Abstract
The analysis of results from CAPRI (Critical Assessment of Predicted Interactions), the first communitywide experiment devoted to protein docking, shows that all successful methods consist of multiple stages. The methods belong to three classes: global methods based on fast Fourier transforms or geometric matching, medium range Monte Carlo methods, and the restraint-guided HADDOCK program. Although these classes of methods require very different amounts of information in addition to the structures of component proteins, they all share the same four computational steps: (1) simplified and/or rigid body search; (2) selecting the region(s) of interest; (3) refinement of docked structures; and (4) selecting the best models. While each method is optimal for a specific class of docking problems, combining computational steps from different methods can improve the reliability and accuracy of results.
Introduction
The challenge for predictive protein docking is to start with the coordinates of the unbound component molecules and to obtain computationally a model of the bound complex [1,2]. Protein docking methods have substantially improved during the past few years. This has been demonstrated by the results of CAPRI (Critical Assessment of Predicted Interactions), the first communitywide experiment devoted to protein docking [3]. In 16 rounds of CAPRI up to 63 participating groups tested their methods in blind predictions of 37 target protein-protein complexes. The predictions were grouped into highly accurate, medium accuracy, acceptable, and incorrect categories on the basis of the fraction of native contacts, the backbone root mean square deviation of the ligand (L_RMS) from the reference ligand structure after superimposing the receptor structures, and the backbone RMSD of the interface residues (I_RMS). The calculation of these measures and the exact definitions of categories are given in the first CAPRI evaluation paper [4]; here we note only that for the highly accurate, medium accuracy, acceptable, and incorrect models the ligand RMSD is given by L_RMS < 1Å, 1Å < L_RMS < 5Å, 5Å < L_RMS < 10Å, and L_RMS > 10Å, respectively. Each participating group was entitled to submit ten predictions for each target. The assessors considered all ten models, and the results for each group include the number of predictions in each of the four categories.
Results for CAPRI rounds 1–11 with 28 targets have been “officially” evaluated [4–6]. Two targets (22 and 23) were cancelled due to the early release of X-ray structures. Table 1 shows the summary of results for the six groups that submitted acceptable or better predictions for at least 10 targets. The table was obtained by summing the results for each group from the three separate CAPRI evaluation meetings [4–6]. The numbers of medium and high accuracy models submitted by these groups are also shown. We note that the Bonvin group also performed extremely well in rounds 3–11 using the HADDOCK program [15], but they did not participate in the first two rounds, and hence their overall score was lower than that of the six groups listed in Table 1.
TABLE 1.
Best performing predictor groups in Rounds 1–11 of CAPRI (26 evaluated targets)
| Predictor group | Approach | Program(s) | Assessment |
||
|---|---|---|---|---|---|
| * | ** | *** | |||
| Weng [7, 21, 35] Boston University, currently UMASS School of Medicine |
FFT search with detailed scoring function including pairwise potentials in the newest version of ZDOCK, refinement with local minimization (RDOCK), and re-scoring with a global potential (ZRANK). |
ZDOCK RDOCK ZRANK |
14 | 7 | 3 |
| Vajda & Camacho [8, 9, 20, 32, 34] Boston University |
FFT search with detailed scoring function including pairwise potentials (PIPER), clustering (ClusPro), refinement using the off-grid stochastic global minimization method SDU, and tests for the “stability” of the minima. |
PIPER SDU ClusPro |
12 | 5 | 4 |
| Abagyan [10, 22] Scripps Institute for Medical Research |
Rigid body pseudo-Brownian Monte Carlo minimization with a grid-based energy function, Monte Carlo refinement of selected conformations with flexible interface side chains. |
ICM-DISCO | 11 | 6 | 3 |
| Baker [11, 23, 24] Univ. of Washington |
Rigid body Monte-Carlo minimization using simplified protein geometry and scoring function, followed by all-atom refinement of low energy clusters with iterative repacking of side chains and possibly adjusting the conformation of backbone segments. |
RosettaDock | 11 | 4 | 5 |
| Wolfson & Nussinov [12, 13, 18, 40] Tel Aviv University |
Geometric docking with matching of local shape features and geometric hashing. FlexDock handles docking with hinge bending on one of the molecules. FireDock performs fast refinement with side chain adjustement. |
PatchDock FlexDock FireDock |
11 | 3 | 1 |
| Eisenstein [14] Weizmann Institute |
FFT with a weighted shape complementarity target function; clustering of good solutions, filtering using a priori information, and small local rigid rotations around selected conformations. |
MolFit | 10 | 1 | 4 |
The number of CAPRI targets, and more generally, the number of test cases for protein-protein docking [16] are rather small for drawing conclusions on a statistically significant basis. Nevertheless, the results so far suggest three observations. First, according to Table 1, all successful methods consist of multiple stages. Second, these methods belong to the three general classes shown in Table 2 that primarily differ in terms of the information which is required in addition to the structures of the component proteins. Third, although the CAPRI rules allow submitting ten models for each target, even the best methods have only about 50% success rate, and thus at this point it is advisable to incorporate experimental information to improve the reliability of predictions.
