Abstract
The assembly of DNA barcode libraries is particularly relevant within species-rich natural communities for which accurate species identifications will enable detailed ecological forensic studies. In addition, well-resolved molecular phylogenies derived from these DNA barcode sequences have the potential to improve investigations of the mechanisms underlying community assembly and functional trait evolution. To date, no studies have effectively applied DNA barcodes sensu strictu in this manner. In this report, we demonstrate that a three-locus DNA barcode when applied to 296 species of woody trees, shrubs, and palms found within the 50-ha Forest Dynamics Plot on Barro Colorado Island (BCI), Panama, resulted in >98% correct identifications. These DNA barcode sequences are also used to reconstruct a robust community phylogeny employing a supermatrix method for 281 of the 296 plant species in the plot. The three-locus barcode data were sufficient to reliably reconstruct evolutionary relationships among the plant taxa in the plot that are congruent with the broadly accepted phylogeny of flowering plants (APG II). Earlier work on the phylogenetic structure of the BCI forest dynamics plot employing less resolved phylogenies reveals significant differences in evolutionary and ecological inferences compared with our data and suggests that unresolved community phylogenies may have increased type I and type II errors. These results illustrate how highly resolved phylogenies based on DNA barcode sequence data will enhance research focused on the interface between community ecology and evolution.
The most difficult challenge for DNA barcoding in plants is discriminating among taxa of highly speciose genera where rates of species identification by using a variety of putative barcodes rarely exceed 70% (1). In some cases of complex recently evolved species groups DNA barcoding may simply be inappropriate as an identification tool (2). This difficulty is especially acute in cases where certain life history traits have affected the rates of molecular evolution in a lineage, which in turn may affect rates of species assignment by DNA barcodes [e.g., generation times (3) and age-of-crown group diversification (4)].
In the absence of a universal barcode region capable of discriminating among all species in all groups of plants, it is clear that DNA barcodes will be most effectively applied in the identification of a circumscribed set of species that occur together in a floristic region or ecological community, rather than in distinguishing among an exhaustive sample of taxonomically closely related species. In these cases only a limited number of closely related species occur in the same region, so identification to genus is all that is required.
It is now generally agreed that a plant barcode will combine more than one locus (5–7) and will include a phylogenetically conservative coding locus (rbcL) with one or more rapidly evolving regions (part of the matK gene and the intergenic spacer trnH-psbA). Although more laborious than a single-locus barcode, multilocus DNA barcodes can also be advantageous in phylogenetic applications owing to increased nucleotide sampling: the conserved coding locus will easily align over all taxa in a community sample to establish deep phylogenetic branches whereas the hypervariable region of the DNA barcode will align more easily within nested subsets of closely related species and permit relationships to be inferred among the terminal branches of the tree.
In this respect a supermatrix design (8, 9) is ideal for using a mixture of coding genes and intergenic spacers for phylogenetic reconstruction across the broadest evolutionary distances, as in the construction of community phylogenies (10). We define a supermatrix as a phylogenetic matrix that may contain a high incidence of missing data and the data content for any one taxon is stochastic (11) (Fig. S1). Confidence of correct sequence alignment is critical in building such complex matrices and testing the robustness and application of these complex data structures in phylogeny reconstruction is a major endeavor (9).
In the field of community ecology, recent investigations have focused on the factors responsible for species assemblages within a specific ecological community (12). In most cases estimates of the phylogenetic relationships among species in an assemblage are the least robust (10). Ideally characters used to generate a phylogeny of a community, e.g., DNA sequence data, should be independent of the functional characters under investigation and reflect true evolutionary relationships among the species actually present in the assemblage. Unfortunately, most community-based studies lack DNA sequence data for all of the taxa and rely on previously published taxon-specific phylogenies (13) organized with programs, such as Phylomatic (14) to construct community phylogenies usually only resolved to the generic and family levels. Despite these drawbacks, such a phylogenetic framework allows one to test the hypotheses that co-occurring species are 1) more closely related than by chance (phylogenetic clustering), 2) more distantly related than by chance (phylogenetic overdispersion) or 3) randomly distributed (10, 15).
