Abstract
The A mating type genes of the mushroom Coprinus cinereus encode two families of dissimilar homeodomain proteins (HD1 and HD2). The proteins heterodimerize when mating cells fuse to generate a transcriptional regulator that promotes expression of genes required for early steps in sexual development. In previous work we showed that heterodimerization brings together different functional domains of the HD1 and HD2 proteins; a potential activation domain at the C terminus of the HD1 protein and an essential HD2 DNA-binding motif. Two predicted nuclear localization signals (NLS) are present in the HD1 protein but none are in the HD2 protein. We deleted each NLS separately from an HD1 protein and showed that one (NLS1) is essential for normal heterodimer function. Fusion of the NLS sequences to the C terminus of an HD2 protein compensated for their deletion from the HD1 protein partner and permitted the two modified proteins to form a functional transcriptional regulator. The nuclear targeting properties of the A protein NLS sequences were demonstrated by fusing the region that encodes them to the bacterial uidA (β-glucuronidase) gene and showing that β-glucuronidase expression localized to the nuclei of onion epidermal cells. These observations lead to the proposal that heterodimerization regulates entry of the active transcription factor complex to the nucleus.
Mating is an essential step in the life cycle of the mushroom Coprinus cinereus. Mating compatibility is determined by multiallelic genes at two complex loci that are designated A and B (1). Provided that mates have different alleles of both sets of genes, somatic cell fusion is sufficient to trigger a developmental program that converts a sterile uninucleate-celled mycelium known as the monokaryon into a fertile binucleate-celled mycelium known as a dikaryon. The dikaryon is an extended vegetative stage between mating cell fusion and karyogamy. Each dikaryotic cell maintains the two haploid nuclei donated by each mating partner and it is only in the fruit body that nuclear fusion occurs and mating is completed. The equal distribution of both nuclei in vegetative cells is maintained by formation of a special structure known as a clamp connection through which one of the daughter nuclei passes each time a tip cell divides. The coordinated activities of both A and B mating type genes are required to form the clamp connection; different A gene alleles lead to development of a clamp cell, different B gene alleles are necessary for its completion by fusion to the adjacent cell.
The A mating type genes, the subject of this report, encode two dissimilar subunits of a heterodimeric transcription factor. The two subunits are distinguished as homeodomain proteins HD1 and HD2 on the basis of conserved but distinctly different homeodomain motifs (2). After mating, HD1 and HD2 proteins heterodimerize to generate a dikaryon-specific transcriptional regulator that triggers the initial steps in sexual development (3). Heterodimerization plays a crucial role in mating partner recognition. The A mating type genes are multiallelic and there are an estimated 160 A mating specificities determined by three pairs of paralogous genes that encode functionally redundant proteins of both classes (the a, b, and d pairs) (4). The HD1 and HD2 proteins present in a cell before mating are unable to heterodimerize, whereas a compatible mating is one that brings together allelic versions of the proteins that can (3). Heterodimerization is mediated by the N-terminal domains of the proteins and serves the essential function of permitting only genetically different mating partners to generate the transcriptional regulator that promotes sexual development.
A similar heterodimerization between two classes of homeodomain proteins has been shown to play a critical role in sexual development in the budding yeast Saccharomyces cerevisiae (for reviews, see refs. 5 and 6). In this species there are just two alleles of the MAT locus and these encode either the a1 or the α2 protein. The proteins heterodimerize when MATa and MATα cells fuse to generate a diploid cell-specific transcription factor that binds operator sites that neither protein can bind alone (7) Heterodimerization is thus crucial in determining DNA binding-site specificity. Although the DNA target site of the C. cinereus A protein heterodimer has yet to be described, it is assumed that, as for a1 and α2 of S. cerevisiae, the combinatorial interaction of both proteins confers operator-site specificity.
Heterodimerization plays another important role in regulating the function of a transcription factor by bringing together different functional domains. We previously have demonstrated this for the A protein heterodimer of C. cinereus. HD1 proteins contribute a potential activation domain present in an essential C-terminal sequence that has been shown to activate transcription of a reporter gene in S. cerevisiae (8). The HD1 homeodomain can be deleted from the C. cinereus HD1 protein without causing impaired function. The HD2 homeodomain, however, is essential, indicating that for the HD1–HD2 heterodimer, this is the critical DNA-binding domain.
