Abstract
Background
Rainbow trout (Oncorhynchus mykiss) are the most-widely cultivated cold freshwater fish in the world and an important model species for many research areas. Coupling great interest in this species as a research model with the need for genetic improvement of aquaculture production efficiency traits justifies the continued development of genomics research resources. Many quantitative trait loci (QTL) have been identified for production and life-history traits in rainbow trout. A bacterial artificial chromosome (BAC) physical map is needed to facilitate fine mapping of QTL and the selection of positional candidate genes for incorporation in marker-assisted selection (MAS) for improving rainbow trout aquaculture production. This resource will also facilitate efforts to obtain and assemble a whole-genome reference sequence for this species.
Results
The physical map was constructed from DNA fingerprinting of 192,096 BAC clones using the 4-color high-information content fingerprinting (HICF) method. The clones were assembled into physical map contigs using the finger-printing contig (FPC) program. The map is composed of 4,173 contigs and 9,379 singletons. The total number of unique fingerprinting fragments (consensus bands) in contigs is 1,185,157, which corresponds to an estimated physical length of 2.0 Gb. The map assembly was validated by 1) comparison with probe hybridization results and agarose gel fingerprinting contigs; and 2) anchoring large contigs to the microsatellite-based genetic linkage map.
Conclusion
The production and validation of the first BAC physical map of the rainbow trout genome is described in this paper. We are currently integrating this map with the NCCCWA genetic map using more than 200 microsatellites isolated from BAC end sequences and by identifying BACs that harbor more than 300 previously mapped markers. The availability of an integrated physical and genetic map will enable detailed comparative genome analyses, fine mapping of QTL, positional cloning, selection of positional candidate genes for economically important traits and the incorporation of MAS into rainbow trout breeding programs.
Background
Rainbow trout (Oncorhynchus mykiss) are the most-widely cultivated cold freshwater fish in the world and are considered by many to be the "aquatic lab-rat". Interests in the utilization of rainbow trout as a model species for genome-related research activities focusing on carcinogenesis, toxicology, comparative immunology, disease ecology, physiology, transgenics, evolutionary genetics, and nutrition have been well documented [1]. Coupling great interest in this species as a research model with the need for genetic improvement for aquaculture justifies the continued development of genome resources facilitating selective breeding.
Genome size estimates derived from molecular weight of DNA per cell for rainbow trout and other salmonids vary from 2.4 to 3.0 × 109 bp [2,3]. As with most salmonids, rainbow trout experienced a recent genome duplication event resulting in a semi-tetraploid state [4]. Our physical mapping experience with BACs from the Swanson library has demonstrated that duplicated loci can be detected by DNA fingerprinting [5]. Additionally, BACs that represent one of two duplicated loci were shown by fluorescent In-situ hybridization (FISH) to distinctly hybridize to a specific chromosome pair [6]. Therefore, it is likely that the vast majority of the duplicated loci contain enough sequence variation to allow correct assembly of a physical map using BAC DNA fingerprinting.
Current genomic resources available for rainbow trout research include multiple bacterial artificial chromosome (BAC) libraries [5,7]; doubled haploid (DH) clonal lines [8-11]; genetic maps [3,12-15]; a large EST database [16,17]; and DNA microarrays [18,19].
Seven rainbow trout BAC libraries were constructed to date. Two libraries constructed in Japan [7] contain average insert sizes of 58 kb and 110 kb, and provide haploid genome coverages of 6.7 fold and 5.3 fold, respectively. However, they have not been arrayed in plates and library screening tools are not available. One BAC library from the Swanson male homozygous line and one from the OSU female homozygous line were commercially constructed by Amplicon Express Inc. in 2002. Both libraries were prepared from partial digestions with HindIII. The OSU BAC library has 96,768 clones with an average insert size of 110 kb (4.5× coverage). The Swanson BAC library has 184,704 clones with an average insert size of 130 kb (10× coverage). HindIII BAC DNA fingerprinting for local physical mapping of 27 Type-I markers in the Swanson library demonstrated the library's utility for identifying duplicated loci and confirmed its 10× coverage [5]. Both libraries have been used for genomic sequencing and genetic mapping of loci of interest [20-27]. An additional 5× genome coverage Swanson DH YY male library (92,160 clones) was constructed at the Children's Hospital Oakland Research Institute (CHORI-220; http://bacpac.chori.org/library.php?id=405) in 2005 using EcoRI partial digestion of genomic DNA with an estimated average insert size of 159 kb. An additional two new libraries were prepared by Amplicon Express in 2008. The two new 5× genome coverage Swanson DH YY male libraries (110,592 clones each) were prepared using BamHI and EcoRI partial genomic digestion to complement the 10X HindIII Swanson library and have estimated insert sizes of 140 kb. The four Swanson DH YY male libraries described above were prepared using genomic DNA from the same Swanson doubled-haploid clonal line that is propagated and maintained at the lab of Gary Thorgaard in Washington State University.
Two genetic maps with improved marker densities were recently developed for rainbow trout by INRA [12] and the NCCCWA [15]. The INRA map is based on a panel of two DH gynogenetic lines. It has more than 900 microsatellites over 31 linkage groups and a total length of 2,750 cM (average resolution of 3 cM). The NCCCWA map is based on a panel of five families that represent the starting genetic material of the NCCCWA selective breeding program. It has 1,124 microsatellite loci over 29 linkage groups and a total length of 2,927 cM (average resolution of 2.6 cM).
The rainbow trout haploid karyotype is composed of 52 chromosome arms, but chromosome numbers can vary among rainbow trout populations in concordance with their native geographic distribution [28]. Therefore, anchoring the genetic linkage groups to the physical chromosome arms was a crucial task accomplished by Phillips et al. [6] using BACs as FISH probes. The range of the haploid chromosome number (N) is between 29 and 32 [28]. The karyotype of the Swanson DH line is composed of 2N = 58 [29]. The offspring of "hybrids" between strains with different chromosome number are viable and they can be used for genetic mapping as two uni-armed (acrocentric) chromosomes from the parent with 2N = 60 will align with a di-armed (metacentric) chromosome from the parent with 2N = 58. A comparative cytogenetic map of the rainbow trout and Atlantic salmon using FISH with BACs that harbor Type-I markers and microsatellites is being developed in a coordinated effort [30]. This cytogenetic map and the comparative genetic map of Danzmann et al. [31] provide a frame-work for future high resolution trout-salmon comparative genome maps.