In a community analysis of this type, Kembel and Hubbell (16) constructed a community phylogeny of the 312 co-occurring tree species (>1 cm in diameter) in the forest dynamics plot on Barro Colorado Island in Panama, based on previously published phylogenies assembled with Phylomatic. They found that tree species in the younger forest and drought-stressed plateaus were phylogenetically clustered, whereas coexisting species in the swamp and slope habitats were more distantly related (overdispersed) than expected, although the average phylogenetic structure was close to random.
Here, we report on the feasibility of generating a multilocus barcode library of three markers (rbcL, matK, and trnH-psbA) for 296 woody plant species currently present in the forest dynamics plot on Barro Colorado Island (BCI) as a tool for rapid and reliable taxonomic identification in ecological studies conducted in the plot. The long history of taxonomic work on BCI (17), the intensive inventory of the species on the island (18), and the inclusion in our study of a taxonomist with expertise on the local flora (R.P.) have been critical for our application of DNA barcoding to the forest dynamics plot. We also undertake an analysis parallel to that of Kembel and Hubbell (16) using the three-locus DNA barcode to generate a community phylogeny of woody species in the BCI plot. Our objective was to determine whether the use of multilocus supermatrices to generate phylogenetic hypotheses at the species-level would improve the statistical power provided by a nearly complete sampling of the actual taxa found in the plot. The ability to reconstruct community phylogenies at the species-level would be a significant advance in testing hypotheses regarding the assembly of these communities, the mechanisms of species coexistence, and the role of trait conservatism in determining the community structure (10, 19–22).
Results
Sequence Recoverability and Quality.
PCR and sequencing success were extremely high for the rbcLa region (93%; Table S1). matK had the lowest overall rate of recovery (69%), but the highest sequence quality for recovered samples. For trnH-psbA, we obtained a very high rate of recovery (94%) although some problems were encountered in sequencing (see below). On average the fraction of bases with quality scores >20 were: matK = 95%, rbcL = 93%, and trnH-psbA = 88%. At least one of the gene regions was generated for 98% of the 296 species. For combinations of two regions, recovery rate declined to the rate of the least successful marker (rbcLa and matK = 69%; rbcLa and trnH-psbA = 93%; trnH-psbA and matK = 69%); all three regions together were recovered 69% of the time (Table S1).
Sequence quality for the rbcLa marker was high for all taxa: 85% of the successfully amplified taxa had >50% contig overlap and <9% of sequences were partial. For matK, 50% of the 205 recovered samples had >50% contig overlap, which was largely because of the excessive length of the amplicon (≈850 bp using KIM3F_KIM1R primers). Another 20% of matK sequences had sequence only in one direction. For trnH-psbA 74% of the 280 samples with sequence data had contigs with >50% contig overlap between sequence reads; the remaining 26% of sequences were interrupted by mononucleotide repeats and had either low overlap between reads or resulted in only partial sequences. Fully 92% of trnH-psbA contigs had full-length sequence when all contigs were included.
Species Assignment and Identification.
The highest success of species-level assignment using BLAST with individual barcodes (Table S2) was obtained with matK (99%), followed by trnH-psbA (95%), and then rbcLa (75%). However, when rates of assignment are incorporated with rates of sequence recovery (i.e., the product of PCR recovery rate times the correct assignment rate), matK is less successful, resolving 69% of the 286 species compared with 70% for rbcLa and 90% for trnH-psbA (Table S2). At the species-level for both matK and trnH-psbA, all failures to assign to the correct species occurred in genera that had numerous species in the plot (Inga, Ficus, and Piper). Assignment to the level of genus ranged from 91% to 100% and all markers provided 100% correct assignment to family (Table S2). The combinations of matK and rbcLa correctly identified 92% of all species and trnH-psbA and rbcLa correctly identified 95% of all species whereas the three-locus barcode identified 98% of the 286 taxa for which sequence data were available.
Only 8% of species exhibited population-level variation in any of the three loci in samples from the plot. This within-species variation was limited to five genera (Inga, Ficus, Ocotea, Nectandra, and Psychotria) with the large majority observed within the trnH-psbA spacer.
Phylogenetic Reconstruction.