TABLE 2.
Classification of docking methods based on the level of a priori information
| Method | Properties |
Examples | |
|---|---|---|---|
| Search | Examples | ||
| Global methods based on Fast Fourier transform (FFT) or geometric hashing |
Global systematic search |
Minimal; smooth potential |
ZDOCK [7], PIPER [20] PatchDock [12] MolFit [14] |
| Medium range methods based on Monte Carlo minimization |
Limited region, stochastic search |
Moderate, mostly side chains, some loops |
RosettaDock [23] ICM-DISCO [22] |
| Restraint-based docking |
Specified by a priori information on interface residues |
Can be substantial | HADDOCK [25] |
Although the results from each group in CAPRI seem quite close to each other, we note that the three classes of methods are most successful for different classes of problems. The global methods were generally the best for those CAPRI targets where neither of the components proteins underwent conformational change of more than 2 Å, particularly if no a priori information on the complex was available, requiring the search of the entire conformational space. The medium-range methods, particularly RosettaDock [11], yielded excellent results for a number of target for which side chain repacking was crucial, e.g., when one the component protein structures was a homology model. HADDOCK produced the best results if sufficient information on a number of the interface residues was available, even when the binding caused large conformational change, possibly including the backbone [15]. Independent of the method, docking is relatively easy for enzyme-inhibitor complexes that usually can be determined with reasonable accuracy, possibly within a few alternative structures [17]. Results are less predictable for antigen-antibody pairs, and are generally poor for small signaling complexes of weakly interacting proteins [17]. The most difficult targets are the transient complexes that have a large interface area and are subject to substantial conformational change. While HADDOCK was able to generate meaningful models in some cases, no acceptable predictions were submitted for several CAPRI targets of this type [4–6].
In this review we focus on two issues. First, we show that although the three classes of methods in Table 2 use the additional information in very different ways, the main computational steps are common and rather similar in essentially all docking methods. Second, we argue that each class of methods is optimal for a specific class of docking problems, but the reliability of results can be further improved by combining computational steps from different methods.
