Abstract
Objective
Megakaryocytic cells (Mks) undergo endomitosis and become polyploid. Mk ploidy correlates with platelet production. We previously showed that nicotinamide (NIC) greatly increases Mk ploidy in cultures of human mobilized peripheral blood (mPB) CD34+ cells. This study aims to examine the generality of NIC effects, NIC’s impact on Mk ultrastructure, and potential mechanisms for the increased ploidy.
Methods
We used electron microscopy to examine Mk ultrastructure and flow cytometry to evaluate NIC effects on Mk differentiation and ploidy in mPB CD34+ cell cultures under diverse megakaryopoietic conditions. Mk ploidy and NAD(H) content were evaluated for NIC and other NAD+ precursors. We tested additional inhibitors of the SIRT1 and SIRT2 histone/protein deacetylases and, after treatment with NIC, evaluated changes in the acetylation of SIRT1/2 targets.
Results
NIC increased ploidy under diverse culture conditions and did not alter Mk ultrastructure. 6.25 mM NIC increased NAD+ levels 5-fold. Quinolinic acid increased NAD+ similar to that for 1 mM NIC, but yielded a much smaller ploidy increase. Similar increases in Mk ploidy were obtained using NIC or the SIRT1/2 inhibitor cambinol, while the SIRT2 inhibitor AGK2 moderately increased ploidy. SIRT1/2 inhibition in cells treated with NIC was evidenced by increased acetylation of nucleosomes and p53. Greater p53 acetylation with NIC was associated with increased binding of p53 to its consensus DNA binding sequence.
Conclusion
NIC greatly increases Mk ploidy under a wide range of conditions without altering Mk morphology. Inhibition of SIRT1 and/or SIRT2 is primarily responsible for NIC effects on Mk maturation.
Keywords: polyploidization, megakaryopoiesis, histone deacetylase, thrombopoiesis, SIRT1, SIRT2, NAD(H), p53
As the precursors to platelets, megakaryocytic cells (Mks) are central to hemostasis. Mk differentiation includes several rounds of endomitosis to form polyploid cells and the formation of cytoplasmic extensions called proplatelets. However, Mk commitment and the mechanisms through which Mks differentiate and mature remain poorly understood. Ex vivo culture of hematopoietic stem and progenitor cells (HSPCs) under conditions that promote Mk commitment, expansion, and maturation would enable the production of progenitors and mature Mks for transplantation therapies to offset thrombocytopenia associated with HSPC transplants following high-dose chemotherapy [1, 2].
CD34+ HSPCs cultured with thrombopoietin (Tpo) yield a high purity of CD41+ Mks [3-5]. However, the ploidy - and the potential for platelet production [6, 7] - of human Mks produced in culture is much lower than that observed in vivo. We previously reported that nicotinamide (NIC) - one form of niacin (vitamin B3) - greatly increases Mk ploidy and proplatelet production in cultures of human mobilized peripheral blood (mPB) CD34+ cells stimulated with Tpo [8]. Evaluating the conditions under which NIC increases Mk ploidy and identifying the mechanism(s) that underlie the effects of NIC would lead to a greater understanding of Mk differentiation and may allow further modulation of Mk maturation.
Nicotinamide has diverse functions in cells. NIC inhibits the activity of the silent information regulator 2 (Sir2) family of histone/protein deacetylases (sirtuins or SIRTs) [9-12]. Sirtuins catalyze a unique NAD+-dependent deacetylation reaction and are important for a wide variety of biological processes including transcriptional silencing, lifespan regulation, and the regulation of apoptosis [13-15]. NIC is also a precursor of NAD+ via the salvage pathway [16, 17]. NAD+ regulates a variety of intracellular activities such as the DNA-binding specificity of p53 [18, 19]. We have previously established that the p53-mediated Mk apoptotic program is intimately linked with terminal maturation and polyploidization, possibly through the downstream effects of p53 on MDM2 and BCL2 expression [20, 21].
The aim of this study was to examine the generality of the NIC effects, NIC’s potential impact on Mk ultrastructure, and possible mechanisms for the profound effect of NIC on Mk ploidy.
Materials and Methods
Unless noted, all reagents were obtained from Sigma-Aldrich (St. Louis, MO).