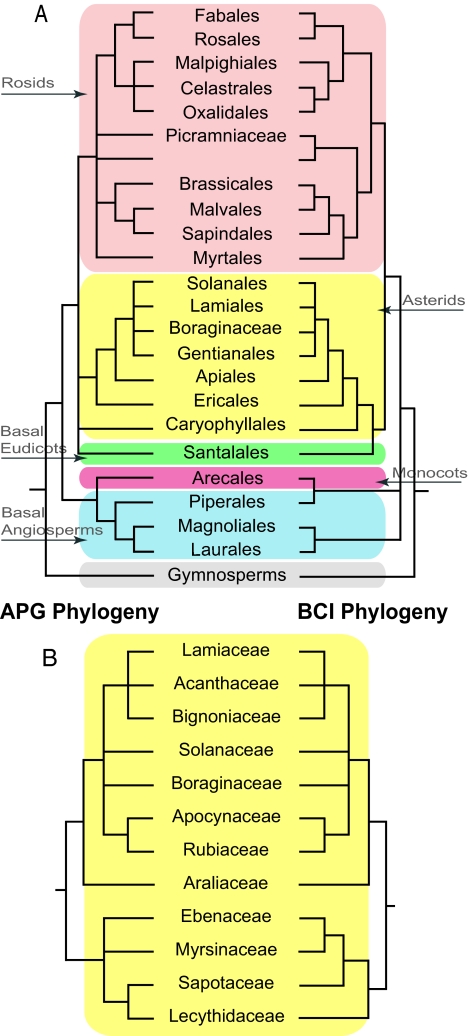
Each of the trees that were generated from the three different combinations of markers (i.e., trnH-psbA + rbcLa, matK + rbcLa, and all three regions combined) differed in topology and the degree of resolution. The three-locus combination provided the most fully resolved and well-supported tree (Fig. 1 and Table S3) thus we focus on the rbcL + matK + trnH-psbA tree in the following discussion. The three-locus tree closely matched the topology of the APG II ordinal-level phylogeny (Fig. 2) with no significant discordances among the 23 orders. In some cases where the APG II tree does not resolve ordinal relationships (e.g., within the asterids and the rosids), the three-locus barcode phylogeny does (e.g., Lamiales, Solanales, and Boraginaceae; Oxalidales, Celastrales, Malpighiales). In several lineages, the barcode phylogeny slightly contradicted the APG II topology (e.g., Piperales and Arecales; Sapindales, Malvales, and Brassicales). Assignment of families within orders was 100% for all marker combinations (Figs. 1 and 2); the topology of families within the major groups of angiosperms as defined on the barcode tree was also concordant with the APG II classification (e.g., asterids; Fig. 2).
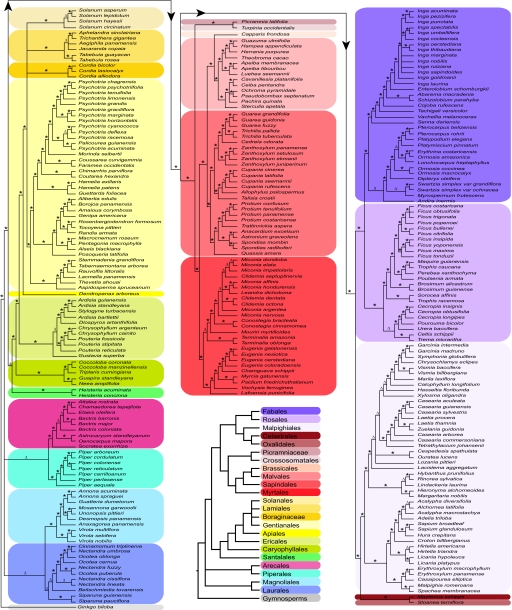
Fig. 1.
Maximum-parsimony tree of 281 species of woody plants in the Forest Dynamics Plot on Barro Colorado Island (BCI) based on a supermatrix analysis of rbcLa, matK, and trnH-psbA sequence data. Color highlights indicate orders represented on BCI. The small tree at the bottom of the central column shows just the ordinal relationships among the species in the BCI flora (see Fig. 2). Nodes with strong ratchet support (>85%) are indicated by an asterisk and nodes with moderate (>70–85%) or weak (>50–70%) support by an open triangle.
Fig. 2.
