLXIP correlated with fractional Rd change (r = 0.51) (Fig. 5A). In general, IRdall subjects with higher SM MLXIP expression before TZD treatment exhibited greater relative insulin-sensitization after TZD treatment [pre-TZD MLXIP expression in IRd+ subjects was 2.6-fold greater than in IRd− subjects (αBonf = 0.05)]. Adipose tissue HLA-DRB1 expression was negatively correlated with fractional Rd change (r = −0.48). HLA-DRB1 plays a central role in antigen presentation and is expressed mainly in antigen presenting cells (APCs). Expression of HLA-DRB1 was 3.4-fold higher in IRd− subjects compared with IRd+ subjects (Fig. 5B). Thus, patients who expressed less HLA-DRB1 in AT before TZD treatment experienced greater relative increases in insulin-sensitivity after TZD treatment.
Fig. 5.
Gene expression predictors of TZD-mediated insulin sensitization. (A) Expression of MLXIP in pre-TZD skeletal muscle was positively correlated with fractional Rd change in IRdall subjects (r = 0.54) and significantly higher in IRd+ vs. IRd− subjects. (B) Expression of HLA-DRB1 in pre-TZD, basal adipose tissue was negatively correlated with fractional Rd change in IRdall subjects (r = −0.47) and significantly lower in IRd+ vs. IRd− subjects. Bars indicate the expression mean and range of fractional Rd change for the IRd+ (red) and IRd− (yellow) subject groups. IRdall subjects (pink, red, and yellow triangles), IRd+ subgroup subjects (pink), IRd− subgroup subjects (yellow), IRdmid subgroup subjects (red).
Discussion
The multidimensionality of our human gene expression dataset allows us to investigate fundamental questions about insulin resistance and TZD-induced sensitization in an unbiased and statistically robust manner. Our results delineate mechanistic differences between normal and insulin-resistant subjects and uniquely identify gene expression signatures that characterize TZD-mediated insulin sensitization and predict TZD responsiveness in insulin resistant subjects. Notably, physiologic and individual proteomic data in the literature support many of the differentially expressed, coordinated pathway models generated by our statistical data analyses. Our findings demonstrate that insulin resistance is characterized by altered basal and insulin-induced expression of genes in skeletal muscle. The fasting-to-feeding transition altered the expression of many genes in insulin sensitive subjects including key regulators of glycolysis and other metabolic pathways. Interestingly, insulin-regulation of many genes was significantly blunted in the insulin-resistant subjects. Our data support the notion that over-nutrition insulin resistance is associated with impaired metabolic flexibility in response to insulin (22). For example, defective insulin-induced regulation of HK2 and PDK4 in insulin resistant subjects (Fig. 2C) would result in decreased glycolytic flux. TZD-induced insulin sensitization in responder (IRd+) subjects was associated with improved insulin-regulated gene expression. Defective insulin-regulated HK2 and PDK4 expression was ameliorated in TZD-treated responder subjects, concomitant with insulin sensitization, but not in TZD-treated nonresponder (IRd−) subjects who did not become more insulin sensitive. Thus, insulin sensitization in the responder subjects was likely associated with improved glycolytic flux and overall metabolic flexibility.
Surprisingly, elevated intramuscular adipocyte marker gene expression in the insulin resistant subjects further increased after TZD treatment. This effect was most significant in responder subjects, which suggests that TZD-enhanced insulin sensitization in muscle is related to increased intramuscular adipocytes. Intramuscular adipocytes have the potential to improve myocyte sensitivity through paracrine signaling and alleviating local hyperlipidemia by storing excess fatty acids (Fig. S3). For example, TZD-induced elevation of adiponectin secretion would increase AMPK activation and lipid oxidation, which is observed in skeletal muscle of TZD-treated subjects (23). Thus, presence of adipocytes in skeletal muscle of insulin resistant subjects may not be a causal factor mediating insulin resistance, but an adaptation that accommodates for excess energy storage.
Chronic low-grade inflammation is associated with and can cause insulin resistance. Macrophages infiltrate adipose tissue of obese animals and humans where they secrete cytokines that interfere with insulin signaling (4, 24). Our pathway analysis results indicate that immunoregulatory genes function together in a network to orchestrate transendothelial migration and antigen presentation in resident leukocytes in adipose tissue from insulin resistant subjects. We found that the activation of proinflammatory pathways correlated with the degree of insulin resistance in our subjects. Furthermore, TZD-mediated repression of inflammatory genes in adipose tissue was relative to TZD-induced insulin sensitization of the subjects, i.e., TZD-induced repression was greater in responder than in nonresponder subjects. Notably, markers of macrophage infiltration were most robust in nonresponder subjects before TZD treatment and may play a role in making insulin-resistance more refractory to TZD treatment.