Human Mk culture
Cultures were initiated in T-flasks with previously frozen mPB CD34-selected cells (AllCells; Berkeley, CA or Fred Hutchinson Cancer Research Center; Seattle, WA) and maintained at a concentration of 100,000 to 300,000 cells/ml as described [8]. Most cultures were supplemented with 100 ng/mL Tpo (Peprotech; Rocky Hill, NJ or Genentech; South San Francisco, CA) and treated with 6.25 mM NIC beginning at day 5 (Tpo + NIC). Selected cultures were also supplemented with 100 ng/mL stem cell factor (SCF, R&D Systems; Minneapolis, MN) or were initiated in 12-well plates coated with fibronectin. For cultures containing Tpo plus 150 ng/mL stromal-derived factor-1α (SDF-1α, R&D Systems), NIC was added beginning at day 2. Cytokine cocktail cultures were supplemented with 1.5 ng/mL interleukin-3 (IL-3, Peprotech), 10 ng/mL IL-6 (Peprotech), and 50 ng/mL SCF. On day 5, the cells were treated with NIC and/or Tpo to generate 4 cultures: cocktail (IL-3, IL-6, SCF), cocktail + 6.25 mM NIC, cocktail + 100 ng/mL Tpo, or cocktail + Tpo + NIC. NAD+ precursors nicotinic acid and quinolinic acid were added on day 5 to cells cultured with Tpo. The SIRT inhibitors cambinol (generously provided by A. Bedalov; Fred Hutchinson Cancer Research Center or purchased from Calbiochem; San Diego, CA) and AGK2 (ChemDiv; San Diego, CA) were added on day 5 to cells cultured with Tpo. The average cell and nuclear volumes were determined by analyzing whole cells and nuclei (released using 3% cetrimide), respectively, using a Multisizer 3 (Beckman Coulter, Fullerton, CA).
Electron microscopy
Cells from Tpo only and Tpo + NIC cultures were removed on day 8 and transferred to fibronectin-coated 35-mm petri dishes (BD Biosciences; San Jose, CA) to induce cell adhesion. After 48 h, cells were fixed for 4 h (2.5% glutaraldehyde/2.5% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4) and processed by the NU Cell Imaging Facility. Briefly, after embedding the cells in Epoxy resin in an inverted capsular mold, ultra-thin sections were cut, stained with uranyl acetate and lead citrate, and examined with a JEOL-1220 transmission electron microscope at an accelerating voltage of 60 kV.
Flow cytometric detection of surface antigens, ploidy, and intracellular proteins
For CD41 expression, Mk apoptosis, and Mk ploidy, cells were prepared for flow cytometry and analyzed as described [8].
For intracellular detection of total and acetylated p53, cells were stained with FITC-conjugated anti-CD41 antibody, fixed with 2% paraformaldehyde, permeabilized with 70% methanol, stained with PE-conjugated-anti-p53 (Santa Cruz Biotechnology, Santa Cruz, CA) and Alexa Fluor 647-conjugated-anti-p53AcK382 (BD Biosciences) antibody, treated with RNase, and stained with 7-AAD. Expression of p53 and p53AcK382 proteins was quantified by subtracting the mean fluorescence intensity (MFI) of unstained CD41+ cells from the MFI of the p53-labeled and p53AcK382-labeled CD41+ cells. A FACSAria flow cytometer and FACSDiva software (BD Biosciences) were used for data acquisition and analysis.
Measurement of intracellular NAD(H) levels
300,000 cells were extracted in 0.6 mL of slightly basic extraction buffer containing 10 mM nicotinamide and 0.05% (w/v) Triton X-100, as described by Wagner and Scott [22]. Standard solutions of NAD+ and NADH were prepared in extraction buffer. To determine NAD(H) (sum of NAD+ and NADH) content, the extracts were subjected to an enzymatic cycling reaction [23] performed in a 96-well plate using a reagent mixture containing 0.1 M bicine, 0.5 M ethanol, 4.17 mM EDTA, 0.83 mg/mL BSA, 0.42 mM MTT, and 1.66 mM phenazine ethosulfate. The reaction was started by adding 2 units of yeast alcohol dehydrogenase. Color was developed in the dark for 30 min and detected at 570 nm. Each sample was run in duplicate. The NADH content was separately determined, after incubating extract samples at 60°C for 30 min to degrade NAD+.