Qualitative/quantitative trait loci (QTL) mapping experiments in rainbow trout have been very successful because of their high fecundity, external fertilization, and ease of gamete handling and manipulation. Many QTL have been identified for production and life-history traits including resistance to the parasite C. shasta [32], resistance to IHNV [33,34] and to IPNV [35], Killer cell-like activity [36], upper thermal tolerance [37,38], embryonic developmental rate [8,39,40], spawning time [41,42], confinement stress response [43] and smoltification [44,45]. The availability of a BAC physical map integrated with the genetic map will facilitate fine mapping of QTL, the selection of positional candidate genes and the incorporation of marker-assisted selection (MAS) into rainbow trout breeding programs. A major shortcoming of QTL studies is that they are limited to the variation present in a limited number of families and typically do not detect loci with small effect. This can be overcome by whole genome association studies and other approaches that capture effects of most QTL that contribute to the population-wide variation in a trait such as genomic selection. Recently we demonstrated the feasibility of low resolution LD association studies in rainbow trout [46]. In the absence of whole genome sequence assembly, the robust integrated physical and genetic map that we aim to construct will provide better resolution than the current genetic maps for ordering of genetic markers and estimating physical distances between markers, thus facilitating whole genome association studies rainbow trout.
Several BAC-based physical maps were constructed in recent years for economically important aquaculture species including tilapia [47], Atlantic salmon [2] and catfish [48,49]. Here we report the construction of the first physical map of the rainbow trout genome using a 10× genome coverage BAC library derived from the Swanson DH clonal line.
Results and Discussion
BAC Fingerprinting and contigs assembly
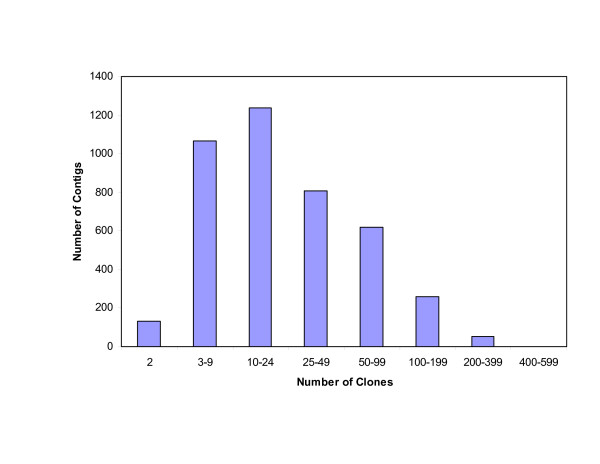
We used the 4-color High-Information-Content Fingerprinting (HICF) SNaPshot method of Luo et al. [50] to fingerprint all the clones from the 10X HindIII BAC library (184,704 clones) and 7,392 clones from the CHORI-220 5X EcoRI library. After editing with FPMiner software (BioinforSoft, Beaverton, OR) 82% BAC fingerprints from the 10X library and 50% from the CHORI-220 library were assembled into physical contigs using FPC software http://www.agcol.arizona.edu/software/fpc/ with a tolerance of 0.5 bp and an initial cutoff of 1 E-70 (1 × 10-70), followed by DQer and several rounds of end-to-end merging and single-to-end merging at progressively lower cutoff stringencies. The current version of the map is composed of 154,439 clones of which 145,060 are assembled into 4,173 contigs and 9,379 remained singletons (Table 1). The average number of BACs per contig is 34.76, and the distribution of the number of BACs per contig is shown in Figure 1. The average number of fingerprinting fragments per BAC is 76.4, and the average insert size for this library is 130 kb [5]. Therefore, each fragment is estimated on average to represent 1.7 kb of genome DNA. The total number of unique fingerprinting fragments (consensus bands) in contigs is 1,185,157, which corresponds to an estimated physical length of 2.0 Gb (75% - 80% of the rainbow trout genome). The average number of consensus bands (CB) per contig is 284, and the estimated contig size is 482 kb. The number of contigs in this assembly is similar to the first generation Atlantic salmon physical map, which resulted in 4,354 contigs and 37,285 singletons with an average contig length of 590 kb from fingerprinting of a 12.5X library [2]. The rainbow trout physical map can be searched and viewed online via WebFPC: http://www.genome.clemson.edu/activities/projects/rainbowTrout
Table 1.
BAC fingerprinting and FPC assembly statistics
| Number of clones fingerprinted | 192,096 | ~10.4× genome coverage |
| Used in FPC assembly | 154,439 | ~8.3× genome coverage |
| Average insert size (Kb) | 130 | |
| Average number of fragments per clone | 76.4 | |
| Estimated average fragment size (Kb) | 1.7 | |
| Number of clones in contigs | 145,060 | ~7.8× genome coverage |
| Number of contigs | 4,173 | |
| Average number of clones per contig | 34.76 | |
| Average contig size in consensus bands | 284 | |
| Estimated average contig size (Kb) | 482 | |
| Number of Q contigs | 811 | 19.4% |
| Number of Q clones | 1,986 | 1.4% |
| Number of CB in contigs | 1,185,157 | |
| Estimate total length in contigs (Kb) | 2,000 | 70% - 80% genome size |
Figure 1.
BAC clones distribution in contigs.
Validation of contigs
The physical map assembly was validated by: 1) comparing contigs assembly to the probe hybridization results and agarose gel fingerprinting contigs of Palti et al. [5]; and 2) anchoring large contigs to the microsatellite-based genetic linkage map of Rexroad et al. [15]. In the first approach we evaluated the contig assignments of 236 clones that were positive by probe hybridization to 27 type I markers (Table 2). Most of the clones (189) that were positive for a single probe and were also assembled into a single contig by Palti et al. [5] also clustered inside a single contig in this physical map, confirming the reliability of this assembly. Only one marker (fabp7b) was truly split into two contigs in the physical map where five clones clustered in contig 2908 and four clustered in contig 1658. The other 10 clones that did not cluster in the major contig of clones positive to each marker did not cluster with other positive clones either, likely representing the fraction of mis-assembled clones. An additional 33 clones (14%) that were positive to the markers of Palti et al. failed our fingerprinting editing criteria and were excluded from the current FPC assembly. Overall, the current assembly of 93% of the clones was in agreement with our previous work and only 10 clones (5%) were likely mis-assembled.
Table 2.