In this report we identify another potential regulatory role for heterodimerization between compatible HD1 and HD2 proteins. Heterodimerization via the N-terminal domains of the proteins is independent of DNA, which implies that it can occur in the cytoplasm. Analysis of the HD1 protein sequence identified two potential bipartite nuclear targeting sequences (NLS) but no corresponding sequences in the HD2 protein (9, 10). In this report we provide evidence that heterodimerization is necessary to target an active transcription factor complex to the nucleus.
MATERIALS AND METHODS
Strains.
The C. cinereus strains used as hosts for DNA-mediated transformations were as follows: LN118, A42B42trp1–1.1;1.6 ade-2; LT2, A6B6 trp1–1.1;1.6; NAB1, AD43B43 m pab-1trp-1.1;1.6.
Transformation.
Competent protoplasts for transformation were prepared according to the method of Binninger et al. (11) and transformed by using the modified technique of Casselton and de la Fuente Herce (12). Plasmids containing mating type genes, pAMT2, b1–1 (9), and pA625, b2–3 (13), and modified genes described below, were cotransformed with plasmid pCc1001 (11) containing the trp-1 gene. Cotransformation frequencies vary from 10 to 80% in C. cinereus. Samples of 50–100 transformants are thus sufficient to determine whether or not the cotransformed gene is expressed. Production of clamp cells by transformants was detected by microscopic examination.
Plasmid Constructs.
Routine cloning and plasmid amplification was in Escherichia coli XL-1 Blue (Stratagene) and DH5α (Bethesda Research Laboratories) as described (14). Deletion of sequences encoding NLS1 and NLS2 was achieved by inverse PCR (15) using pAMT2 as template (9).The four oligonucleotide primer sequences (Genosys, The Woodlands, TX), homologous to regions flanking the NLS coding domains, were as follows: 5′-CGCGAACGATGGGGGCGAAG-3′ and 5′-CTGCCTTCACCTTCC-3′ for the ΔNLS1; 5′-CGCCGAGTCAGTCGGGGTGA-3′ and 5′-GCTACCAGCAGCGAACATTG-3′ for the ΔNLS2. Inverse PCR was performed with VENT polymerase (New England Biolabs) under the following conditions: 95°C for 1 min, 57°C for 1 min, and 72°C for 4 min for 19 cycles. The ΔNLS1 plasmid with the ΔNLS2 oligonucleotide primers were used under the same conditions to achieve the double NLS deletion molecule.
pTS8 was constructed as follows: the NLS coding domain of b1–1 was amplified by standard PCR using the oligonucleotide 5′-GGGGGGATCCCGCGCTCGCCACGCG-3′ as the 5′ primer and 5′-CCCCCCATGGTTAGGCCGGGGGAATGTCCCC-3′ as the 3′ primer. The primers include a BamHI site (5′ primer) and a NcoI site (3′ primer). PCR amplification from pAMT2 (9) template was performed with Taq polymerase in the presence of 1.5 mM MgCl2 under the following conditions: 95°C for 45 s, 58°C for 1 min, and 72°C for 1 min for 30 cycles. Purified product was subcloned into pTAG (Ingenius, Abingdon, UK) under manufacturer’s conditions. The b1–1 NLS coding domain was recovered as a BamHI–NcoI fragment and ligated with pRTL2-GUS/NIa (16) BamHI–NcoI fragments of 3.9 kb and 1.7 kb (where GUS is β-glucuronidase). pTS20, a full-length b2–3-GUS construct, was created by cloning a BglII–BamHI PCR fragment into the BglII–BamHI backbone of pRTL2-GUS/NIA. PCR was achieved by using 5′-GGGGGAGATCTATGCAGGAACGACCAAACGG-3′ and 5′-GGGGGATCCTGTCAAGCCATACGCGGG-3′ as 5′ and 3′ primers, respectively. PCR was carried out as for pTS8 but with an extension time of 2 min.
pGEMT:b2–3:b1–1NLS was constructed as follows: the full coding length and promoter of b2–3 was amplified by using pA625 (13) as template and the oligonucleotides 5′-CTCGAGTCCGGTCAAT-3′ and 5′-GGGGAATTCCAGGTCAGTCGAATCCACGG-3′ as 5′ and 3′ primers, respectively. The 3′ primer included the insertion of a EcoRI site. The b1–1 NLS coding domain was amplified by using pAMT2 (9) as template and the following oligonucleotides as the 5′ and 3′ primers, respectively: 5′-GGGGAATTCGCGATCGATTCAGATAAATTG-3′ and 5′-CCCCCCCGCGGTTATTACGTAACGGCCGGGGGAATGTCCCC-3′. The 5′ primer inserts a EcoRI site before the coding sequence and the 3′ primer includes a stop codon after the NLS sequence. PCR for both b2–3 and b1–1 was performed as for pTAG:b1–1NLS described above except the annealing temperature was lowered to 56°C. Both amplified b2–3 and b1–1 NLSs were cloned into pGEM-T (Promega) under manufacturer’s conditions. The b1–1 NLS was excised as a ApaI–EcoRI fragment and cloned into pGEM-T:b2–3.