Western analysis
Nuclear lysates were prepared according to the manufacturer’s protocol (NE-PER Kit; Pierce Biotechnology; Rockford, IL). Thirty micrograms of total protein per sample was denatured, separated by SDS-PAGE Ready-Gel (BioRad; Hercules, CA), and transferred onto nitrocellulose membranes (BioRad). Membranes were blocked in non-fat dry milk for 1 h and incubated overnight at 4°C with primary antibody for acetylated lysine residues (Cell Signaling), Nmnat1 (Abcam), or AcK382p53 (Cell Signaling). After washing, membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse- or anti-rabbit-IgG antibody (Cell Signaling). Bound antibodies were detected using chemiluminescence. The membranes were then stripped and reprobed with antibodies for nucleosomes (Chemicon), actin (Abcam), or total p53 (Santa Cruz Biotechnology). Secondary antibodies were applied and detection was performed as described above. Densitometry was performed using ImageQuant 5.2 software (GE Healthcare).
Electrophoretic mobility shift assays (EMSAs)
Nuclear lysates were used for EMSAs as described [20].
Results
NIC increases Mk maturation without altering normal Mk ultrastructure
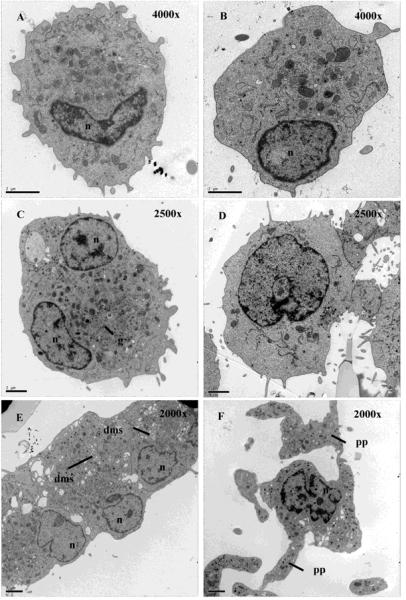
The ultrastructure of Mks cultured with Tpo (Fig 1A,B) was similar to that of Mks cultured with Tpo + NIC (Fig 1C,D). Alpha-granules, dense granules, and mitochondria were visible in cells from both conditions. However, NIC-treated cells were larger and generally had a more complex nuclear structure, both of which are indicative of greater Mk maturation. NIC-treated cells also showed evidence of a demarcation membrane system (Fig 1E). NIC-treated Mks had a greater number of short cytoplasmic projections and generally had a larger number of small cytoplasmic microparticles surrounding each cell. Proplatelets were present on some cells cultured with Tpo + NIC (Fig 1F), and the morphology of the proplatelets was similar to that obtained by Cramer et al. [24]. Thus, NIC-treated Mks exhibit normal ultrastructure.
Figure 1.
Mks cultured with NIC exhibit normal ultrastructure. Transmission electron microscopy of Mks cultured with Tpo only (A-B) and Tpo + NIC (C-F) at day 8. The images in A-D were acquired such that the magnification gave one cell per field of view. Due to the larger cell size with Tpo + NIC, it was necessary to use a lower magnification (2500x) compared to that for Tpo only (4000x). (E) An image of 3 cells exhibiting a demarcation membrane system (2000x). (F) A proplatelet-bearing Mk (2000x). n = nucleus, g = granules, dms = demarcation membrane system, pp = proplatelet. Scale bar = 2 μm.