Assembly of BACs positive to gene probes and the agarose HindIII fingerprinting of Palti et al. [5].
| Gene | Major Contig | No. of Clones | Other Contigsa | Failedb |
| 1. a1-mg-1 | 4410 | 5 | 1 | |
| 2. CXC-R4 | 1145 | 8 | 1 (348) | 1 |
| 3. DAB | Singleton | 1 | ||
| 4. fabp7b | 2908 | 5 | 4 (1658), 1 (566) | 2 |
| 5. GH2-1 | 228 | 10 | 2 | |
| 6. GH2-2 | 560 | 4 | ||
| 7. GTPBP-Gi-1 | 2715 | 5 | 1 | |
| 8. GTPBP-Gi-2c | 4594 | 7 | 1 (247) | |
| 9. GTPBP-Gi-3c | 4594 | 3 | 1 | |
| 10. GTPBP-Gi-4 | 608 | 5 | ||
| 11. Hep-1 | 336 | 17 | 1 (9737) | 4 |
| 12. Hep-2 | 12 | 7 | ||
| 13. HSZFP238-a | 1304 | 16 | 2 | |
| 14. HSZFP238-b | 3300 | 2 | ||
| 15. ID1B | 1492 | 16 | 2 | |
| 16. ID1C-1 | 2759 | 3 | 2 | |
| 17. Irp-1A-1 | 2251 | 9 | 2 | |
| 18. Irp-1A-2 | 1106 | 7 | 1 (761) | 3 |
| 19. MHCIa-1 | 3093 | 8 | 1 (450), 1 (1607), 1 (1937) | 3 |
| 20. MHCIa-2 | 84 | 9 | 1 (1304) | 1 |
| 21 MHCIa-3 | 959 | 8 | 1 (3816) | |
| 22. NPY-1 | 5683 | 4 | ||
| 23. NPY-2 | 3205 | 2 | ||
| 24. RP-S16-a | 2361 | 5 | 2 | |
| 25. RP-S16-b | 1657 | 3 | ||
| 26. SCAR163 | 3249 | 9 | 1 | |
| 27. TAP1 | 260 | 11 | 3 | |
| Total: | 189 (93%) | 14 (7%) | 33 (14%) | |
a The contig numbers are in parenthesis.
b Clones that were not fingerprinted successfully and did not pass the editing step of the analysis.
c GTPBP-Gi-2 and GTPBP-Gi-3 appear tightly linked on contig 4594, but the respective positive BACs do not overlap. This may be caused by local tandem duplication of this locus.
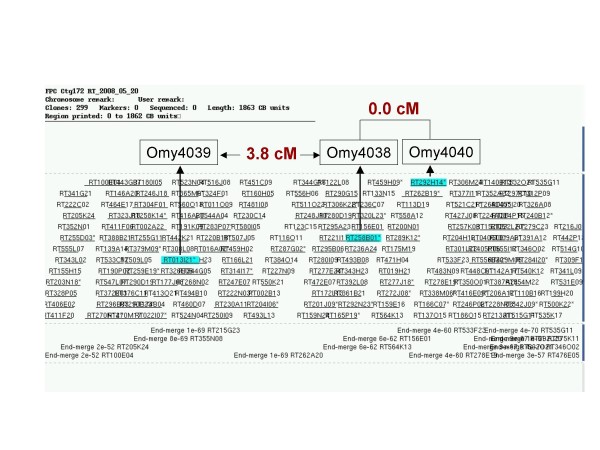
In the second validation approach, 11 of the largest contigs were anchored to the genetic linkage map using 25 microsatellite markers isolated from BAC end sequences (BES) and reported here for the first time, and three microsatellites that were previously isolated from other clones in the contigs (Tables 3 and 4). Two to four markers were developed per contig from clones that were distal to each other on the contig as illustrated in Figure 2. All of the 28 markers were placed on the rainbow trout genetic map by two-point linkage analysis. Markers from 9 of the 11 contigs displayed close genetic linkage of 0 cM - 10 cM. The other two contigs (138 and 450) were likely mis-joined as for each contig two of the three markers were closely linked and the third marker was mapped to another linkage group (Table 4). Those two were also the largest contigs with 334 and 431 clones and estimated length of 3.7 Mb and 4.6 Mb, respectively. The ratio of physical to genetic linkage distances varied among the contigs we sampled, which is similar to other vertebrate genomes [48,51]. We will be able to better investigate this relationship in the rainbow trout genome when we will have a comprehensive and robust integration between the physical and genetic maps. In terms of number of contigs, 9 of 11 (82%) are in agreement with the genetic map. In terms of genome coverage in number of markers, 24 of 26 (92%) are in agreement between the physical and genetic maps. This 8%-18% error rate is higher then the 5% estimated for the catfish physical map of Quiniou et al. [48] or the 4% rate detected in the 3-color HICF physical map of the maize genome [52]. However, the whole assembly error rate for this trout physical map is likely lower than the estimate of 18% or even 8% as this validation was heavily skewed toward the largest contigs, and indeed the two mis-joined contigs were also the largest contigs.
Table 3.
Genetic linkage mapping of microsatellites isolated from BAC end sequences.