Nuclear Localization Assay.
The pTS8 and pTS20 plasmids were purified using a Qiagen Midiprep kit before expression in onion epidermal cells. Inner epidermal peels from onion bulbs were placed on solid Chu medium (Sigma). Peels were then subject to bombardment by pTS8, pTS20, pRTL2-GUS, and pRTL2-GUS/NIa (16) after precipitation on gold particles as detailed by the delivery system manufacturer. Bombardment was carried out by using a Bio-Rad Model PDS, 1000/He Biolistic particle delivery system at a vacuum of 24 inches (1 inch = 2.54 cm) of mercury (1 mmHg = 133 pa). with delivery on rupture of a 1,100 psi disc (1 psi = 6.89 kPa). Plates were incubated for 15–18 h in the dark at room temperature. To stain for GUS activity, plates were incubated for 1 h at 37°C in 100 mM sodium phosphate, pH 7/1 mM EDTA/1% Triton X-100/5 mM potassium ferricyanide/5 mM potassium ferrocyanide/1 mM 5-bromo-4-chloro-3-indoyl β-d-glucuronide. Cells were subsequently stained in 4′,6-diamidino-2-phenylindole (2 μg/ml)/50 mM Tris⋅HCl, pH 7, and mounted on microscopy glass slides. Cells were viewed and photographed with a Zeiss Axiophot photomicroscope loaded with Kodak Ektachrome film.
RESULTS
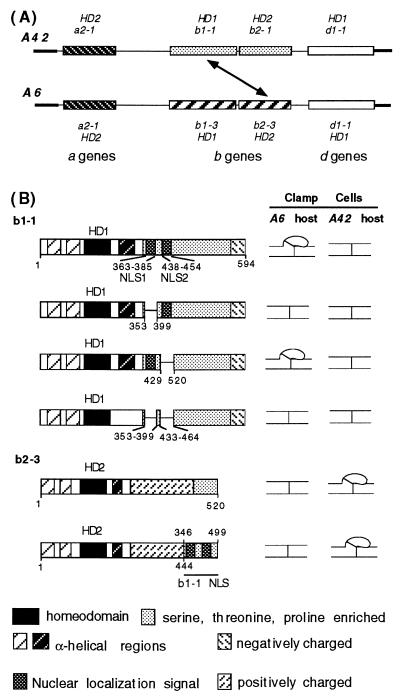
The A locus of C. cinereus derives some 160 specificities from three pairs of multiallelic HD1 and HD2 genes. It requires alleles of only one pair of these genes to be different for mating partners to be able to generate an active HD1–HD2 heterodimer capable of activating A regulated development. It is rare to find all six genes present in any one locus (17). As seen for A6 and A42 (Fig. 1A), only the b gene pair is complete, and there is a solo a gene and a solo d gene. The alleles of the two solo genes are identical in both A6 and A42 and thus they can play no part in determining mating compatibility between A42 and A6 monokaryons. Two A protein heterodimers can be formed after cell fusion, b1–1 with b2–3 and b1–3 with b2–1 (3), each of which is sufficient to trigger A regulated clamp-cell development. We chose the b1–1 and b2–3 genes for the experiments we now describe and this is the compatible gene interaction indicated by a diagonal arrow in Fig. 1A.
Figure 1.
Effect of deleting or adding predicted NLSs on b1–1–b2–3 heterodimer function. (A) Organization of the A42 and A6 mating type loci of C. cinereus. A42 and A6 contain representative members of three pairs of functionally redundant genes (a, b, and d), each pair encoding an HD1 and an HD2 protein. For simplicity, a nonfunctional pseudogene in the A42 locus is not shown. The diagonal arrow indicates one of two possible gene combinations that activate A- regulated development because their encoded proteins can heterodimerize when present in the same cell. (B) Wild-type and manipulated genes from which the NLSs were deleted (b1–1) or added (b2–3) were introduced into A6 and A42 host cells by DNA-mediated transformation and transformants were examined for signs of A- regulated development. unfused clamp cell indicates A-regulated development observed; simple septum indicates no A-regulated development observed. Predicted structural features of the HD1 and HD2 proteins are indicated by different motifs defined in the key.