Multistage approach to docking
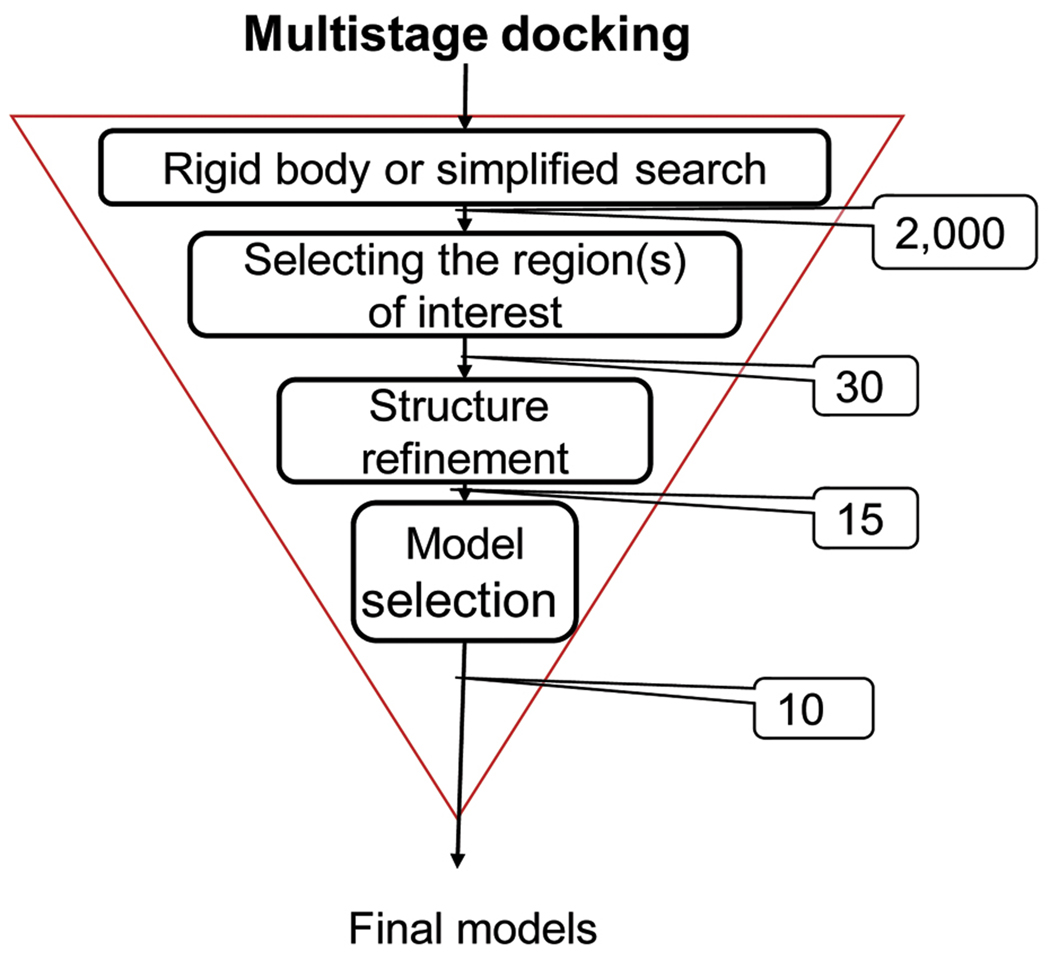
The four steps that seem to occur in most docking algorithms are as follows: (1) simplified and/or rigid body search; (2) selecting the region(s) of interest; (3) refinement of docked structures; and (4) global discrimination, i.e., selecting the best models. Figure 1 shows these steps and the typical number of conformations retained in each step for the most general case which starts with a global search for the orientation and position of one component protein relative to the other. However, as shown in Table 2, the search may be restricted to a region of the conformational space, simplifying the selection of structures in Steps 2 and 4.
Figure 1.
Computational steps in multistage docking for the most general case, which includes a global search over the entire rotational/translational space for the orientation and position of one component protein relative to the other. Although all methods start with a rigid or simplified search, in medium-range and restraint-guided methods the search is restricted to a region of the conformational space, simplifying the selection of the region of interest, as well as the final model selection.
Rigid body and/or simplified geometry searches
Due to computational constraints, truly global searches over the entire rotational/translational space are routinely carried out only by rigid body methods that use either fast Fourier transforms (FFT) [7, 8] or geometric matching [12,18]. FFT based methods systematically evaluate billions of docked conformations on a grid using correlation-type scoring functions. The original scoring function, based only on shape complementarity [19], has been expanded to include electrostatic and solvation terms, and more recently structure-based interaction potentials [20, 21], substantially improving the accuracy of the method. In all scoring functions the shape complementarity term allows for overlaps, thereby accounting for the differences between bound and unbound (separately crystallized) structures.