| Marker | Clone | Contig | Chr. | Forward/Reverse Primers | GenBank |
| OMY4002 | 170E02 | 58 | 14 | AGGTTATTTCCATTTCCCGC/GAGGAGTCCCAGAGGAAAGG | GF100674 |
| OMY4003 | 378C10 | 58 | 14 | GACTTCTGCTCTGTCGGTCC/GACAGGTAGCCAAAACTCCG | GF100675 |
| OMY4005 | 116G20 | 260 | 2 | TCATAAGTCATATGGTGACTATCATTT/GCAAATGCATTGACATCTCG | GF100676 |
| OMY4006 | 162L19 | 260 | 2 | GATACACCCCTGCTGTTCGT/ACCCACCAAGCCACTCTCT | GF100677 |
| OMY4007 | 146D09 | 260 | 2 | AACGCATAGGAGGGAGGATT/AAAATATTGTGGCCAGCAGC | GF100678 |
| OMY4008 | 198M04 | 84 | 18 | ATGCTTTTGCAATTTCCTGG/ATGTTCATGCTGACCGACTG | GF100698 |
| OMY4009 | 227H04 | 791 | 22 | CGCTGGAATGTTTTCATCTG/ATTTCACAAATGGCCAGGAG | GF100679 |
| OMY4010 | 383M11 | 930 | 11 | TGATCATGGCACCCATACTG/TCACCCTGGTGGCCTACTAC | GF100680 |
| OMY4011 | 154C16 | 930 | 11 | CAGTATGTCCTGTGAGGCCC/TCCACTTTAAGGGCATTTGG | GF100681 |
| OMY4012 | 194O09 | 100 | 27 | AGCAAGCTCAATGAAGCACA/GAGCCCAGAGGTGAGATCAG | GF100682 |
| OMY4013 | 251I01 | 100 | 27 | AGCGGACTGGGCTGTAATAA/ATGGACCAACTGAGCCTGAC | GF100683 |
| OMY4015 | 318K03 | 138 | 27 | GTGGGCATTTTTGCTGACTT/CCGTTGATACATTTTGGCAG | GF100684 |
| OMY4017 | 300B08 | 138 | 1 | TCATCTTCGACAGCATGGAG/GAAGGCCAAAGAAGCATGAA | GF100685 |
| OMY4018 | 207I23 | 138 | 1 | CCTGTTTTGAAAATGGGACC/ACCAACCGCCATAGTAGCAG | GF100686 |
| OMY4020 | 377G20 | 168 | 10 | TGTCCCTCAAAGTGCTACCC/CAGATGTGGGAACTTGAGCA | GF100687 |
| OMY4022 | 207O04 | 168 | 10 | ACAAAGACCACAACGGCATT/TTGGCATTTACATATGTCCCC | GF100688 |
| OMY4023 | 241D02 | 336 | 3 | ATCTCCAAGCCCTGAGGAAT/TTTTTGGTCCCCACAAGAAT | GF100689 |
| OMY4024 | 203O06 | 336 | 3 | GAGCCAGTAATTCATTCGCC/GCAGGACAATCGTTTTAGGG | GF100690 |
| OMY4025 | 278E22 | 336 | 3 | ATGACCCTGACGGGATGTTA/AGCTCCACACACAACACAGC | GF100691 |
| OMY4026 | 178K11 | 450 | 22 | GTCGCAAAAGGCACTAAAGG/TGTGGCAGGTGCTGTTAGAG | GF100692 |
| OMY4027 | 204J15 | 450 | 22 | ATGCCAAAGAAATGGACAGG/TGGCCTCCCTTGTCATTAAA | GF100693 |
| OMY4028 | 220L14 | 450 | 1 | TCCCAGTGGATGGGACTTAG/GTGGGTGTCACATGTGTGGT | GF100694 |
| OMY4038 | 218N06 | 172 | 6 | GGGGAAATTCAACCCACTTT/ATGGCGAATTGGCTAGACTG | GF100695 |
| OMY4039 | 275M02 | 172 | 6 | ACTCTCCCCTGTCCTCCATT/CTAGTATCGACCCCTGCGAA | GF100696 |
| OMY4040 | 386B11 | 172 | 6 | TGAAGGGGGCTGATTAGTTG/ACAGCGTTCCATAGCGAGAT | GF100697 |
All clones were from the 10X HindIII YY male Swanson doubled haploid BAC library (RT).
Table 4.
Validation of physical map assembly by linkage mapping of microsatellites isolated from clones that were part of 11 of the largest contigs in the rainbow trout physical map.
| Contig | No. of Clones (Q)a | Contig Length (Kb) | No. of Markers | rb | LOD | Chr. |
| 58 | 174 (3) | 1,938 | 2 | 0.000 | 29.2 | 1 |
| 84c | 190 (3) | 2,300 | 2 | 0.014 | 19.1 | 18 |
| 100 | 313 (2) | 3,300 | 2 | 0.101 | 20.6 | 27 |
| 168 | 306 (3) | 2,934 | 2 | 0.009 | 28.7 | 10 |
| 172 | 299 (6) | 3,167 | 3 | 0.040 | 8.1 | 6 |
| 260c | 224 (1) | 2,692 | 4 | 0.039 | 31.3 | 2 |
| 336 | 136 (0) | 1,722 | 3 | 0.005 | 46.3 | 3 |
| 791c | 124 (0) | 1,722 | 2 | 0.000 | 25.2 | 22 |
| 930 | 112 (1) | 1,394 | 2 | 0.006 | 49.7 | 11 |
| Mis-joined Contigs | ||||||
| 138a | 431 (4) | 4,590 | 2 | 0.043 | 16.0 | 1 |
| 138b | 431 (4) | 4,590 | 1 | N/A | 32.0 | 27 |
| 450a | 334 (4) | 3,709 | 2 | 0.006 | 38.6 | 22 |
| 450b | 334 (4) | 3,709 | 1 | N/A | 39.4 | 1 |
a Number of Q-clones in parenthesis. Clones that were assigned to a contig but may be false positives (e.g. chimerical clones) are marked by FPC as Q-clones.
b Two point linkage recombination between markers from the two most distant clones in the contig.
c Additional markers that were previously isolated from other BACs in the contigs were linked to the new BAC end sequence microsatellites by two point linkage analysis and included in this table. Contig 84: OMM3090/MHCI-A [27]; Contig260: OMM3079/TAP1 [23]; Contig 791: OMM3183/TLR8a (manuscript in preparation).
Figure 2.
Example of a rainbow trout contig anchored to a genetic linkage group using microsatellites isolated from BAC end sequences. The contig shown is number 172 containing 299 clones. The 3 markers were mapped to Chromosome 6. Genetic distances between markers were calculated from a two-point linkage analysis. The clones from which the markers were isolated (Table 3) are "hidden" within the highlighted clones. Clone groups with more then 90% DNA fingerprints overlap are represented by the largest BAC in the group, which was not the actual BAC from which the microsatellites shown here were isolated.