One NLS Sequence Is Essential for A-Regulated Development.
Fig. 1B illustrates some predicted structural features of the b1–1 and b2–3 proteins. These include the α-helical regions that constitute the N-terminal dimerization domains, the HD1 and HD2 homeodomains, an α-helical region just C-terminal to the homeodomains in both proteins, and a C-terminal domain in both proteins that is relatively rich in serine, threonine, and proline. In HD1 proteins the C terminus contains an essential negatively charged domain, the predicted activation domain. A truncated version of the b1–1 gene, lacking 120 bp at the 3′ end, was used in this study. This has no detectable effect on b1–1 protein function in the assay we used (9), and the truncated gene was chosen because it simplified cloning procedures.
Our assay for function of b1–1 was to introduce the gene into host strains having either the A6 or the A42 mating type genes by DNA-mediated transformation. In the A6 host cells the protein of the introduced b1–1 gene can heterodimerize with the protein of the resident b2–3 gene and promote formation of unfused clamp cells. In the self A42 host, the b1–1 protein is unable to heterodimerize with the resident b2–1 protein, and no clamp cell development occurs. Similarly, the b2–3 protein function can be assayed by introduction into the same two host strains. In this case, the b2–3 protein promotes clamp cell development in the A42 host where it meets the b1–1 protein, but not in the self A6 host, which has the b1–3 protein.
There are two predicted bipartite nuclear location sequences in the b1–1 protein at amino acid positions 363–385 (NLS1) and 438–454 (NLS2) (9). We made three internal deletion constructs of the b1–1 gene that allowed us to test whether one or both predicted NLS sequences were essential for b1–1 function. Deletion of NLS1 or both NLS sequences led to complete loss of clamp cell development, whereas deletion of only NLS2 had no effect (Fig. 1B).
b1–1 Deletions Are Compensated by Adding NLS Sequences to b2–3.
We tested the affect of adding NLS sequences to the b2–3 protein. The b2–3 gene was truncated to make a version of b2–3 that lacked the C-terminal 74 amino acids that we have previously shown to be nonessential in the clamp cell assay (10). The region of b1–1 that encodes both NLS sequences was then added to generate the protein illustrated in Fig. 1B. The modified protein was fully functional in an A42 host, indicating that the additional amino acid sequence did not interfere with its regulatory role. It is of interest to note that this protein was still recognized as incompatible in the self A6 host.
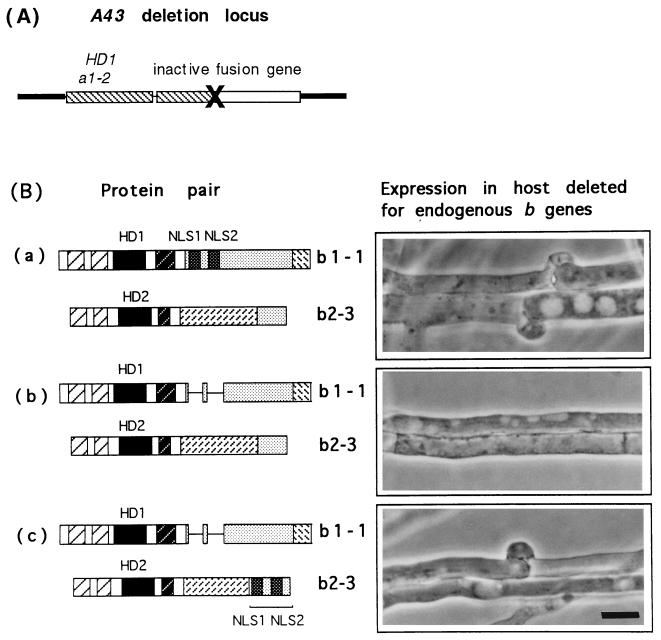
After engineering genes encoding an HD1 protein lacking both predicted NLSs and an HD2 protein to which these had been added, we now asked whether these two proteins could heterodimerize to generate a functional regulator of development. The host strain used in this experiment was generated by Pardo et al. (4) to demonstrate the functional independence of the a, b, and d genes present in each A mating type locus. As shown in Fig. 2A, the endogenous A locus in this strain (NAB1) contains two genes, an HD1 gene belonging to the a gene pair (a1–2) and a totally inactive chimeric gene. Neither the b1–1 nor the b2–3 protein can promote clamp cell development in this host because neither can form a heterodimer with a1–2. Another feature of this host strain is that it has a self-compatible mutation in the B locus that leads to constitutive activation of the B-regulated steps in dikaryotic growth (4, 18). If A-regulated clamp cell formation is induced in this host strain by a compatible b1–1–b2–3 protein interaction, the clamp cells are fused by the constitutive B gene function.