ICM-DISCO [22] and Rosetta Dock [23] start with rigid body Monte Carlo minimization runs in the rotational/translational space from random initial structures around the known or hypothetical receptor binding site, and thus generally explore only certain regions of the conformational space. In the first stage of RosettaDock the proteins are represented as backbones plus side chain centroids, and the search is guided by a residue-scale interaction potential. Benefiting from the simplified protein representation, the method was recently extended to account for loop flexibility [24]. However, due to the increased computational burden loop search was feasible only in local rather than global docking [24], further restricting the search region. HADDOCK (High Ambiguity Driven biomolecular DOCKing) starts with rigid body energy minimization from completely random initial states, typically retaining 1000 complex structures [25]. However, HADDOCK utilizes extra information in the form of a number of active residues (which are supposed to be part of the interface) and passive residues (surface neighbors of active residues). Ambiguous interaction restraints are defined between any atom of the active residues and all atoms of active and passive residues on the partner protein. The interaction restraints are incorporated into the scoring function and guide the search toward regions of the conformational space in which the restraints are satisfied.
HADDOCK applications generally involve ambiguous interaction restraints based on 10 to 25 active residues on the two sides of the interface. These residues may be selected using biochemical and/or biophysical information such as chemical shift perturbation data or the results of mutagenesis experiments, but predicted interface residues were also used for some of the CAPRI targets [15].
Selecting the region(s) of interest
Since the rigid body searches rely on “soft” scoring functions that allow for overlaps, the accuracy is limited. The refinement of structures requires some level of protein flexibility, and due to the higher computational costs the number of structures must be reduced. ICM and HADDOCK simply retain a few hundred low energy conformations. In RosettaDock generally the centers of low energy clusters are selected [23]. In the web-based docking server ClusPro [26] we cluster the low energy conformations and rank the clusters according to their size [27]. Lorenzen and Zhang [28] compared the performance of clustering algorithms for selecting near-native docking conformations among structures generated by four FFT-based protein–protein docking methods and showed that although the performance of clustering depends on the quality and structural distribution of the decoys, the ranking based on clustering is better than that by the inherent scoring functions. Large scale docking studies by Vakser and co-workers have shown that the number of distinct energy basins is generally small and correlated with known binding modes [29].
Refinement of docked structures
In ICM-DISCO the retained solutions are further optimized with flexible interface ligand side chains using a biased probability Monte Carlo procedure [22]. In RosettaDock the Monte Carlo minimization in translational and rotational coordinates is integrated with repacking the interface side chains using a backbone-dependent rotamer library. More recently the method has been extended to dock proteins with backbone conformational changes by combining the rigid-body search with modeling of some variable loops [11, 24]. Although the method gave excellent results for some CAPRI targets, the search had to be restricted to even smaller regions of the rotational/translational space, and this was a disadvantage in others [11]. Chaudhury and Gray [30] also improved the ability of RosettaDock to consider backbone flexibility by adding ensemble docking and induced fit capabilities.
After global docking one may have to refine structures in many clusters, and hence efficiency is important. The FireDock refinement algorithm shows that it is enough to remove a few side chain clashes to substantially improve ranking in rigid body docking [13]. SDU (Semi-Definite programming based Underestimation), an efficient stochastic optimization algorithm [31], is based on the assumption that the free energy is a funnel-like function within the region defined by each cluster [32]. In HADDOCK the refinement starts with simulated annealing procedures that allow the interface side chains and the backbone to move, and proceeds with energy minimization and molecular dynamics simulations in a shell of TIP3P water molecules [25].
Final model selection
Since the accuracy of energy functions is limited, selecting the best predictions is not at all trivial, and it is not always clear how the individual predictors select the 10 models for CAPRI submission. It appears that the lowest energy structures are selected from the ICM-DISCO runs. In RosettaDock and HADDOCK, clusters of low energy structures are chosen. Following the refinement by SDU we rank the clusters according to the energies of their lowest energy structures [32]. However, the energy function is not globally discriminatory in any of these methods. This is generally not a major problem in HADDOCK, because the search is restrained by the additional information, but improvement is needed in the other two classes of methods. Analyzing structures generated by semi-global runs of RosettaDock, London and Schueler-Furman [33] applied a machine learning algorithm to distinguish ensembles of low-energy conformations around the native conformation from other low-energy ensembles. By applying recursive feature elimination, the starting 42 features were reduced to seven, all with well defined biophysical interpretation, thereby reducing the possibility of overfitting, a serious problem for machine learning methods. The resulting classifier, FunHunt, identified the native orientation in 50/52 protein complexes in a test set, and showed that the energy decrease of trajectories toward near-native orientations is significantly larger than for other orientations.