NIC increases ploidy under diverse Mk culture conditions
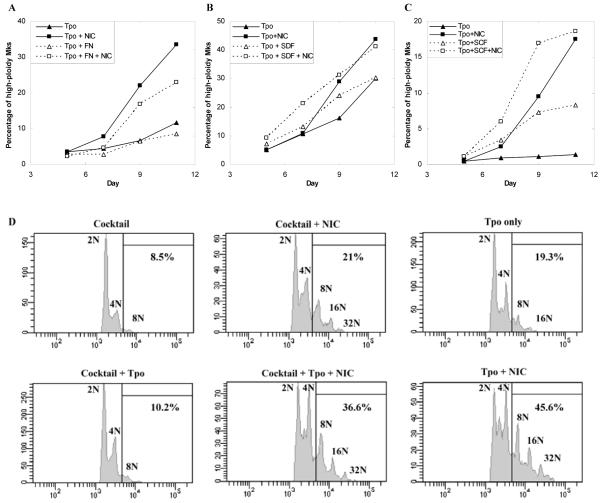
Since our previous cultures with NIC were carried out using Tpo as the only cytokine, we wanted to evaluate the generality of NIC effects. We first examined culture on fibronectin (FN), as well as additional supplementation with SDF-1α or SCF. Culture on fibronectin delayed the increase in ploidy for Mks supplemented with Tpo (Fig. 2A). However, NIC increased the percentage of high-ploidy (≥ 8N) Mks to a similar extent in the presence or absence of fibronectin. SDF-1α has previously been shown to increase Mk ploidy when added early in culture [25]. Adding 150 ng/mL SDF-1α resulted in a more rapid increase in ploidy compared to cultures with Tpo only, but the final ploidy was unchanged (Fig. 2B). Also, NIC increased ploidy to a similar extent in cultures with or without SDF-1α. SCF has been shown to synergize with Tpo to increase the number of Mks produced from umbilical cord blood (CB) or mPB CD34+ cells [26, 27]. SCF increased Mk ploidy in cultures with or without NIC (Fig. 2C). Furthermore, NIC substantially increased Mk ploidy in cultures with or without SCF.
Figure 2.
NIC increases Mk ploidy under diverse culture conditions. (A-C) The percentage of CD41+ cells with DNA content ≥ 8N (high-ploidy Mks) in cultures initiated with mPB CD34+ cells. (A) Cells were grown in fibronectin (FN)-coated or control well plates in media containing 100 ng/mL Tpo. On day 5, 6.25 mM NIC was added to a subset of the wells. Entire wells were sacrificed to determine Mk ploidy. (B) Cells were cultured with 100 ng/mL Tpo with or without 150 ng/mL SDF-1α beginning at day 0. Where indicated, 6.25 mM NIC was added at either day 2 (SDF-1α culture) or day 5. (C) Cells were cultured with 100 ng/mL Tpo with or without 100 ng/mL SCF beginning at day 0. At day 5, the indicated cultures were supplemented with 6.25 mM NIC. (D) mPB CD34+ cells were cultured with IL-3, IL-6, and SCF (Cocktail). Beginning on day 5, either 100 ng/mL Tpo, 6.25 mM NIC, or Tpo + NIC was added to the cultures. The histograms show the DNA content (stained with PI) of CD41+ cells at day 11. (A-D) The data shown are representative of two biological experiments.
We also examined the ability of NIC to increase Mk ploidy in the absence of Tpo. mPB CD34+ cells were cultured with IL-6, IL-3, and SCF [28, 29]. This cytokine combination yielded ca. 14% CD41+ cells by day 11 compared to 80% CD41+ cells in cultures with Tpo only (data not shown). On day 11, the percentage of high-ploidy Mks was ca. 2-fold lower in cocktail cultures compared to those with Tpo only, and reached only 9% high-ploidy Mks by day 11 (Fig. 2D). Addition of NIC to cocktail cultures beginning at day 5 increased the percentage of high-ploidy Mks at day 11 by more than 2-fold (Fig. 2D), indicating that NIC can act independently of Tpo. In contrast, addition of Tpo to cocktail cultures beginning at day 5 had a relatively small effect on the percentage of high-ploidy Mks. Adding both NIC and Tpo to cocktail cultures beginning at day 5 increased the percentage of high-ploidy Mks by a much greater extent (Fig. 2D), showing the synergy between Tpo and NIC. Together, these data demonstrate the versatility of NIC and its ability to increase the ploidy of Mks cultured under a variety of Mk-promoting conditions for donor samples that exhibit a wide range of Mk ploidy in cultures with Tpo only (Fig. 2 A-D). Adding NIC to cultures with Tpo alone increased the percentage of high-ploidy cells by 3.9(± 0.5)-fold (n = 29; p < 10-15, using a paired t-test) for the donor samples used in this study.