Comparison of phylogenetic relationships based on barcode sequence data versus the Angiosperm Phylogeny Group (APG II; 13). (A) Comparison of the phylogenetic relationships of 23 orders of flowering woody plants found in the forest dynamics plot on Barro Colorado Island between the maximum parsimony analysis of the barcode sequence data on the right-hand side and the Angiosperm Phylogeny Group (APG II, 2003) on the left-hand side. The major groupings of angiosperms are indicated by the color shading. (B) Comparison of the family-level relationships within the asterid clade between the maximum parsimony analysis of the barcode sequence data on the right-hand side and the Angiosperm Phylogeny Group (APG II, 2003) on the left-hand side.
To assess and quantify branch support we defined four separate categories of parsimony ratchet values for each node of the tree: 85% or greater = strong; 70–85% = moderate; 50–70% = weak; 50% or less = poor or no support. The most strongly supported tree was the three-locus supermatrix tree constructed by using MP (Table S3). All 23 orders were supported as monophyletic (ratchet values >70%) with 19 of these orders strongly supported. We were able to obtain good sequence data in 56 of the total 57 families in the plot (with poor quality sequence for one sample in the Rhamnaceae). Of these 56 families 53 either had only one genus represented on BCI or were supported as being monophyletic with 46 (89%) demonstrating strong support (medium support level = 98%). The nonmonophyletic families were the Bignoniaceae, Euphorbiaceae, and Salicaceae. Of the 47 genera with more than one species on BCI 35 (74%) were monophyletic (nonmonophyletic genera were in the Laurales, Arecales, Ericales, Myrtales, Sapindales, Malpighiales, and Fabales); 91% of these monophyletic genera were strongly supported (median support of 97%). The total fraction of nodes throughout the tree that were supported at each level was: strong = 85.7%; moderate = 1.8%; weak = 10.2%; poor = 2.2%. In contrast, in the most fully resolved of the ML bootstrap analyses (the three-locus supermatrix) the fraction of nodes that were supported at each level was: strong = 54%; medium = 11.3%; weak = 16%; poor = 21%.
In a comparison of the three-locus barcode tree with that of the most up-to-date Phylomatic tree for all taxa on BCI, the barcode tree has 97.5% of the nodes resolved, whereas only 48.4% of the nodes of the Phylomatic tree are resolved. Within the large plant families present on BCI, this disparity between the barcode tree and the phylomatic tree in resolved nodes ranges from 100% versus 19.35% in the Rubiaceae (32 taxa) to 82.35% versus 50.0% in the Fabaceae (35 taxa).
Community Phylogenetic Structure Analyses.
Our pruned version of the Kembel and Hubbell (16) Phylomatic tree yielded the same set of inferences as they concluded in their original work (table 5 in ref. 16; Table S4). Specifically, we found phylogenetic clustering in the “high plateau” and “young” habitats by using the Net Relatedness Index (NRI) metric and in the “young” habitat using the Nearest Taxon Index (NTI) metric; we also found phylogenetic overdispersion in the “swamp” and “slope” habitats by using the NRI metric (Table S4 and Fig. S2).
Using the barcode phylogeny, we found significant phylogenetic structuring for eight of the 14 inferences made (Table S4). When comparing our barcode phylogeny to the less resolved Phylomatic tree, nine of 14 inferences were dissimilar. Analyses based on the barcode phylogeny identified significant phylogenetic structure in five cases for which the pruned Kembel and Hubbell phylogeny did not. In two cases (NRI values on the “slope” and “young” habitats) significant phylogenetic structure was revealed in both analyses, but the opposite inference was made. In the remaining two cases, the pruned Kembel and Hubbell phylogeny demonstrated significant phylogenetic structuring and the barcode phylogeny did not. Only two of the five cases of significant phylogenetic structuring reported in Kembel and Hubbell and confirmed with our pruned Phylomatic tree were supported by the barcode phylogeny. When directly comparing the NRI and NTI values from the 1,250 individual subplots within the seven habitat types by using paired t tests, eight of the 14 comparisons were significantly different between the pruned Kembel and Hubbell phylogeny and the barcode tree (Table S4).
Discussion
Species Identification.
We have demonstrated here that a three-locus DNA barcode provides accurate species identifications for 98% of the 281 taxa on BCI for which we had data. The three loci, rbcLa, matK, and trnH-psbA, correctly identified species at 75%, 99%, and 95% success, respectively (Table S2). As expected rbcLa demonstrated insufficient sequence variation to distinguish among closely related species (5, 23). matK was principally handicapped in our study by poor PCR recovery. Other investigations have shown higher rates of recovery for matK (7, 24), which suggests that the success of this locus may be improved in the future.