Strengths of the methods and of their combinations
Each class of methods in Table 2 has its specific strengths and limitations. In principle, global methods can be used without any information beyond the structures of the component proteins. However, at present experimental information is generally required for reliable docking results. The usage pattern of the docking server ClusPro proves this point [26]. Since its release in December 2004, the server has performed close to 20,000 docking calculations for more than 2000 users, resulting in over 100 publications. In most studies the server was used to generate putative complex conformations, and the best models were selected and validated using a variety of experimental techniques, including site-directed mutagenesis, cross-linking, FRET, enzymatic proteolysis, or radiolytic protein footprinting. The advantage of this approach is that the putative models can be effectively used to design the most appropriate validation experiments. To emphasize the importance of such validation we note that, due to the rigid body step, global methods tend to fail if the bound and unbound protein structures substantially differ, whereas selecting final models for weak complexes is very unreliable.
Side chains can be relatively easily adjusted within the Monte Carlo steps of the translational/rotational search. As implemented in ICM-DISCO, and particularly in RosettaDock, the repacking of side chains allows for some conformational change upon binding and improves the accuracy of models. According to the CAPRI results, both programs generated respectable predictions for a few targets that were beyond the scope of the FFT-based methods. However, the CAPRI results also showed that without any information on the binding mode the search may be performed in a wrong region of the conformational space. The success of extending RosettaDock for dealing with backbone flexibility so far appears to be inconclusive, as the increased degrees of freedom tends to increase the number of false positive predictions, and due to the increased computational efforts the search becomes even more local [11]. HADDOCK is the ideal docking method if substantial and reliable information is available from mutagenesis, mass spectrometry, or NMR. With appropriate restraints, the program can provide good results even for proteins with substantial change in side chain and backbone conformations. However, restraints based on incorrect information are likely to lead to incorrect structures. Nevertheless, in the latest rounds of CAPRI, HADDOCK was able to yield good results for several targets using the information which was available in the literature, and neural network based predictions of the interface residues [15].
The accuracy and reliability of docking results can be improved by combining different classes of methods. For example, we have studied the 30 clusters generated by FFT-based docking by starting RosettaDock runs from random points around the cluster centers, and observing whether a certain fraction of trajectories converge to a small region within the cluster [34]. A cluster was considered stable if such a strong attractor existed and contained a low energy structure. It was shown that all clusters close to the native structure are stable, and that restricting considerations to stable clusters eliminates around half of the false positives. In similar spirit, Pierce and Weng [35] refined global docking predictions from ZDOCK using RosettaDock, and selected the best models based on their ZRANK score. Refining docking benchmark predictions from ZDOCK led to improved structures of top ranked hits in 20 of 27 cases, and an increase from 23 to 27 cases with hits in the top 20 predictions. In addition, the ZRANK energy function was optimized using the refined models. With the new energy function, the numbers of cases with hits ranked at number one increased from 12 to 19 and from 7 to 15 for two different ZDOCK versions. These results show that combinations of independently developed docking protocols (ZDOCK/ZRANK and RosettaDock) can substantially improve protein docking results.
Lorenzen and Zhang [36] refined initial docking estimates of protein complex structures, generated by an FFT-based method, using a Monte Carlo approach including rigid-body moves and side-chain optimization. During the simulation they gradually shifted from a smoothed van der Waals potential, which prevented trapping in local energy minima, to the standard Lennard-Jones potential. Following the simulation, the conformations were clustered to obtain the final predictions. The refinement procedure was able to generate near-native structures (interface RMSD <2.5 Å) as first model in 14 of 59 cases in the benchmark set. More generally, improving model accuracy using Monte Carlo methods enables the use of potentials that are more accurate but also more sensitive to structural errors. It may also be useful to combine HADDOCK with Monte Carlo methods if the extra information used is not fully reliable. In such cases the dependence of results on the restraints was tested by randomly removing 25% of data in docking trials [15]. Since RosettaDock has a highly accurate scoring function (at least in a neighborhood of the native state) and performs complete repacking of side chains, it may be less biased to generate candidate models using HADDOCK, and to explore the “stability’’ of these models by Monte Carlo simulations without any restraints.