FPC identified 1,986 questionable (Q) clones in 811 contigs in this physical map (Table 1). Q-clones are the result of false overlaps between DNA fingerprinting patterns, which can be caused by the presence of chimerical clones in the BAC library, cross-contamination between neighboring wells, large repetitive regions of the genome or duplicated regions that are frequent in the trout genome. The occurrence of Q-clones in this assembly (1.4%) is lower than the 4%-11% reported for other HICF projects [48,49,52]. However, the fraction of contigs with Q-clones in this assembly (19.4%) is similar to the catfish physical map assembly of Xu et al. [49]. The initial high cutoff stringency and relatively deep genome coverage that we used likely contributed to the lower fraction of Q-clones in this assembly. The quality of this physical map was validated, but it could still benefit from better computational tools for identifying Q-clones. Clearly, the assembly of physical maps can be significantly improved by identifying the specific Q-clones in each contig, which in turn will enable evaluation of their location within the contig and relationship to the neighboring clones. As a proof of concept, the computational approach for improving physical maps assembly that is currently being developed by Frenkel et al. [53] was tested on the two large mis-joined contigs that we identified in this assembly and correctly identified the specific Q-clones causing the mis-joining of the contigs and how they should be split into smaller contigs that would also be in good agreement with the genetic map (data not shown). Contig 260 that was also analyzed by this approach was found to be an intact contig, which is also in agreement with our results (Table 4). Taken together the results of this analysis illustrate that better handling of Q-clones by the assembly software can dramatically improve physical maps.
Conclusion
The production and validation of the first physical map of the rainbow trout genome is described in this paper. We are currently integrating this map with the genetic map using more than 200 microsatellites isolated from BAC end sequences and by identifying BACs that harbor more than 300 previously mapped markers. The availability of an integrated physical and genetic map will enable detailed comparative genome analyses, fine mapping of QTL, positional cloning, selection of positional candidate genes for economically important traits and the incorporation of MAS into rainbow trout breeding programs. A comprehensive integrated map can also provide a minimal tiling path for genome sequencing and a framework for whole genome sequence assembly.
Methods
BAC libraries and DNA fingerprinting
BAC clones were obtained from the DH Swanson YY male 10X HindIII library [5] and the CHORI-220 Swanson 5X EcoRI library http://bacpac.chori.org/library.php?id=405. All the clones from the 10X HindIII BAC library (184,704 clones) and 7,392 clones from the CHORI-220 library, were fingerprinted. We used the 4-color HICF SNaPshot method of Luo et al. [50]. For each 384-well plate from the library four 96-well blocks were inoculated using a 96-pin replicator. For plate orientation and fingerprinting quality, wells E7 and H12 were replaced in each 96-well block with a control BAC clone. The cultures were grown on an orbital shaker at 37 C and 400 rpm for 22 hr. The BAC DNA was purified using the Qiagen R.E.A.L. 96 prep kit (Qiagen, Valencia, CA). Each BAC was simultaneously digested with four 6-bp recognizing restriction endonucleases generating 3' recessed ends and a 4-bp recognizing restriction endonuclease producing blunt ends. Each of the four recessed 3' ends was labeled with a different fluorescent dye using the SNaPshot kit (Applied Biosystems, Foster City, CA). Restriction fragments were sized with a capillary DNA analyzer ABI3730XL (Applied Biosystems) using an internal GeneScan-1200 LIZ v. 1 size standard. Fragment size-calling was performed with the GeneMapper v. 3.7 (Applied Biosystems). Outputs of size-calling files were automatically edited with the FPMiner program http://bioinforsoft.com/ using the program's default setting. This software package was used to distinguish peaks corresponding to restriction fragments from peaks generated by background noise in the profile of each BAC fingerprint and to remove vector restriction fragments from the profiles. The program also removed sub-standard profiles that could negatively affect contigs assembly. The files generated by FPMiner were used in the FPC contig assembly.
Contigs assembly
Contigs were assembled from fragments within size range of 70-1,000 bp using FPC program version 8.5.3 [52,54]. FPC parameters were adjusted for the HICF method as previously described [48,50,52]. An initial assembly was performed with a tolerance of 0.5 bp and a Sulston score of 1 × 10-70. Contigs with more than 15% Q-clones were re-assembled by setting the DQer function to 15% and the "Step" value to 2. This was followed by several rounds of end-to-end merging and single-to-end merging at progressively lower cutoff stringencies (Table 5). The "Best of" function was set to 50 builds.
Table 5.
FPC parameters used to assemble the physical map.
| Sulton Score | Step in the Assembly Build | Merge Function |
| 1.00E-70 | Initial assembly | -- |
| 1.00E-70 | Dqer, 15%, Step: 2 | -- |
| 1.00E-65 | Single-to-End | 35 |
| 1.00E-55 | Single-to-End | 35 |
| 1.00E-68 | End-to-End, 2 | 35 |
| 1.00E-60 | End-to-End, 2 | 35 |
| 1.00E-50 | End-to-End, 2 | 35 |
| 1.00E-45 | Single-to-End | 35 |
| 1.00E-38 | Single-to-End | 35 |
| 1.00E-40 | End-to-End, 2 | 35 |
| 1.00E-35 | End-to-End, 2 | 35 |
| 1.00E-50 | End-to-End, 1 | 35 |
| 1.00E-40 | singletons to contigs | -- |
| 1.00E-35 | singletons to contigs | -- |
BAC end sequencing and markers development
The 10X HindIII Rainbow trout BAC library [5] was used for BAC-end sequencing (BES). BAC culture was conducted using standard protocols and end sequencing with SP6 and T7 primers was done using standard Sanger technique. The raw, untrimmed files were processed by PHRED software [55]. The PHRED quality score cut-off value was set at 20 for the acquisition of Q20 values. The BESs were trimmed of vector sequences (pBeloBAC11 vector [56]) and filtered of E. coli sequences. Microsatellites and other simple sequence repeats (SSR) were analyzed using Tandem repeat Finder software [57]. We examined ten classes of SSRs by using a maximum period size of 10. BESs harboring at least 50 base pairs (bp) flanking sequences on either side of the microsatellites were selected for PCR primer design. Primers for BESs containing microsatellites were designed using Primer3 software [58]. The primer product size range was chosen between 150 and 450 nucleotides. The optimum size of primers was set to 20 nucleotides (range from 18 to 27 nucleotides) with an optimum melting temperature of 60.0°C (range from 57 to 63°C).