Figure 2.
Predicted NLSs are essential for b1–1–b2–3 heterodimer function and are protein-unspecific. (A) Organization of the partial null A locus of strain NAB1 that lacks both members of the b gene pair and in which the function of the b1–1–b2–3 heterodimer was assayed. (B) b2–3 and b1–1 were introduced by transformation into the NAB host and transformants screened for the development of clamp connections. Predicted proteins encoded by wild-type b1–1 and b2–3 (a), b1–1 ΔNLS and wild-type b2–3 (b), and b1–1 ΔNLS and modified b2–3 with added NLSs (c). (Bar = 10 μm.)
We introduced either the b2–3 wild-type gene or the b2–3 gene to which the NLS coding sequences had been added into the NAB1 host. Presence of the gene in transformants was confirmed by a genetic test described by Pardo et al. (4). We then introduced a second gene into the transformed host strain encoding either the unmodified b1–1 gene or the gene from which the NLS sequences had been deleted and looked for the development of fused clamp connections (Fig. 2B). The two controls for this experiment are shown in Fig. 2 Ba and Bb. Transformants expressing the unmodified b1–1 and b2–3 genes produced mycelia with clamp connections (Fig. 2Ba), whereas transformants containing the b1–1 gene lacking the predicted NLSs and the wild-type b2–3 gene produced no clamp connections (Fig. 2Bb). Transformants expressing both modified genes, b1–1 lacking the NLSs and b2–3 with added NLSs, produced typical clamp connections (Fig. 2Bc). The amino acid sequences added to b2–3 can, therefore, compensate their deletion from b1–1. This experiment shows that the loss of function of b1–1 lacking the NLSs is not caused by protein instability. It must be present in the cell to dimerize with the modified b2–3 protein to which the NLSs were added.
The b1–1 NLS Sequences Target a Reporter Gene to the Nucleus.
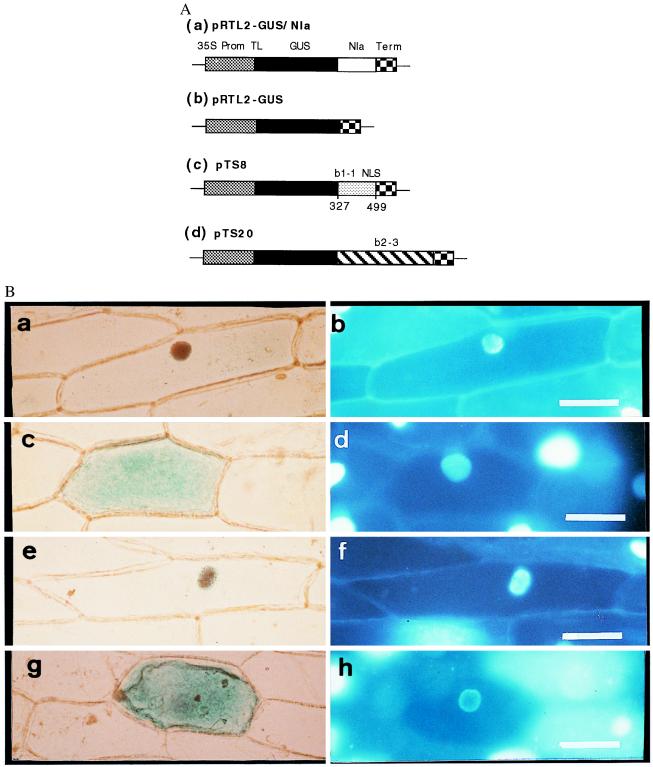
To test the ability of the b1–1 sequences to confer nuclear localization, we adopted the experimental approach of McGonigle et al. (19) and generated a translational fusion of a C. cinereus sequence containing both NLSs and the bacterial uidA gene that encodes GUS driven by a cauliflower mosaic virus 35S promoter (Fig. 3). We used the same two control constructs as McGonigle et al. (19): a translational fusion of the GUS gene to the plant potyviral Nla protein that contains a bipartite NLS sequence and has been shown to localize to the nucleus and an unfused GUS gene that has been shown to localize in the cytoplasm (16, 20, 21). The three constructs were introduced into Allium cepa (onion) epidermal cells by using microprojectile bombardment (22, 23). Subcellular localization of GUS enzyme activity was assayed after 24 h. 4′,6-Diamidino-2-phenylindole staining was used to identify the nucleus in cells exhibiting GUS activity.