NIC increases the level of intracellular NAD(H) in cultured Mks by up to 5-fold
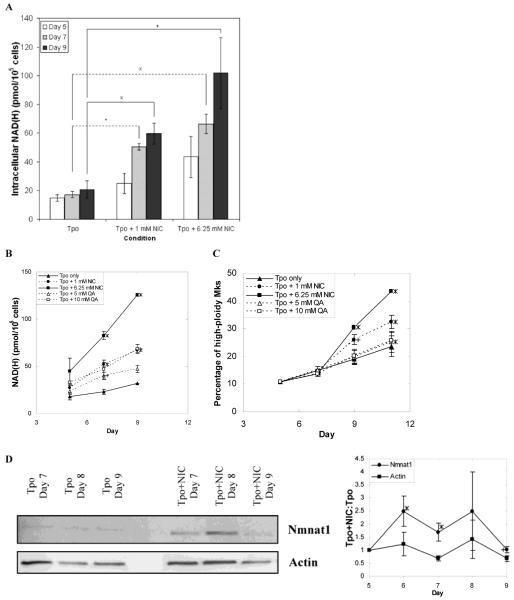
NIC may directly modulate Mk differentiation or it may act via an intermediary such as NAD+, the level of which has been shown to increase greatly in diverse human cell types after NIC addition [18, 30, 31]. In order to explore this latter possibility, we first examined if NIC increases NAD+ levels in Mks. NAD(H) levels increased only slightly with Mk maturation in cultures with Tpo only (Fig. 3A). In contrast, NIC rapidly increased the NAD(H) content - by 60% for 1 mM NIC and 3-fold for 6.25 mM four hours after NIC addition. By day 9, the NAD(H) content was ca. 3-fold higher for 1 mM NIC and almost 5-fold higher for 6.25 mM NIC compared to Tpo alone (Fig. 3A). For comparison, at day 9 the average volume of cells treated with 6.25 mM NIC was ca. 30% greater than cells cultured with Tpo alone and the average nuclear volume was 50% greater with NIC (data not shown). For all conditions, the level of NADH was much lower (ca. 10-20%) than that of NAD+ (data not shown). Thus, the increase in NAD(H) was predominantly due to an increase in NAD+. The increase in NAD(H) with NIC was dose-dependent (Fig. 3A), and the relative increase in NAD(H) for 1 mM vs. 6.25 mM NIC compared to Tpo alone was similar to the relative increase in the fraction of high-ploidy Mks (Fig. 3B,C).
Figure 3.
Effects of NIC and quinolinic acid (QA) on intracellular NAD(H) content, Mk ploidy, and Nmnat1 levels. (A) Intracellular NAD(H) levels in cells cultured with Tpo only, Tpo + 1 mM NIC, or Tpo + 6.25 mM NIC were measured using a cycling enzymatic assay on days 5 (4 h after NIC addition), 7, and 9. Data shown are the mean ± SEM of n = 7 (day 7) or n = 5 (days 5 and 9) experiments. Based on a paired t-test, values of p < 0.005 (x), p < 0.05 (*) and p < 0.1 (+) are indicated in comparison to the Tpo only treatment. (B-C) mPB CD34+ cells were cultured with 100 ng/mL Tpo. Beginning on day 5, cells were treated with either NIC or QA. (B) NAD(H) levels were measured on days 5 (4 h after addition), 7, and 9. (C) The percentage of high-ploidy Mks was determined on days 5, 7, 9 and 11. (B-C) Data shown are the mean ± SEM of 2 experiments. Based on a paired t-test, values of p < 0.05 (*) and p < 0.1 (+) are indicated in comparison to the Tpo only treatment. (D) Western blot (representative of five biological experiments) of nuclear lysates prepared from primary Mks cultured with 100 ng/mL of Tpo only or supplemented with 6.25 mM NIC on day 5 were probed for nuclear NAD processing enzyme Nmnat1 and actin (loading control). The densitometry values for Nmnat1 or actin expression in cells grown with NIC were divided by the densitometry values for Nmnat1 or actin expression in cells grown with Tpo only. The ratios shown correspond to the mean ± SEM of protein expression in whole cell or nuclear lysates harvested from 5 biological experiments. Based on a paired t-test, values of p < 0.05 (*), and p < 0.1 (+) are indicated for the various time points in comparison to actin band density.