Conclusions
The analysis of docking predictions for the 28 CAPRI targets evaluated so far shows that similar success rates have been achieved by three classes of methods that require very different amount of information in addition to the structures of the component proteins. In spite of this substantial difference, all methods include very similar computational steps. However, each method is optimal for a specific class of docking problems. Global methods can provide valid predictions without any additional information, although experimental validation is highly recommended even in this case. Due to the repacking of side chains, Monte Carlo based methods can yield highly accurate models, but the search is restricted to a neighborhood of the starting structures. Finally, HADDOCK is the ideal method if substantial and reliable interaction information is available to guide the search. We suggest that reliable docking results can be obtained for a broader class of problems by combining computational steps from different methods. With increasing computing power such combined approaches become increasingly feasible, and can more efficiently utilize the information from a given set of experimental data.
At present the major unsolved problem in docking is the treatment of proteins with substantial backbone conformational change. In spite of attempts to introduce loop prediction and backbone adjustment in Monte Carlo based methods, it appears that the most successful method is still HADDOCK, of course assuming that appropriate interaction information is available. In order to avoid futile attempts for docking flexible proteins using rigid methods, it is necessary to develop methods that can predict protein flexibility [37–39]. In addition, predicting the hinge regions in one of the component proteins [40] enables the use of special methods such as FlexDock that deals with localized backbone flexibility by docking the rigid parts of the flexible molecule, and builds consistent configurations of the entire protein from these candidate parts [13].
Acknowledgments
This work has been supported by grant GM061867 from the National Institutes of Health
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Ritchie DW. Recent progress and future directions in protein-protein docking. Curr Protein Pept Sci. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- 2.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Principles of flexible protein-protein docking. Proteins. 2008;73:271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, Vakser I, Wodak SJ. CAPRI: A Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 4.Mendez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein–protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 5.Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 6. Lensink MF, Méndez R, Wodak SJ. Docking and scoring protein complexes: CAPRI. Proteins. (3rd Edition) 2007;69:704–718. doi: 10.1002/prot.21804. The official assessment of rounds 6–12 of CAPRI quantifies the achievements of the challenge, summarizes the methods tested and concludes that the docking field is making steady progress. The paper also gives an overview of the current state of the art in protein-protein docking.
- 7. Wiehe K, Pierce B, Tong WW, Hwang H, Mintseris J, Weng Z. The performance of ZDOCK and ZRANK in rounds 6–11 of CAPRI. Proteins. 2007;69:719–725. doi: 10.1002/prot.21747. Experience of the group that was the best overall performer in rounds 1–11 of the CAPRI docking experiment. The newest version of the ZDOCK program includes a pairwise potential in its scoring function, and ZRANK is a new development.
- 8. Shen Y, Brenke R, Kozakov D, Comeau SR, Beglov D, Vajda S. Docking with PIPER and refinement with SDU in rounds 6–11 of CAPRI. Proteins. 2007;69:734–742. doi: 10.1002/prot.21754. The paper describes PIPER, the first FFT-based docking program with a scoring function which includes a pairwise interaction potential, and SDU, a refinement program which takes into account the funnel-like behavior of the free energy function in a neighborhood of the native state. The group was the second best overall performer in rounds 1–11 of the CAPRI docking experiment.