Figure 3.
Subcellular localization of GUS activity directed by sequences from the A mating type proteins of C. cinereus. (A) Constructs introduced into onion cells. Coding regions were expressed under the control of the cauliflower mosaic 35S promoter and nopaline synthase terminator. (a) Control in which GUS was fused to the plant potyviral Nla protein that contains a bipartite NLS. (b) Control with GUS sequence alone. (c) GUS fused to amino acids 327–499 from b1–1. (d) GUS fused to b2–3. (B) GUS activity (a, c, e, and g) and 4′,6-diamidino-2-phenylindole-stained nuclei (b, d, f, and h) in transiently transformed onion epidermal cells. GUS activity localized to the nucleus by the Nla protein (a and b), GUS alone localized to the cytoplasm, (c and d) GUS activity localized to the nucleus by the predicted b1–1 NLSs (e and f), and GUS activity localized to the cytoplasm when fused to b2–3 (g and h). (Bar = 100 μm.)
In our experiments the controls behaved as expected; expression of the Nla-GUS fusion in onion cells led to predominantly nuclear GUS staining (Fig. 3 a and b) whereas expression of the unfused GUS protein led to cytoplasmic staining (Fig. 3 c and d). When we introduced the GUS-b1–1NLS fusion, GUS staining was predominantly in the nucleus (Fig. 3 e and f). We thus demonstrated that the predicted bipartite NLS sequences in the b1–1 protein were able to confer nuclear localization.
We considered the possibility that the HD2 protein might contribute sequences capable of directing it to the nucleus independently of its HD1 partner. We therefore made a fusion between the entire b2–3 HD2 gene and the GUS reporter gene and looked for GUS expression in onion cells. All GUS activity localized to the cytoplasm (Fig. 3 g and h). Failure of the HD2 protein to localize GUS activity to the nucleus suggests that it has no NLSs or that these are masked in the undimerized protein. The HD2 protein may thus rely on heterodimerization with its HD1 partner to enter the nucleus.
DISCUSSION
Nuclear localization is essential for a protein to control transcription. Two NLSs in the C. cinereus HD1 proteins were predicted (9) on the basis of similarity to the bipartite NLS of nucleoplasmin (24–26), but we noted that no similar sequences were present in HD2 proteins (8). An analysis of the corresponding HD1 and HD2 proteins encoded by the bE and bW mating type genes of the basidiomycete Ustilago maydis identified a typical bipartite NLS in the HD1 bE protein (27) but not in the HD2 bW protein (28). Without an NLS, a protein will not be actively transported into the nucleus. In this study, we set out to test the idea that HD2 proteins can localize to the nucleus only when dimerized to an HD1 partner.
By separately deleting the two predicted NLSs from an HD1 protein, we were able to show that only one sequence was essential for heterodimer function. By fusing the HD1 NLSs to the C terminus of an HD2 protein, we showed that this essential function was protein-unspecific. A heterodimer formed from two modified proteins, an HD1 protein lacking NLSs and an HD2 protein containing NLSs, was as effective in promoting A regulated development as a heterodimer formed from the two unmodified proteins. At the present time, we do not have a reporter system that would permit us to visualize the proteins within C. cinereus cells. To overcome this, we generated a translational fusion between a C. cinereus sequence encoding the two NLSs and the bacterial uidA gene that encodes GUS. We showed that the HD1 NLSs were necessary and sufficient to direct the fusion protein to the nuclei of onion cells.
It is not unusual to have more than one NLS in a protein and both of those predicted in the C. cinereus HD1 proteins may contribute to efficient nuclear import, even if only one is essential. The MATα2 homeodomain protein of S. cerevisiae has been reported to contain two NLSs, one of which appears to be in the homeodomain (29, 30). There is no consensus NLS but a richness in lysine and arginine residues is characteristic (31). The homeodomain has been implicated in nuclear localization of the POU-domain protein Tst-1/Oct6 (32) and several other POU homeodomain sequences in mammalian transcription factors can be seen to contain a conserved cluster of basic residues with potential NLS properties (32). It has been suggested that DNA-binding and nuclear localization functions of the homeodomain may have coevolved (30). We think it unlikely, however, that either homeodomain in the C. cinereus protein plays an important role in nuclear localization despite being rich in basic residues. The homeodomain of the C. cinereus HD1 proteins is known to be totally dispensable (8, 13), and in this study, we showed that the HD2 protein, and thus homeodomain, was unable to localize the GUS reporter protein to the nucleus of onion cells.