We also supplemented cultures with other NAD+ precursors. Nicotinic acid (NA), the other form of niacin, has been shown to increase NAD+ content in several cell types [32, 33]. The NAD(H) content increased only slightly after addition of 3 or 6.25 mM NA to cells cultured with Tpo, and there was little if any effect of NA on CD41 expression or the percentage of high-ploidy Mks (data not shown). Thus, NA is not an effective NAD(H) precursor in cultured human Mks and does not increase Mk ploidy. NAD+ can also be synthesized from tryptophan via quinolinic acid (QA) [34], and QA increased intracellular NAD+ levels in cultured glial cells [32]. QA increased NAD(H) levels in a dose-dependent manner (Fig. 3B). Despite the similar NAD(H) levels, Mks in cultures treated with 1 mM NIC exhibited much greater ploidy than those with 10 mM QA, which was just slightly higher than that of Mks in cultures with Tpo alone (Fig. 3C). QA is incorporated into NAD+ via the de novo synthesis pathway, which is distributed throughout the cell [34]. In contrast, NIC is incorporated into NAD+ via the salvage pathway, which in yeast cells is primarily localized to the nucleus [35, 36]. Thus, the differential effects of NIC compared to QA could be due to differences in the location of NAD+ synthesis. Nicotinamide mononucleotide adenylyltransferase 1 (Nmnat1) localizes exclusively to the nucleus [37] and is essential for NAD+ biosynthesis by catalyzing the formation of NAD+ from nicotinamide mononucleotide and ATP [38]. Cultured Mks treated with NIC expressed ca. 2-fold higher levels of Nmnat1 than cells treated with Tpo only (Fig. 3D), which is consistent with increased nuclear NAD(H) content in NIC-treated cells.
NIC increases Mk ploidy at least in part through SIRT inhibition
NIC has been extensively characterized as an inhibitor of sirtuins, which were originally identified as NAD+-dependent Class III histone deacetylases [39-41]. NIC is a potent inhibitor of SIRT1 [42], which deacetylates a wide range of histones and non-histone proteins [41]. NIC also inhibits SIRT2, which deacetylates tubulin [11, 43], histone H4 [44, 45], and a growing number of proteins [46]. We had previously concluded that inhibition of SIRT1 and SIRT2 was not responsible for NIC-mediated increases in Mk ploidy. This was based on our finding that the yeast Sir2p inhibitors sirtinol (inhibits mammalian SIRT1 [47-49]) and splitomicin (inhibits SIRT1 and SIRT2 [50, 51]) did not affect Mk ploidy [8]. However, using these compounds in Mk cultures can be problematic due to splitomicin instability at pH 7.3-7.4 [45, 52] and sirtinol toxicity in Mk cultures at doses below those reported to be effective in mammalian cells [8, 47-49]. Therefore, we evaluated the effects of two recently described SIRT inhibitors.
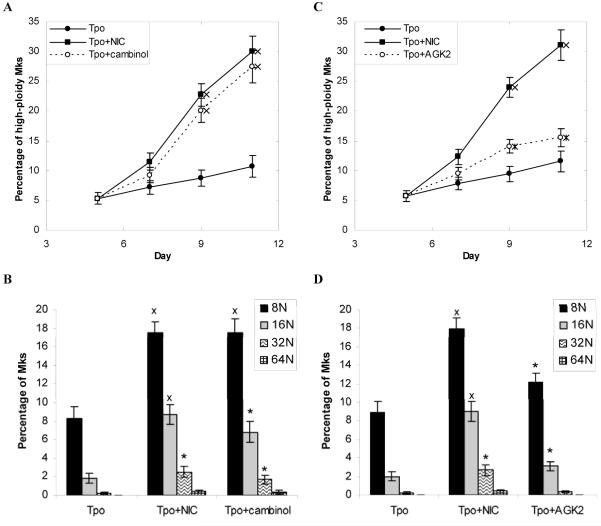
Cambinol is a small molecule that inhibits both SIRT1 and SIRT2 [45]. When added on day 5 at 10 μM, cambinol increased the fraction of high-ploidy Mks to a similar extent as NIC (Fig. 4A). The Mk ploidy distributions (Fig. 4B) and mean ploidy values (4.2 ± 0.2 for NIC and 3.9 ± 0.2 for cambinol) were also similar for 10 μM cambinol and 6.25 mM NIC. Adding 3.125 mM NIC plus 5 μM cambinol slightly increased the fraction of high-ploidy cells compared to 6.25 mM NIC or 10 μM cambinol alone, but the difference was not statistically significant (Fig. S1A). Synergistic addition of 3.125 mM NIC plus 5 μM cambinol had a greater effect on the fraction of Mks with ploidy ≥ 16N (Fig. S1B); the increase was statistically significant in comparison to 10 μM cambinol (p = 0.03), but not to 6.25 mM NIC (p = 0.14). No further increase in ploidy was obtained by adding 6.25 mM NIC plus 10 μM cambinol (Fig. S1 C,D). Cambinol (10 μM) and NIC had similar effects on the fraction of CD41+ cells (Fig. S2A) and total-cell fold-expansion (Fig. S2B). NIC and cambinol also had similar effects on the fractions of viable cells and apoptotic Mks, although the viability tended to be slightly higher with cambinol (Fig. S2C,D).