Decreased mitochondrial capacity, gene expression, and mass are observed in adipose tissue from insulin resistant humans and rodents and are improved after TZD treatment (6, 22, 25–28). Our studies of s.c. adipose tissue extend previous findings and show that fatty acid and BCAA oxidation, TCA cycle, and oxidative phosphorylation pathways are down-regulated in proportion to insulin resistance, i.e., TZD-induced insulin sensitization was positively correlated with expression of genes regulating mitochondrial activity. Our findings are evidence that the magnitude of insulin resistance in insulin resistant individuals is mechanistically linked to the magnitude of dysfunctional mitochondrial capacity driving pathogenic lipotoxicity, futile triacylglycerol cycling, and generation of reactive oxygen species.
Clinical or transcriptional signatures that differentiate insulin-resistant TZD responder and nonresponder subjects have not heretofore been characterized. We have identified distinctions between TZD responders and nonresponders and have identified gene expression predictors of TZD-mediated insulin sensitization. In skeletal muscle, insulin resistant responder subjects exhibited greater post-TZD normalization of insulin-induced glycolytic flux than insulin resistant nonresponder subjects, indicating that responders recover metabolic flexibility as they become more insulin sensitive. This may be due to their higher pre-TZD expression level of MLXIP which was predictive of post-TZD insulin sensitization. MLXIP is a metabolic sensor that shuttles between mitochondria and the nucleus where it is a transcription factor activating HK2 and other glycolytic target genes (21). In adipose tissue, pre-TZD HLA-DRB1 expression was correlated with improved insulin sensitivity after TZD treatment. Expression of HLA-DRB1, an antigen-presenting cell gene, might be mechanistically involved in determining the dichotomous anti-inflammation and insulin sensitization responses exhibited by TZD-treated responder and nonresponder subjects in our study.
In conclusion, this study describes system-wide mechanistic differences between normal and insulin-resistant subjects and identifies transcriptional signatures that differentiate insulin-resistant TZD responder from nonresponder subjects.
Materials and Methods
Human Subject Studies.
Our subject cohort includes subjects from three clinical studies. See SI Text for details about the study subjects, drug treatment, biopsy and hyperinsulinemic euglycemic clamp protocols (23, 29, 32).
Microarray Studies.
We generated gene expression profiles from tissue biopsy RNA using Affymetrix Human Genome U133 Plus 2.0 oligonucleotide microarrays (Affymetrix, Inc., Santa Clara, CA). See SI Text for specific details.
Statistical Analyses.
Supplementary Material
Acknowledgments.
We thank Dr. J. Denis Heck, Sriti Misra, and Lana Bordcosh at the DNA & Protein MicroArray Facility, University of California, Irvine for conducting the RNA microarray analyses and Jerrold Olefsky for support in the study, interpretation and manuscript preparation. Funding was provided by the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant K01-DK62025 (D.D.S.); the Eunice Kennedy Shriver National Institute of Child Health and Human Development and NIH cooperative agreement U54-HD012303 as part of the Specialized Cooperative Centers Programs in Reproduction and Infertility Research (J.M.O., J.G.Y., and C.H.C.); NIH Grants P01-DK074868 (D.D.S., J.M.O., J.G.Y., and C.H.C.) and R01-DK033651 (J.M.O., J.G.Y., and C.H.C.); National Heart, Lung and Blood Institute Grant R33-HL087375 (S.S.); National Institute of General Medical Sciences Grant U54-GM69338 (S.S.); NIDDK Grant P01-DK074868 (S.S.); and the University of California Discovery Program Project #bio03–10383 with matching funds from Pfizer, Inc. (D.D.S., J.G.Y., C.H.C., and S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903032106/DCSupplemental.