The trnH-psbA spacer alone showed the highest rates of species identification (90%) of the three loci when both sequence recovery and correct assignment were taken into account. By combining all three loci correct species assignment was improved to an impressive 98%. The taxonomic distribution of the 296 species on BCI, which includes a high proportion of single-species families and genera (Dataset S1), almost certainly yields higher rates of correct assignment than would be the case for communities comprised of more taxonomically dense samples of species.
In summary, our results show that DNA barcodes can be very effective in the context of a clearly circumscribed floristic sample or plant community, and that additional data, such as geography and morphology may be required to obtain higher rates of species identification in other contexts. The importance of a solid taxonomic foundation (17) for such applications of DNA barcoding cannot be overemphasized.
Community Phylogenies.
Our application of an aligned supermatrix of the three barcode loci permitted the construction of a well-resolved phylogeny of the woody plant community in the forest dynamics plot on BCI. The ability to build such well-resolved phylogenies adds extra value to the development of DNA barcode libraries in general and supports future endeavors to collect DNA barcodes in ecologically important environments, especially in the tropics.
The high level of correspondence of the topology of the BCI barcode phylogeny to the APG II classification is a clear reflection of the central role that the plastid gene rbcL has played in our overall understanding of the evolution of the angiosperms (13, 25). In this respect the selection of rbcL as a component of the plant DNA barcode was not simply fortuitous (5). However, the ability of a 540-bp portion, which was selected for use as a plant barcode, of the complete 1,428-bp gene to resolve higher level relationships across a broad section of the angiosperms was surprising.
The successful alignment of matK through back-translation across the entire taxonomically diverse sample set, as well as, the use of a supermatrix alignment for the hypervariable trnH-psbA spacer also account for the success of our phylogenetic reconstruction. Supermatrix methodology allows for broad taxonomic sampling, where alignment across all phylogenetic markers and for all taxa is problematic. Thus, despite the sparseness of our alignment matrix (94% of the cells were missing data) the topological arrangements closely approximated expected relationships according to APG II. The importance of including in a supermatrix one gene that is present for all taxa and readily aligns across the full matrix, e.g., rbcL in our analysis, has been shown in other studies using supermatrices for building phylogenies (8, 9).
The most advantageous aspects of generating community phylogenies from DNA barcode sequences are the refinements and improvements provided in the resolution of the terminal branches of the tree. Trees produced by Phylomatic may in principle result in equally resolved terminal branches if the species have been included in previously published molecular phylogenetic investigations (14). This situation is unlikely for a majority of communities and it is particularly unlikely for tropical communities where phylogenetic information at the species-level is rare. The ability of the barcode phylogenies both to resolve the terminal branches of phylogenies and to provide refined measures of branch lengths is because the phylogenies are built from sequence data of individuals representing the species in the community.
Highly variable intergenic spacers, which present some disadvantages when used as a DNA barcode because of problems with alignment and length variation (26), provide a significant amount of sequence variation when reconstructing phylogenies within a family or a genus (27, 28). In such taxonomically narrow phylogenies, intergenic spacers contain few indels and are likely to have many more point mutations than a coding region, thereby providing more phylogenetic information. Our application of a supermatrix approach to the analysis of community phylogenies has allowed us to avoid the alignment problems of intergenic spacers while taking advantage of the high levels of sequence variation found within them. The combination of the slower evolving coding gene rbcL with the faster evolving spacer trnH-psbA and coding matK gene in a supermatrix provides the appropriate complement of sequence evolution for constructing phylogenies of the set of disparate taxa which occur in tropical plant communities, such as BCI. If greater phylogenetic resolution is required for a particular group of taxa, the analysis can always be enhanced with more extensive and targeted sequence data from additional genes, such as the nuclear internal transcribed spacer (29).
Community Structure.
The primary use of phylogenies in plant community ecology has been to determine whether coexisting species are more or less closely related than expected by chance, which would be evidence for the overriding influence of habitat filtering as a determinant of community membership. Conversely, the coexistence of distantly related species would be evidence for the overriding influence of biotic interactions in determining community membership.