Figure 4.
SIRT inhibitors increase Mk ploidy. mPB CD34+ cells were cultured with 100 ng/mL Tpo. On day 5 cultures were supplemented with (A-B) 10 μM cambinol (n = 12) or (C-D) 10 μM AGK2 (n = 11). For comparison, replicate cultures were either supplemented with 6.25 mM NIC on day 5 or maintained with Tpo alone. Flow cytometry was used to determine (A,C) the percentage of CD41+ cells with DNA content ≥ 8N (high-ploidy Mks) and (B,D) the high-ploidy Mk distribution on day 11. Data shown represent the mean ± SEM. Based on a paired t-test, values of p < 0.0005 (x) and p < 0.05 (*) are indicated for the various time points in comparison to the Tpo only culture.
The similar results for NIC and cambinol suggest that NIC increases Mk ploidy by inhibiting SIRT1 and/or SIRT2. In an effort to decouple the effects of SIRT1 vs. SIRT2 inhibition, we evaluated AGK2, which has been shown to decrease SIRT2 activity in vitro without significantly affecting SIRT1 or SIRT3 activity at concentrations up to 10 μM [53]. When added at 10 μM, AGK2 moderately increased the percentage of high-ploidy Mks (Fig. 4C). At day 11, the AGK2-mediated increase in high-ploidy cells was about 30% as great as that for NIC. The Mk ploidy distribution (Fig. 4D) and mean ploidy value (4.3 ± 0.2 for NIC, 3.3 ± 0.1 for AGK2, and 3.0 ± 0.1 for Tpo only) at day 11 for 10 μM AGK2 were intermediate between those for Tpo only and 6.25 mM NIC. It is unlikely that higher AGK2 concentrations would be more effective because the ploidy difference between 5 and 10 μM AGK2 was small (data not shown) and because 10 μM AGK2 inhibited cell growth to a similar extent as 6.25 mM NIC (Fig. S2F). AGK2 (10 μM) had similar effects on the fraction of CD41+ cells (Fig. S2E) as NIC, but AGK2 yielded a slower decline in cell viability (Fig. S2G) and a slower increase in the fraction of apoptotic Mks (Fig. S2H).
SIRT inhibition by NIC increases acetylation of nucleosomes and p53
SIRT1 and SIRT2 have been extensively characterized as Class III histone deacetylases. Genomic DNA is packaged into chromatin via repeating units of nucleosomes - octamers comprised of equimolar amounts of histones H2A, H2B, H3, and H4. Acetylation of lysine residues on nucleosome fragments was increased by up to 3-fold in cells treated with NIC (Fig. S3). Small increases in acetylation were observed as soon as one day after NIC addition. This general increase in histone acetylation is consistent with inhibition of SIRT1 and/or SIRT2 by NIC.