Genotyping
The NCCCWA mapping panel of 5 families was genotyped with microsatellites as previously described [15]. A total of 25 microsatellite markers isolated from BAC end sequences (Table 3) and three microsatellites that were previously isolated from BAC clones [23,24,27] were genotyped using the tailed protocol of Boutin-Ganache et al. [59]. Primers were obtained from commercial sources (Alpha DNA, Montreal, Quebec, Canada). Three oligonucleotide primers were used in each DNA amplification reaction (Forward: 5' GAGTTTTCCCAGTCACGAC-primer sequence 3'; reverse: 5' GTTT-primer sequence 3'; fluorescent labeled primer with FAM: 5' GAGTTTTCCCAGTCACGAC 3'). Primers were optimized for amplification by varying annealing temperatures and MgCl2 concentrations. PCR reactions (12 μl total volume) included 50 ng DNA, 1.5-2.5 mM MgCl2, 2 pmol of forward primer, 6 pmol of reverse primer, 1 pmol of fluorescent labeled primer, 200 μM dNTPs, 1× manufacturer's reaction buffer, and 0.5 unit Taq Polymerase (ABI, Foster City, CA, USA). Amplifications were conducted in an MJ Research DNA Engine thermal cycler model PTC 200 (MJ Research, Waltham, MA) as follows: an initial denaturation at 95°C for 10 min, 30 cycles consisting of 94°C for 60 sec, annealing temperature for 45 sec, 72°C extension for 45 sec; followed by a final extension of 72°C for 10 min. PCR products were visualized on agarose gels after staining with ethidium bromide. Three μl of each PCR product was added to 20 μl of water, 1 μl of the diluted sample was added to 12.5 μl of loading mixture made up with 12 μl of HiDi formamide and 0.5 μl of Genscan 400 ROX internal size standard. Samples were denatured at 95°C for 5 min and kept on ice until loading on an ABI 3730 DNA Analyzer (ABI, Foster City, CA, USA). Output files were analyzed using GeneMapper version 3.7 (ABI, Foster City, CA, USA), formatted using Microsoft Excel and stored in a Microsoft Access database.
Linkage analysis
The 28 microsatellites were placed on the rainbow trout genetic map by two-point linkage analysis as previously described [15,60]. Genotype data combined for both sexes were formatted using MAKEPED of the LINKAGE [61] program and checked for inconsistencies with Mendelian inheritance using PEDCHECK [62]. RECODE [63] and LNKTOCRI [64] were used to assemble the data into CRIMAP [65] format. Genotype data were added to that of Rexroad et al [15] and MULTIMAP [66] was used to conduct two-point linkage analyses to identify the closest markers from the published map having the highest LOD Scores.
Authors' contributions
YP designed the study, conducted quality control analyses for the BAC fingerprints, collected microsatellite genotypes, validated contigs assembly and wrote the manuscript draft; MCL participated in the study design, supervised DNA extractions and BAC fingerprinting and assembled the physical map; YH conducted DNA extractions and BAC fingerprinting; FMY provided bioinformatics support for the fingerprints analyses and physical map assembly; CG obtained BAC end sequences, identified microsatellites and designed PCR primers; RLV participated in the study design and developed the genetic linkage analysis pipeline; GHT and PAW developed and maintained the Swanson DH line and provided blood from the clonal line as a source of genomic DNA for the BAC libraries; CER participated in the study design and conducted the genetic linkage analysis. All authors reviewed and contributed to the manuscript.
Acknowledgments
Acknowledgements
This project was supported by National Research Initiative Grant no. 2007-35616-17875 from the USDA Cooperative State Research, Education, and Extension Service Animal Genome program. We thank Renee Fincham, Roseanna Long, Kristy Shewbridge and Brian Smith for their technical support in the microsatellites genotyping, and Zeev Frankel and Abraham Korol from Haifa University for evaluating the assembly of three large contigs using new computational tools for physical map assembly.
Contributor Information
Yniv Palti, Email: yniv.palti@ars.usda.gov.
Ming-Cheng Luo, Email: mcluo@ucdavis.edu.
Yuqin Hu, Email: yqhu@ucdavis.edu.
Carine Genet, Email: carine.genet@jouy.inra.fr.
Frank M You, Email: frank.you@ars.usda.gov.
Roger L Vallejo, Email: roger.vallejo@ars.usda.gov.
Gary H Thorgaard, Email: thorglab@wsu.edu.
Paul A Wheeler, Email: pwheeler@wsu.edu.
Caird E Rexroad, III, Email: caird.rexroadiii@ars.usda.gov.
References
- Thorgaard GH, Bailey GS, Williams D, Buhler DR, Kaattari SL, Ristow SS, Hansen JD, Winton JR, Bartholomew JL, Nagler JJ, et al. Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 2002;133:609–646. doi: 10.1016/S1096-4959(02)00167-7. [DOI] [PubMed] [Google Scholar]
- Ng SH, Artieri CG, Bosdet IE, Chiu R, Danzmann RG, Davidson WS, Ferguson MM, Fjell CD, Hoyheim B, Jones SJ, et al. A physical map of the genome of Atlantic salmon, Salmo salar. Genomics. 2005;86:396–404. doi: 10.1016/j.ygeno.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Young WP, Wheeler PA, Coryell VH, Keim P, Thorgaard GH. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics. 1998;148:839–850. doi: 10.1093/genetics/148.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW, Thorgaard GH. Tetraploidy and the evolution of salmonid fishes. In: Turner BJ, editor. Evolutionary Genetics of Fishes. New York: Plenum Press; 1984. pp. 1–46. [Google Scholar]
- Palti Y, Gahr SA, Hansen JD, Rexroad CE. Characterization of a new BAC library for rainbow trout: evidence for multi-locus duplication. Anim Genet. 2004;35:130–133. doi: 10.1111/j.1365-2052.2004.01112.x. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Nichols KM, DeKoning JJ, Morasch MR, Keatley KA, Rexroad C, 3rd, Gahr SA, Danzmann RG, Drew RE, Thorgaard GH. Assignment of rainbow trout linkage groups to specific chromosomes. Genetics. 2006;174:1661–1670. doi: 10.1534/genetics.105.055269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Asakawa S, Minagawa S, Shimizu N, Hirono I, Aoki T. Construction and characterization of BAC libraries for three fish species; rainbow trout, carp and tilapia. Anim Genet. 2001;32:200–204. doi: 10.1046/j.1365-2052.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- Robison BD, Wheeler PA, Sundin K, Sikka P, Thorgaard GH. Composite interval mapping reveals a major locus influencing embryonic development rate in rainbow trout (Oncorhynchus mykiss) J Hered. 2001;92:16–22. doi: 10.1093/jhered/92.1.16. [DOI] [PubMed] [Google Scholar]