Of the seven habitat types in the BCI forest dynamics plot, our analyses demonstrated that five contained nonrandom phylogenetic assemblages. Thus, in general we can conclude that the coexistence of species in this forest is structured by abiotic and biotic interactions. In particular, both the low plateau and slope habitats had assemblages where the coexisting species were more closely related than expected by chance. This result suggests that habitat filtering plays a dominant role in structuring these communities. The high plateau and the mixed and young habitats were phylogenetically overdispersed and thus, in these habitats biotic interactions are likely to play a dominant role in determining species coexistence. Along streams and in swamps species assemblages were no different from those expected by chance, which suggests that phylogenetic relatedness may not play a role in determining the structure of communities in these habitats.
Our reexamination of the phylogenetic structure of subplots in the different BCI forest habitats revealed the power of a more finely resolved barcode phylogeny, which increased the statistical power to reject the null expectation and showed that the less resolved Phylomatic trees are prone to falsely accepting a null hypothesis of no effect (Type II error). For example, in five cases the barcode phylogeny found significant phylogenetic structure whereas the Phylomatic phylogeny found none. Furthermore, of the five cases of significant phylogenetic structuring detected by the Phylomatic tree, only one was supported by the barcode analyses and most of the barcode results tended to find more phylogenetic overdispersion.
The popularity and success of the Phylomatic approach to building community phylogenies has been based upon the ability of the program to produce rapid estimates of phylogenetic relatedness of complex lists of species within a community. It is now clear that studies of community evolution will be substantially improved by the production of more finely resolved phylogenetic trees and that DNA barcodes stand poised to serve as an efficient and effective approach to building community phylogenies. Such studies will have a bearing on the factors that govern not only local plant community assembly and dynamics, but also on understanding niche conservation and the dynamics of species composition at landscape and global scales (30, 31).
Materials and Methods
Tissue Sampling and DNA Extraction, Amplification, and Sequencing.
The 1,000 × 500 m Forest Dynamics Plot on Barro Colorado Island (BCI) in Panama was established in 1982 and all tree stems >1 cm diameter at breast height (dbh) have been mapped and identified to species in repeated censuses (32). DNA barcodes were generated for one to four tagged individuals for each of the 296 species (1,035 total samples) recorded from the plot in the last census in 2006 representing 23 orders, 57 families, and 181 genera. The standing plant serves as a living voucher for the life of each tagged individual. In addition, an herbarium voucher for each species sampled for this study is deposited at STRI and US (see Dataset S1).
Tissue samples were field collected at BCI and preserved through either flash freezing or silica gel desiccation. Approximately 50 mg of each sample of leaf material was placed within a well of a 2-mL polypropylene 96-well matrix screen-mate plate (Matrix Technologies) and transferred to the Laboratories of Analytical Biology at the Smithsonian Institution for DNA extraction and sequencing. Details on extraction, PCR, and robotic sequencing are provided in SI Materials and Methods and Table S5.
Sequence Editing, Alignment, and Assembly into a Supermatrix.
Recovered trace files for each of the three markers were imported into Sequencher 4.8, trimmed, and assembled into contigs. Each of the three markers was handled differently in alignment. Alignment of rbcLa was unambiguous because of the absence of indel variation and all sequences were readily assembled. For matK we used transAlign (33) to perform alignment via back-translation. The matK sequences were then aligned with each other and concatenated onto the rbcLa alignment using MacClade (34) to produce a two-gene alignment for all taxa.
The trnH-psbA sequences were partitioned by family or order to build eighteen separate taxonomically structured sequence files, which were each then aligned by using Muscle (35). In cases where only one species per family was present in the plot, that individual was aligned with another family in the same order. When only a single species was represented for an order the sequence was not included in the phylogenetic alignment because the rbcLa sequence would accurately place the taxon on the tree. The individual sets of aligned trnH-psbA sequences were then assembled into a supermatrix by sequentially concatenating them with the rbcLa + matK alignment by using MacClade (Fig. S1). The resulting matrix had >94% of the cells consisting of missing data or gaps. For more details on the supermatrix construction see SI Materials and Methods.
Rates of Sequence Assignment to Species.