- 9.Comeau SR, Kozakov D, Brenke R, Shen Y, Beglov D, Vajda S. ClusPro: Performance in CAPRI rounds 6–11 and the new server. Proteins. 2007;69:781–785. doi: 10.1002/prot.21795. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Recio J, Abagyan R, Totrov M. Improving CAPRI predictions: Optimized desolvation for rigid-body docking. Proteins. 2005;60:308–313. doi: 10.1002/prot.20575. [DOI] [PubMed] [Google Scholar]
- 11. Wang C, Schueler-Furman O, Andre I, London N, Fleishman SJ, Bradley P, Qian B, Baker D. RosettaDock in CAPRI rounds 6–12. Proteins. 2007;69:758–763. doi: 10.1002/prot.21684. Describes the extension of RosettaDock to account for the flexibility of backbone in loop regions, and demonstrates that the method improves docking results for some of the CAPRI targets. However, the need for more extensive computations further reduces the region of the conformational space which can be explored, and hence may reduce the quality of results.
- 12. Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Automatic prediction of protein interactions with large scale motion. Proteins. 2007;69:764–773. doi: 10.1002/prot.21759. Describes the web-based implementation of the FlexDock geometric docking algorithm which is capable of accounting for hinges in the flexible molecule. The algorithm is highly efficient and yielded the best performing server in CAPRI rounds 6–12.
- 13.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalsman N, Eisenstein M. Inherent limitations in protein-protein docking procedures. Bioinformatics. 2007;23:421–426. doi: 10.1093/bioinformatics/btl524. [DOI] [PubMed] [Google Scholar]
- 15. de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. HADDOCK (High Ambiguity Driven biomolecular DOCKing) is a relative newcomer in CAPRI, but performed extremely well when participating, in spite of using restraints based on literature data and the predictions of interface residues, rather than restraints based on experiments.
- 16. Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. This is the third installment of the benchmark set that became the standard in protein-protein docking and facilitates the comparison of docking methods. The 124 unbound-unbound test cases in Benchmark 3.0 are classified into 88 rigid-body cases, 19 medium-difficulty cases, and 17 difficult cases, based on the degree of conformational change at the interface upon complex formation. This is 48% increase relative to Benchmark 2.0. However, some of the binary complexes are actually parts of larger multiprotein structures. Such cases may not properly test the performance of docking algorithms, and hence the benchmark set needs further analysis by the predictor groups.
- 17.Vajda S. Classification of protein complexes based on docking difficulty. Proteins. 2005;60:176–180. doi: 10.1002/prot.20554. [DOI] [PubMed] [Google Scholar]
- 18.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Geometry-based flexible and symmetric protein docking. Proteins. 2005;60:224–231. doi: 10.1002/prot.20562. [DOI] [PubMed] [Google Scholar]
- 19.E. Katchalski-Katzir E, I. Shariv I, M. Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. PIPER is the first FFT-based docking program with a scoring function which includes a structure-based interaction potential. The corresponding energy term is converted to a sum a few correlation functions using the eigenvalue-eigenvector decomposition of the matrix of interaction energy coefficients, which enables energy evaluation by the fast Fourier transform (FFT) correlation approach.
- 21. Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z. Integrating statistical pair potentials into protein complex prediction. Proteins. 69:511–520. doi: 10.1002/prot.21502. Describes the current version of the successful ZDOCK algorithm with a new scoring function which includes a structure-based interaction potential. It uses an optimized alphabet of 12 atom types, but is still less efficient than the eigenvalue-eigenvector decomposition used in PIPER.
- 22.Fernandez-Recio J, Totrov M, Abagyan R. ICM-DISCO docking by global energy optimization with fully flexible side-chains. Proteins. 2003;52:113–117. doi: 10.1002/prot.10383. [DOI] [PubMed] [Google Scholar]
- 23. Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. This is the main publication describing the successful RosettaDock docking method based on a Monte Carlo minimization algorithm. Due to the detailed and fairly accurate scoring function and the repacking of interface side chains, RosettaDock can achieve remarkable accuracy, and set a new standard in protein-protein docking for cases in which the information on the binding mode helped to chose appropriate starting conformations for the complex.
- 24.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 25.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a protein-protein docking approach based on biochemical and/or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 26. Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. Describes the first web-based protein-protein docking server. Since its release in December 2004, the server has performed close to 20,000 docking calculations for more than 2000 users, resulting in over 100 publications.