- Robison BD, Wheeler PA, Thorgaard GH. Variation in development rate among clonal lines of rainbow trout (Oncorhynchus mykiss) Aquaculture. 1999;173:131–141. doi: 10.1016/S0044-8486(98)00481-5. [DOI] [Google Scholar]
- Young WP, Wheeler PA, Fields RD, Thorgaard GH. DNA fingerprinting confirms isogenicity of androgenetically derived rainbow trout lines. J Hered. 1996;87:77–80. doi: 10.1093/oxfordjournals.jhered.a022960. [DOI] [PubMed] [Google Scholar]
- Quillet E, Dorson M, Le Guillou S, Benmansour A, Boudinot P. Wide range of susceptibility to rhabdoviruses in homozygous clones of rainbow trout. Fish & Shellfish Immunology. 2007;22:510–519. doi: 10.1016/j.fsi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Guyomard R, Mauger S, Tabet-Canale K, Martineau S, Genet C, Krieg F, Quillet E. A type I and type II microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) with presumptive coverage of all chromosome arms. BMC Genomics. 2006;7:302. doi: 10.1186/1471-2164-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols KM, Young WP, Danzmann RG, Robison BD, Rexroad C, Noakes M, Phillips RB, Bentzen P, Spies I, Knudsen K. A consolidated linkage map for rainbow trout (Oncorhynchus mykiss) Anim Genet. 2003;34:102–115. doi: 10.1046/j.1365-2052.2003.00957.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Danzmann RG, Gharbi K, Howard P, Ozaki A, Khoo SK, Woram RA, Okamoto N, Ferguson MM, Holm L-E, et al. A Microsatellite Linkage Map of Rainbow Trout (Oncorhynchus mykiss) Characterized by Large Sex-Specific Differences in Recombination Rates. Genetics. 2000;155:1331–1345. doi: 10.1093/genetics/155.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroad CE, 3rd, Palti Y, Gahr SA, Vallejo RL. A second generation genetic map for rainbow trout (Oncorhynchus mykiss) BMC Genet. 2008;9:74. doi: 10.1186/1471-2156-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroad CE, Lee Y, Keele JW, Karamycheva S, Brown G, Koop B, Gahr SA, Palti Y, Quackenbush J. Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenet Genome Res. 2003;102:347–354. doi: 10.1159/000075773. [DOI] [PubMed] [Google Scholar]
- Govoroun M, Le Gac F, Guiguen Y. Generation of a large scale repertoire of Expressed Sequence Tags (ESTs) from normalised rainbow trout cDNA libraries. BMC Genomics. 2006;7:196. doi: 10.1186/1471-2164-7-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rise ML, von Schalburg KR, Brown GD, Mawer MA, Devlin RH, Kuipers N, Busby M, Beetz-Sargent M, Alberto R, Gibbs AR, et al. Development and Application of a Salmonid EST Database and cDNA Microarray: Data Mining and Interspecific Hybridization Characteristics. Genome Res. 2004;14:478–490. doi: 10.1101/gr.1687304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M, Kenney B, Rexroad CE, Yao J. Development of a 37 k high-density oligonucleotide microarray: a new tool for functional genome research in rainbow trout. Journal of Fish Biology. 2008;72:2187–2206. [Google Scholar]
- Coulibaly I, Danzmann RG, Palti Y, Vallejo R, Gahr SA, Yao J, Rexroad CE. Mapping of genes in a region associated with upper temperature tolerance in rainbow trout. Animal Genetics. 2006;37:598–599. doi: 10.1111/j.1365-2052.2006.01529.x. [DOI] [PubMed] [Google Scholar]
- Coulibaly I, Gahr SA, Palti Y, Yao J, Rexroad CE. Genomic structure and expression of uncoupling protein 2 genes in rainbow trout (Oncorhynchus mykiss) BMC Genomics. 2006;7:203. doi: 10.1186/1471-2164-7-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahr SA, Palti Y, Rexroad CE. Genomic characterization of a novel pair of ID genes in the rainbow trout (Oncorhynchus mykiss) Anim Genet. 2004;35:317–320. doi: 10.1111/j.1365-2052.2004.01142.x. [DOI] [PubMed] [Google Scholar]
- Palti Y, Rodriguez MF, Gahr SA, Hansen JD. Evolutionary history of the ABCB2 genomic region in teleosts. Developmental & Comparative Immunology. 2007;31:483–498. doi: 10.1016/j.dci.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Palti Y, Rodriguez MF, Vallejo RL, Rexroad CE., 3rd Mapping of Toll-like receptor genes in rainbow trout. Anim Genet. 2006;37:597–598. doi: 10.1111/j.1365-2052.2006.01527.x. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Zimmerman A, Noakes MA, Palti Y, Morasch MR, Eiben L, Ristow SS, Thorgaard GH, Hansen JD. Physical and genetic mapping of the rainbow trout major histocompatibility regions: evidence for duplication of the class I region. Immunogenetics. 2003;55:561–569. doi: 10.1007/s00251-003-0615-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Wiens G, Purcell M, Palti Y. Characterization of Toll-like receptor 3 gene in rainbow trout (Oncorhynchus mykiss) Immunogenetics. 2005;57:510–519. doi: 10.1007/s00251-005-0013-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez MF, Gahr SA, Rexroad CE, 3rd, Palti Y. A polymerase chain reaction screening method for rapid detection of microsatellites in bacterial artificial chromosomes. Mar Biotechnol (NY) 2006;8:346–350. doi: 10.1007/s10126-005-5064-7. [DOI] [PubMed] [Google Scholar]
- Thorgaard GH. Chromosomal differences among rainbow trout populations. Copeia. 1983;1983:650–662. [Google Scholar]
- Phillips RB, Morasch MR, Wheeler PA, Thorgaard GH, Quattro JM. Rainbow Trout (Oncorhynchus mykiss) of Idaho and Alaskan Origin (2n = 58) Share a Chromosome Fusion Relative to Trout of California Origin (2n = 60) Copeia. 2005;2005:661–664. [Google Scholar]
- Danzmann R, Davidson E, Ferguson M, Gharbi K, Koop B, Hoyheim B, Lien S, Lubieniecki K, Moghadam H, Park J, et al. Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon) BMC Genomics. 2008;9:557. doi: 10.1186/1471-2164-9-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzmann RG, Cairney M, Davidson WS, Ferguson MM, Gharbi K, Guyomard R, Holm LE, Leder E, Okamoto N, Ozaki A. A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic char and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae) Genome. 2005;48:1037–1051. doi: 10.1139/g05-067. [DOI] [PubMed] [Google Scholar]