The fact that the HD2 protein failed to localize GUS activity to the nucleus indicates that it lacks an NLS or that this is masked in the undimerized protein. Our experiments do not permit us to distinguish between these alternatives but do support the conclusion that HD2 proteins only enter the nucleus when dimerized to an HD1 partner. The need to dimerize with a suitable partner to localize to the nucleus is likely to be a general mechanism for regulating transcription factor function during development. Of particular relevance to the experiments described herein is the interaction between the Drosophila homeodomain proteins Extradenticle (EXD) and Homothorax (HTH) (33). EXD and HTH, like the C. cinereus A proteins, have been shown to dimerize in vitro, independently of DNA. Genetic experiments indicate that EXD is dependent on HTH for function. In cells that do not express hth, EXD is found exclusively in the cytoplasm but is translocated to the nucleus when HTH is present. In Arabidopsis, the APETALA3 (AP3) and PISTILLATA (PI) MADS box proteins are required to specify petal and stamen identity in the flower (34, 35). Evidence suggests that the proteins form a complex in the cytoplasm and that both contribute sequences necessary for colocalization to the nucleus (19).
An interesting question is why the HD1 protein might localize to the nucleus independently of its HD2 partner. We think that it is likely that HD1 proteins have a transcriptional role that is independent of HD2 proteins. In S. cerevisiae, the corresponding a1–α2 heterodimer plays an essential role in regulating diploid cell-specific gene transcription, but the MATα2 protein binds different DNA target sites in conjunction with the general transcription factor MCM1p to regulate transcription of haploid cell-specific genes (5, 6, 36). Although the HD1 homeodomain of the C. cinereus A proteins is dispensable for heterodimer function, its sequence is highly conserved in heteroallelic and paralogous versions of the proteins (4, 37). This is consistent with an alternative regulatory function analogous to that of the MATα2 protein of S. cerevisiae (8). If HD2 proteins have no independent regulatory role, it would be efficient to localize these to the nucleus only when an appropriate HD1 partner is present, thereby preventing any promiscuous binding to DNA. The A locus of C. cinereus may contain up to three genes encoding HD2 proteins that are undimerized in unmated cells and, even in mated cells, may not all find a compatible dimerization partner.
The acquisition of NLSs by the b2–3 HD2 protein did not overcome the need to choose a compatible HD1 protein partner to activate A regulated development. When the modified b2–3 protein was introduced into a B6 host that contained only the incompatible b1–3 protein as potential partner, A regulated development did not occur. The N-terminal domains that identify compatible dimerization partners are not required for transcriptional regulation (3) but, if undimerized, might cause steric hindrance to DNA binding.
We can now suggest a model to explain how the function of the mating type protein heterodimer may be regulated in C. cinereus and other basidiomycete fungi. Cell fusion between compatible mates brings together two classes of homeodomain proteins that can heterodimerize in the cytoplasm via an N-terminal domain (3, 28, 38). NLSs on the HD1 protein permit the heterodimer to localize to the nucleus. Once in the nucleus, the heterodimer binds specific operator sites to bring about A-regulated changes in gene transcription. The two components of the heterodimer contribute different functional domains (8); on DNA, the HD2 protein provides the critical binding activity and the HD1 protein contributes the activation domain. This model is attractive because it encompasses several different mechanisms that are known to regulate the activity of transcription factors.
Acknowledgments
We thank Vivian Irish for the gift of pRTL2-GUS/Nla and pRTL2-GUS and Laura Rossini for her help with the transient expression assay for GUS activity in onion cells. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) (awarded to L.A.C. and E.J.C.M.). R.H.H. was supported by a Summer Studentship from the Gatsby Charitable Foundation. L.A.C. is a BBSRC Senior Research Fellow.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: NLS, nuclear localization sequence; GUS, β-glucuronidase; HD1 and HD2, homeodomain proteins 1 and 2, respectively.