References
- 1.Despres JP, et al. Hyperinsulinemia as an independent risk factor for ischemic heart disease. The New Eng J Med. 1996;334:952–957. doi: 10.1056/NEJM199604113341504. [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109:433–438. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 3.Sinha R, et al. Assessment of skeletal muscle triglyceride content by (1)H nuclear magnetic resonance spectroscopy in lean and obese adolescents: Relationships to insulin sensitivity, total body fat, and central adiposity. Diabetes. 2002;51:1022–1027. doi: 10.2337/diabetes.51.4.1022. [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzawa Y. The metabolic syndrome and adipocytokines. FEBS Lett. 2006;580:2917–2921. doi: 10.1016/j.febslet.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 6.Dahlman I, et al. Down-regulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-alpha. Diabetes. 2006;55:1792–1799. doi: 10.2337/db05-1421. [DOI] [PubMed] [Google Scholar]
- 7.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 8.Olefsky JM. Treatment of insulin resistance with peroxisome proliferator-activated receptor gamma agonists. J Clin Invest. 2000;106:467–472. doi: 10.1172/JCI10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi M, et al. Effect of pioglitazone on atherogenic outcomes in type 2 diabetic patients: A comparison of responders and non-responders. Diabetes Res Clin Pract. 2007;77:389–398. doi: 10.1016/j.diabres.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Suter SL, Nolan JJ, Wallace P, Gumbiner B, Olefsky JM. Metabolic effects of new oral hypoglycemic agent CS-045 in NIDDM subjects. Diabetes Care. 1992;15:193–203. doi: 10.2337/diacare.15.2.193. [DOI] [PubMed] [Google Scholar]
- 11.Lapsys NM, et al. Expression of genes involved in lipid metabolism correlate with peroxisome proliferator-activated receptor gamma expression in human skeletal muscle. J Clin Endocrinol Metab. 2000;85:4293–4297. doi: 10.1210/jcem.85.11.6973. [DOI] [PubMed] [Google Scholar]
- 12.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 13.Arner P. The adipocyte in insulin resistance: Key molecules and the impact of the thiazolidinediones. Trends Endocrinol Metab. 2003;14:137–145. doi: 10.1016/s1043-2760(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 14.Fruebis J, et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci USA. 2001;98:2005–2010. doi: 10.1073/pnas.041591798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006;116:1767–1775. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyoshima Y, et al. Leptin improves insulin resistance and hyperglycemia in a mouse model of type 2 diabetes. Endocrinology. 2005;146:4024–4035. doi: 10.1210/en.2005-0087. [DOI] [PubMed] [Google Scholar]
- 17.Yamauchi T, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 18.Hsiao A, Ideker T, Olefsky JM, Subramaniam S. VAMPIRE microarray suite: A web-based platform for the interpretation of gene expression data. Nucleic Acids Res. 2005;33:W627–632. doi: 10.1093/nar/gki443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Araki T, et al. Insulin resistance index as a predictor for pioglitazone treatment in type 2 diabetes. Osaka City Med J. 2005;51:19–25. [PubMed] [Google Scholar]
- 21.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: Mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muoio DM, Newgard CB. Mechanisms of disease: Molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 23.Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM. Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis; thiazolidinedione treatment reverses these defects. Diabetes. 2006;55:2277–2285. doi: 10.2337/db06-0062. [DOI] [PubMed] [Google Scholar]
- 24.Neels JG, Olefsky JM. Inflamed fat: What starts the fire? J Clin Invest. 2006;116:33–35. doi: 10.1172/JCI27280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogacka I, Xie H, Bray GA, Smith SR. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 2005;54:1392–1399. doi: 10.2337/diabetes.54.5.1392. [DOI] [PubMed] [Google Scholar]
- 26.Choo HJ, et al. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–791. doi: 10.1007/s00125-006-0170-2. [DOI] [PubMed] [Google Scholar]
- 27.Rong JX, et al. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes. 2007;56:1751–1760. doi: 10.2337/db06-1135. [DOI] [PubMed] [Google Scholar]
- 28.Wilson-Fritch L, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu JG, et al. The effect of thiazolidinediones on plasma adiponectin levels in normal, obese, and type 2 diabetic subjects. Diabetes. 2002;51:2968–2974. doi: 10.2337/diabetes.51.10.2968. [DOI] [PubMed] [Google Scholar]
- 30.Frias JP, Yu JG, Kruszynska YT, Olefsky JM. Metabolic effects of troglitazone therapy in type 2 diabetic, obese, and lean normal subjects. Diabetes Care. 2000;23:64–69. doi: 10.2337/diacare.23.1.64. [DOI] [PubMed] [Google Scholar]
- 31.Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann N Y Acad Sci. 1959;82:420–430. doi: 10.1111/j.1749-6632.1959.tb44923.x. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe RR. Tracers in metabolic research: Radioisotope and stable isotope/mass spectrometry methods. Lab Res Methods Biol Med. 1984;9:1–287. [PubMed] [Google Scholar]
- 33.Desbuquois B, Aurbach GD. Use of polyethylene glycol to separate free and antibody-bound peptide hormones in radioimmunoassays. J Clin Endocrinol Metab. 1971;33:732–738. doi: 10.1210/jcem-33-5-732. [DOI] [PubMed] [Google Scholar]
- 34.Hsiao A, Worrall DS, Olefsky JM, Subramaniam S. Variance-modeled posterior inference of microarray data: Detecting gene-expression changes in 3T3–L1 adipocytes. Bioinformatics. 2004;20:3108–3127. doi: 10.1093/bioinformatics/bth371. [DOI] [PubMed] [Google Scholar]
- 35.Glantz SA. 6th Ed. New York: McGraw-Hill Medical Pub Division; 2005. Primer of Biostatistics; p. 520. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.