- 27.Kozakov D, Clodfelter KH, Vajda S, Camacho CJ. Optimal clustering for detecting near-native conformations in protein docking. Biophys J. 2005;89:867–875. doi: 10.1529/biophysj.104.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lorenzen S, Zhang Y. Identification of near-native structures by clustering protein docking conformations. Proteins. 2007;68:187–194. doi: 10.1002/prot.21442. (2007) Comparison of clustering algorithms for picking up near-native docking conformations generated by four FFT-based protein-protein docking methods. It shows that the ranking based on clustering is better than that by the inherent scoring functions.
- 29. O’Toole N, Vakser IA. Large-scale characteristics of the energy landscape in protein-protein interactions. Proteins. 2008;71:144–152. doi: 10.1002/prot.21665. Large scale docking studies show that the number of distinct energy basins is generally small and correlated with the known binding modes, which provides the conceptual basis for retaining a moderate number of the largest clusters for refinement.
- 30.Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J Mol Biol. 2008;381:1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paschalidis ICh, Shen Y, Vakili P, Vajda SSDU. A Semi-definite programming-based underestimation method for global optimization in molecular docking. IEEE Transactions for Aut Control. 2007;52:664–676. doi: 10.1109/TAC.2007.894518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen Y, Paschalidis ICh, Vakili P, Vajda S. Protein docking by the underestimation of free energy funnels in the space of encounter complexes. PLoS Comput Biol. 2008;4:e1000191. doi: 10.1371/journal.pcbi.1000191. Given a set of local minima, SDU constructs an underestimating function, select new points the conformational space close to the minimum of the underestimator as starting points for local minimization, updates the set of local minima, and iterates these steps until convergence.
- 33. London N, Schueler-Furman O. Funnel hunting in a rough terrain: learning and discriminating native energy funnels. Structure. 2008;16:269–279. doi: 10.1016/j.str.2007.11.013. A machine learning algorithm was used to distinguish ensembles of low-energy conformations around the native conformation from other low-energy ensembles. The resulting classifier, FunHunt, identified the native orientation in 50/52 protein complexes in a test set, and showed that the energy decrease of trajectories toward near-native orientations is significantly larger than for other orientations.
- 34. Kozakov D, Schueler-Furman O, Vajda S. Discrimination of near-native structures in protein-protein docking by testing the stability of local minima. Proteins. 2008;72:993–1004. doi: 10.1002/prot.21997. Since structures at narrow minima loose more entropy, some of the nonnative states can be detected by determining whether or not a local minimum is surrounded by a broad region of attraction on the energy surface. The analysis is based on starting Monte Carlo Minimization (MCM) runs from random points around each minimum, and observing whether a certain fraction of trajectories converge to a small region within the cluster. It was shown that using global docking, dividing the conformational space into clusters and determining the stability each cluster, the combined approach is less dependent on a priori information than exploring the potential conformational space by Monte Carlo minimizations alone.
- 35. Pierce B, Weng Z. A combination of rescoring and refinement significantly improves protein docking performance. Proteins. 2008;72:270–279. doi: 10.1002/prot.21920. Shows that refining global docking predictions from ZDOCK using RosettaDock, and selecting the best models based on their ZRANK score substantially improves model accuracy.
- 36.Lorenzen S, Zhang Y. Monte Carlo refinement of rigid-body protein docking structures with backbone displacement and side-chain optimization. Protein Sci. 2007;16:2716–2725. doi: 10.1110/ps.072847207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. J Am Chem Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 38.Garzón JI, Kovacs J, Abagyan R, Chacón P. DFprot: a webtool for predicting local chain deformability. Bioinformatics. 2007;23:901–902. doi: 10.1093/bioinformatics/btm014. [DOI] [PubMed] [Google Scholar]
- 39.Dobbins SE, Lesk VI, Sternberg MJ. Insights into protein flexibility: The relationship between normal modes and conformational change upon protein-protein docking. Proc Natl Acad Sci USA. 2008;105:10390–10395. doi: 10.1073/pnas.0802496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: automated prediction of hinges in protein structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]