The rate at which each barcode marker could be assigned to the correct species was determined by using BLASTn (short nearly exact search; 36). All recovered sequences at each of the three markers were formatted as both database and query; all query sequences were compared against the entire database library of sequences. For barcode sequences that varied within a species, all variant haplotypes were included within the database and queried in the BLASTn searches. For species lacking intraspecific variation only a single individual was included. A sequence was counted as being correctly assigned when that species had the highest Bit-Score among all candidates; a sequence was not counted as correctly assigned when the correct species was either tied with another species, or received a lower score. All barcodes were tested singly and in combination.
Phylogenetic Reconstruction.
We reconstructed a community phylogeny for the woody species of the BCI plot using maximum likelihood (ML) and maximum parsimony (MP) algorithms with three different marker combinations: rbcL + matK, rbcL + trnH-psbA, and rbcL + matK + trnH-psbA. For each combination either 281 or 277 of the 296 species were included depending on the available sequences. ML analyses were conducted using RAxML (37) and the MP analysis using PAUP v.4.0 (38), both run through the CIPRES supercomputer cluster (www.phylo.org). Implementation of the MP ratchet (39) was used to assess support for trees. To assess phylogenetic structure the trees were compared with the topology of APG II (13) using Mesquite (34) by directly projecting ordinal-level trees and family-level trees (within the asteroid clade) in opposition. For more details on phylogeny construction see SI Materials and Methods (S1).
Community Phylogenetic Structure Analyses.
One of the 121 equally parsimonious trees of the three-locus MP analysis of 281 BCI taxa was selected to quantify the phylogenetic structure of tree assemblages in different habitats in the forest dynamics plot. Proportional branch lengths from the MP barcode phylogeny were applied to the community analyses, which were designed to emulate the original phylogenetic structuring-habitat analyses of this plot performed by Kembel and Hubbell (16). To facilitate a direct comparison between the two approaches, we pruned their Phylomatic phylogeny to include only the taxa found in our MP barcode phylogeny and retained their original branch lengths produced by using the Phylocom algorithm bladj (40).
For both the barcode and Phylomatic phylogenies, we quantified by using Phylocom (40) the Net Relatedness Index (NRI: 10) and the Nearest Taxon Index (NTI: 10) for each 400-m2 subplot (n = 1,250 subplots). Positive NRI and NTI values indicates phylogenetic clustering; negative NRI and NTI values indicate phylogenetic overdispersion.
Because the NRI and NTI values in the 1,250 subplots were spatially autocorrelated, we estimated the mean NRI and NTI values within habitats by using simultaneous spatial autoregression analyses. Next, we used the habitat defined for each 400m2 subplot (41) to determine, using t tests, whether each habitat tended to contain subplots that were on average phylogenetically clustered, overdispersed, or random. Finally, for each subplot, we compared the barcode NRI and NTI values to those values calculated from the Phylomatic phylogeny by using paired t tests. For more details on the community structure analyses see SI Materials and Methods (S1).
Supplementary Material
Acknowledgments.
We thank Ida Lopez for assistance with the figures, Lee Weigt and Ken Wurdack for advice in the lab, Jamie Whitaker for sequence data management, Kyle Harms for providing the habitat maps for Barro Colorado Island, and Steve Hubbell for the insights and energy that he has put into the Forest Dynamics Plot. This work supported by National Science Foundation Grant DEB 043665, the Smithsonian Institution, and a Tupper Postdoctoral Fellowship in Tropical Biology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ981925–GQ982412).
This article contains supporting information online at www.pnas.org/cgi/content/full/0909820106/DCSupplemental.