References
- 1.Casselton L A. In: The Filamentous Fungi, Fungal Development. Smith J E, Berry D R, editors. Vol. 3. London: Edward Arnold; 1978. pp. 275–297. [Google Scholar]
- 2.Kües U, Richardson W V J, Tymon A M, Mutasa E S, Göttgens B, Gaubatz S, Gregoriades A, Casselton L A. Genes Dev. 1992;4:568–577. doi: 10.1101/gad.6.4.568. [DOI] [PubMed] [Google Scholar]
- 3.Banham A H, Asante-Owusu R N, Göttgens B, Kingsnorth C S, Thompson S A J, Mellor E J C, Casselton L A. Plant Cell. 1995;7:773–783. doi: 10.1105/tpc.7.6.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardo E H, O’Shea S F, Casselton L A. Genetics. 1996;144:87–945. doi: 10.1093/genetics/144.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herskowitz I. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson A D. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 7.Goutte C, Johnson A D. J Mol Biol. 1993;233:359–371. doi: 10.1006/jmbi.1993.1517. [DOI] [PubMed] [Google Scholar]
- 8.Asante-Owusu R N, Banham A H, Bönhart H, Mellor E J C, Casselton L A. Gene. 1996;172:25–31. doi: 10.1016/0378-1119(96)00177-1. [DOI] [PubMed] [Google Scholar]
- 9.Tymon A M, Kües U, Richardson W V J, Casselton L A. EMBO J. 1992;11:1805–1816. doi: 10.1002/j.1460-2075.1992.tb05232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kües U, Asante-Owusu R N, Mutasa E S, Tymon A M, Pardo E H, O’Shea S F, Göttgens B, Casselton L A. Plant Cell. 1994;6:1467–1475. doi: 10.1105/tpc.6.10.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binninger D, Skrzynia C, Pukkila P J, Casselton L A. EMBO J. 1987;6:835–840. doi: 10.1002/j.1460-2075.1987.tb04828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casselton L A, de la Fuente Herce A. Curr Genet. 1989;16:35–40. [Google Scholar]
- 13.Kües U, Göttgens B, Richardson W V J, Stratmann R, Casselton L A. EMBO J. 1994;13:4054–4059. doi: 10.1002/j.1460-2075.1994.tb06722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Imai Y, Matsushima Y, Sugimura T, Terada M. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Restropo M A, Freed D D, Carrington J C. Plant Cell. 1990;2:987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kües U, Tymon A M, Richardson W V J, May G, Gieser P T, Casselton L A. Mol Gen Genet. 1994;243:45–52. doi: 10.1007/BF00279749. [DOI] [PubMed] [Google Scholar]
- 18.Swamy S, Uno I, Ishikawa T. J Gen Microbiol. 1984;130:3219–3224. [Google Scholar]
- 19.McGonigle B, Bouhidel K, Irish V F. Genes Dev. 1996;10:1812–1821. doi: 10.1101/gad.10.14.1812. [DOI] [PubMed] [Google Scholar]
- 20.van der Krol A R, Chua N-H. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrington J C, Freed D D, Leinicke A J. Plant Cell. 1991;3:953–962. doi: 10.1105/tpc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein T M, Wolf E D, Wu R, Sanford J C. Nature (London) 1987;327:70–73. [Google Scholar]
- 23.Varagona M J, Schmidt R J, Raikhel N V. Plant Cell. 1992;4:1213–1227. doi: 10.1105/tpc.4.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 25.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Bustos J, Heitman J, Hall M N. Biochim Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- 27.Kronstad J W, Leong S A. Genes Dev. 1990;4:1384–1395. doi: 10.1101/gad.4.8.1384. [DOI] [PubMed] [Google Scholar]
- 28.Kämper J, Reichmann T, Romeis M, Bölker M, Kahmann R. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- 29.Hall M N, Hereford L, Herskowitz I. Cell. 1984;36:1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- 30.Hall M N, Craik C, Hiraoka Y. Proc Natl Acad Sci USA. 1990;87:6954–6958. doi: 10.1073/pnas.87.18.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jans D A. In: Protein Targeting. Hurtley S M, editor. Oxford: IRL; 1996. pp. 25–62. [Google Scholar]
- 32.Sock E, Enderich J, Rosenfeld M G, Wegner M. J Biol Chem. 1996;271:17512–17518. doi: 10.1074/jbc.271.29.17512. [DOI] [PubMed] [Google Scholar]
- 33.Rieckhoff G E, Casares F, Ryoo H D, Abu-Shar M, Mann R S. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 34.Bowman J L, Smyth D R, Meyerowitz E M. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowman J L, Smyth D R, Meyerowitz E M. Development (Cambridge, UK) 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Smith D, Johnson A D. Cell. 1992;68:133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- 37.Casselton L A, Kües U. In: in The Mycota, Growth Differentiation and Sexuality. Wessels J G H, Meinhardt F, editors. I. Berlin: Springer; 1994. pp. 307–321. [Google Scholar]
- 38.Magae O Y, Novotny C, Ullrich R. Biochem Biophys Res Commum. 1995;211:1071–1076. doi: 10.1006/bbrc.1995.1920. [DOI] [PubMed] [Google Scholar]