- Nichols KM, Bartholomew J, Thorgaard GH. Mapping multiple genetic loci associated with Ceratomyxa shasta resistance in Oncorhynchus mykiss. Dis Aquat Organ. 2003;56:145–154. doi: 10.3354/dao056145. [DOI] [PubMed] [Google Scholar]
- Barroso RM, Wheeler PA, LaPatra SE, Drew RE, Thorgaard GH. QTL for IHNV resistance and growth identified in a rainbow (Oncorhynchus mykiss)xYellowstone cutthroat (Oncorhynchus. Aquaculture. 2008;277:156–163. [Google Scholar]
- Rodriguez MF, LaPatra S, Williams S, Famula T, May B. Genetic markers associated with resistance to infectious hematopoietic necrosis in rainbow and steelhead trout. Aquaculture. 2004;241:93–115. [Google Scholar]
- Ozaki A, Sakamoto T, Khoo S, Nakamura K, Coimbra MR, Akutsu T, Okamoto N. Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss) Mol Genet Genomics. 2001;265:23–31. doi: 10.1007/s004380000392. [DOI] [PubMed] [Google Scholar]
- Zimmerman A, Evenhuis J, Thorgaard G, Ristow S. A single major chromosomal region controls natural killer cell-like activity in rainbow trout. Immunogenetics. 2004;55:825–835. doi: 10.1007/s00251-004-0645-6. [DOI] [PubMed] [Google Scholar]
- Perry GM, Danzmann RG, Ferguson MM, Gibson JP. Quantitative trait loci for upper thermal tolerance in outbred strains of rainbow trout (Oncorhynchus mykiss) Heredity. 2001;86:333–341. doi: 10.1046/j.1365-2540.2001.00838.x. [DOI] [PubMed] [Google Scholar]
- Perry GM, Ferguson MM, Sakamoto T, Danzmann RG. Sex-linked quantitative trait loci for thermotolerance and length in the rainbow trout. J Hered. 2005;96:97–107. doi: 10.1093/jhered/esi019. [DOI] [PubMed] [Google Scholar]
- Nichols KM, Broman KW, Sundin K, Young JM, Wheeler PA, Thorgaard GH. Quantitative trait loci × maternal cytoplasmic environment interaction for development rate in Oncorhynchus mykiss. Genetics. 2007;175:335–347. doi: 10.1534/genetics.106.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin K, Brown KH, Drew RE, Nichols KM, Wheeler PA, Thorgaard GH. Genetic analysis of a development rate QTL in backcrosses of clonal rainbow trout, Oncorhynchus mykiss. Aquaculture. 2005;247:75–83. [Google Scholar]
- O'Malley KG, Sakamoto T, Danzmann RG, Ferguson MM. Quantitative trait Loci for spawning date and body weight in rainbow trout: testing for conserved effects across ancestrally duplicated chromosomes. J Hered. 2003;94:273–284. doi: 10.1093/jhered/esg067. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Danzmann RG, Okamoto N, Ferguson MM, Ihssen PE. Linkage analysis of quantitative trait loci associated with spawning time in rainbow trout (Oncorhynchus mykiss) Aquaculture. 1999;173:33–43. [Google Scholar]
- Drew RE, Schwabl H, Wheeler PA, Thorgaard GH. Detection of QTL influencing cortisol levels in rainbow trout (Oncorhynchus mykiss) Aquaculture. 2007;272:S183–S194. [Google Scholar]
- Haidle L, Janssen J, Gharbi K, Moghadam H, Ferguson M, Danzmann R. Determination of Quantitative Trait Loci (QTL) for Early Maturation in Rainbow Trout (Oncorhynchus mykiss) Marine Biotechnology. 2008;10:579–592. doi: 10.1007/s10126-008-9098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols KM, Edo AF, Wheeler PA, Thorgaard GH. The Genetic Basis of Smoltification-Related Traits in Oncorhynchus mykiss. Genetics. 2008;179:1559–1575. doi: 10.1534/genetics.107.084251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Vallejo RL, Silverstein JT, Welch TJ, Wiens GD, Hallerman EM, Palti Y. Suggestive Association of Major Histocompatibility IB Genetic Markers with Resistance to Bacterial Cold Water Disease in Rainbow Trout (Oncorhynchus mykiss) Mar Biotechnol. 2008;10:429–437. doi: 10.1007/s10126-007-9080-7. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Kidd C, Tomasino E, Davis JT, Wishon C, Stern JE, Carleton KL, Howe AE, Kocher TD. A BAC-based physical map of the Nile tilapia genome. BMC Genomics. 2005;6:89. doi: 10.1186/1471-2164-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiniou S, Waldbieser G, Duke M. A first generation BAC-based physical map of the channel catfish genome. BMC Genomics. 2007;8:40. doi: 10.1186/1471-2164-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Wang S, Liu L, Thorsen J, Kucuktas H, Liu Z. A BAC-based physical map of the channel catfish genome. Genomics. 2007;90:380–388. doi: 10.1016/j.ygeno.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Luo M-C, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OD, Dvorak J. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics. 2003;82:378–389. doi: 10.1016/s0888-7543(03)00128-9. [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Smith DW, Kohlenberg JB, Schork NJ. Large-Scale Integration of Human Genetic and Physical Maps. Genome Research. 2004;14:1199–1205. doi: 10.1101/gr.1475304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, Kim H, Wing RA, Messing J, Soderlund C. Whole-Genome Validation of High-Information-Content Fingerprinting. Plant Physiol. 2005;139:27–38. doi: 10.1104/pp.105.061978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. Improving The Efficiency Of Contig Assembly For Physical Mapping. Plant & Animal Genome XVII, the international conference on the status of plant and animal genome research. 2009. p. W282.
- Nelson W, Soderlund C. Integrating sequence with FPC fingerprint maps. Nucl Acids Res. 2009;37:e36. doi: 10.1093/nar/gkp034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, Kang HL, Simon MI, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques. 2001;31:24–26. [PubMed] [Google Scholar]
- Kongchum P, CE Rexroad I, Hallerman EM, David L, Palti Y. Single nucleotide polymorphism identification, genetic mapping and tissue expression of the rainbow trout TLR9 gene. Animal Genetics. 2009 doi: 10.1111/j.1365-2052.2009.01924.x. [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- LNKTOCRI++ http://www.filewatcher.com/b/ftp/ftp.ebi.ac.uk/pub/software/linkage_and_mapping/MULTIMA/lnktocri/C%2B%2B.0.0.html
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]