References
- 1.Seberg O, Petersen G. How many loci does it take to DNA barcode a crocus? PLoS ONE. 2009;4(2):e4598. doi: 10.1371/journal.pone.0004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spooner DM. DNA barcoding will frequently fail in complicated groups: An example in wild potatoes. Am J Bot. 2009;96:1177–1189. doi: 10.3732/ajb.0800246. [DOI] [PubMed] [Google Scholar]
- 3.Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 4.Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- 5.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007;2(6):e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fazekas AJ, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE. 2008;3:e2802. doi: 10.1371/journal.pone.0002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bininda-Edmonds ORP, Sanderson MJ. Assessment of the accuracy of matrix representation with parsimony analysis supertree construction. Syst Biol. 2001;50:565–579. [PubMed] [Google Scholar]
- 9.Smith SA, Beaulieu JM, Donoghue MJ. Mega-phylogeny approach for comparative biology: An alternative to supertree and supermatrix approaches. BMC Evol Biol. 2009;9:37. doi: 10.1186/1471-2148-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 11.McMahon MM, Sanderson MJ. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst Biol. 2006;55:818–836. doi: 10.1080/10635150600999150. [DOI] [PubMed] [Google Scholar]
- 12.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton: Princeton Univ Press; 2001. [DOI] [PubMed] [Google Scholar]
- 13.Angiosperm Phylogeny Group II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 14.Webb CO, Donoghue MJ. Phylomatic: Tree assembly for applied phylogenetics. Mol Ecol Notes. 2005;5:181–183. [Google Scholar]
- 15.Westoby M. Phylogenetic ecology at world scale, a new fusion between ecology and evolution. Ecology. 2006;87:S163–S165. doi: 10.1890/0012-9658(2006)87[163:peawsa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Kembel SW, Hubbell SP. The phylogenetic structure of a neotropical forest tree community. Ecology. 2006;87:S86–S99. doi: 10.1890/0012-9658(2006)87[86:tpsoan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Croat TB. Flora of Barro Colorado Island. Stanford, CA: Stanford Univ Press; 1978. [Google Scholar]
- 18.Foster RB, Hubbell SP. The floristic composition of the Barro Colorado Island forest. In: Gentry AH, editor. Four Neotropical Forests. New Haven, CT: Yale Univ Press; 1990. [Google Scholar]
- 19.Webb CO. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 20.Swenson NG, Enquist BJ, Thompson J, Zimmerman JK. The influence of spatial and size scales on phylogenetic relatedness in tropical forest communities. Ecology. 2007;88:1770–1780. doi: 10.1890/06-1499.1. [DOI] [PubMed] [Google Scholar]
- 21.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecol Lett. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- 22.Wright SJ, et al. Relationships among ecologically important dimensions of plant trait variation in seven neotropical forests. Ann Bot. 2007;99:1003–1015. doi: 10.1093/aob/mcl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newmaster SG, Fazekas AJ, Ragupathy S. DNA barcoding in land plants: Evaluation of rbcL in a multigene tiered approach. Can J Bot. 2006;84:335–441. [Google Scholar]
- 24.Lahaye RM, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA. 2008;105:2923–2928. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soltis DE, Soltis PE, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 26.Devey DS, Chase MW, Clarkson JW. A stuttering start to plant DNA barcoding: Microsatellites present a previously overlooked problem in non-coding plastid regions. Taxon. 2009;58:7–15. [Google Scholar]
- 27.Shaw J, et al. The tortoise and the hare II: Comparison of the relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- 28.Small RL, Lickey EB, Shaw J, Hauk WD. Amplification of noncoding chloroplast DNA for phylogenetic studies in lycophytes and monilophytes with a comparative example of relative phylogenetic utility from Ophioglossaceae. Mol Phylogenet Evol. 2005;36:509–522. doi: 10.1016/j.ympev.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 29.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donoghue MJ. A phylogenetic perspective on the distribution of plant diversity. Proc Natl Acad Sci USA. 2008;105S:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crisp MD, et al. Phylogenetic biome conservatism on a global scale. Nature. 2009;458:754–756. doi: 10.1038/nature07764. [DOI] [PubMed] [Google Scholar]
- 32.Condit R. Tropical Forest Census Plots: Methods and Results from Barro Colorado Island, Panama, and a Comparison with Other Plots. New York: Blackwell; 1998. [Google Scholar]
- 33.Bininda-Edmonds ORP. transAlign: Using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics. 2005;6:156. doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddison WP, Maddison DR. Mesquite: A modular system for evolutionary analysis. Version 2.0. 2007. http://mesquiteproject.org.
- 35.Edgar R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 2007;1:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 38.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2003. Version 4. [Google Scholar]
- 39.Nixon KC. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 40.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of community phylogenetic structure and character evolution. Version 3.41. 2007. www.phylodiversity.net/phylocom. [DOI] [PubMed]
- 41.Harms KE, Condit R, Hubbell SP, Foster RB. Habitat associations of trees and shrubs in a 50-ha neotropical forest plot. J Ecol. 2001;89:947–959. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.