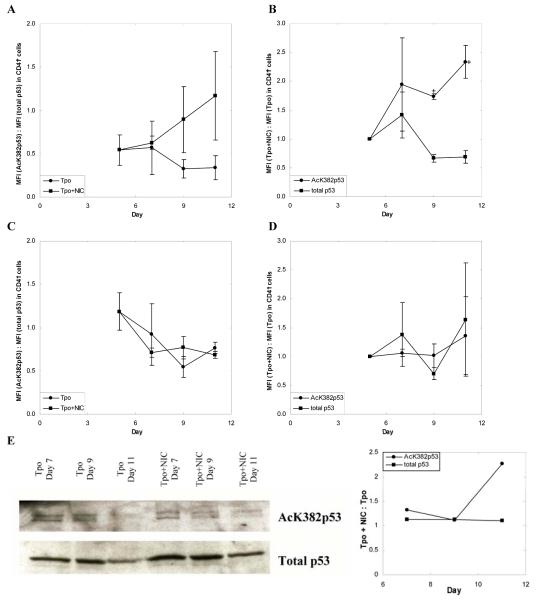
One of the best characterized SIRT1 targets is p53 [54, 55] and a recent study also associated SIRT2 with p53 deacetylation [46]. Among CD41+ cells, the ratio of AcK382p53 to total p53 steadily increased with time in cultures with NIC, but decreased with time in the absence of NIC (Fig. 5A). By day 11, the ratio of AcK382p53 to total p53 was 3.4-fold higher in Mks cultured with NIC (Fig. 5A). The large error bars for these data may be attributed to shifts in arbitrary fluorescence units and donor-to-donor variability between experiments. To more accurately capture the relative effects of NIC on p53 acetylation, the ratio between the mean fluorescence intensity (MFI) of CD41+ cells grown with NIC compared to that for cells maintained with Tpo alone was calculated for cells stained with antibodies against either AcK382p53 or total p53. Among CD41+ cells at day 11, the MFI for AcK382p53 staining was 2.3-fold higher for cells cultured with NIC (Fig. 5B). Conversely, the MFI for total p53 staining was 30% lower for cells cultured with NIC (Fig. 5B), resulting in a 3-fold greater fraction of acetylated p53 in cultures with NIC. In contrast, among the CD41- cell population, there was little difference between the ratio of AcK382p53 to total p53 for cells cultured with or without NIC (Fig. 5C). Interestingly, for both culture conditions, the ratio of AcK382p53 to total p53 decreased with time in a manner similar to that for CD41+ cells in cultures with Tpo only (Fig. 5A), although the overall level of the ratio was greater for CD41- cells. Among CD41- cells, the MFI of AcK382p53 staining was approximately the same for cells cultured with or without NIC, and did not change appreciably with time (Fig. 5D). The same was true for the MFI of total p53 staining (Fig. 5D). Greater p53 acetylation in Mks cultured with NIC was also evident from Western blots (Fig. 5E). Cells treated with NIC had sustained expression of acetylated p53, while cells supplemented with Tpo alone experienced a drop in acetylated p53 expression after day 9. Although this assessment is based on a single time point (day 11), this was a reproducible finding. These Western-blot data should be viewed, however, as supporting the more accurate and sensitive flow cytometry data (Fig. 5B), rather than as stand-alone evidence. The reason for this statement is that Western analysis is based on lysates from a mixed population of cells (CD41+ and CD41- cells); the presence of CD41- cells (although a small fraction: 5.9% and 6.3% in Tpo-treated Mks and 10.2% and 10% in Mks treated with Tpo+NIC for days 9 and 11, respectively), based on the data of Figs. 5A-D, would result in smaller observable differences of acetylated p53 by Western analysis. Furthermore, quantitative Western analysis is considerably less sensitive than flow-cytometric analysis. Overall, these data suggest that SIRT inhibition by NIC specifically increases p53 acetylation in Mks.
Figure 5.
NIC increases acetylation of p53. mPB CD34+ cells were cultured with 100 ng/mL Tpo. On day 5 cultures were supplemented with 6.25 mM NIC. (A-D) Intracellular flow cytometry was used to detect expression of AcK382p53 and total p53 in (A-B) CD41+ and (C-D) CD41- cells. Mean fluorescence intensity (MFI) was calculated by subtracting the MFI of an unstained sample from the MFI of a stained sample to correct for background fluorescence. Data shown represent the mean ± SEM of two experiments. (A,C) The MFI of cells stained with an antibody against AcK382p53 was divided by the MFI of cells stained with an antibody against total p53. This ratio is shown for cells maintained in Tpo with or without NIC. (B,D) The MFI of cells grown with NIC was divided by the MFI of cells grown with Tpo only. This ratio was calculated for cells stained either with an antibody against AcK382p53 or against total p53. Based on a paired t-test, values of p < 0.1 (*) are indicated for the various time points. (E) Nuclear lysates of unselected cells were prepared at all time points shown and loaded onto SDS-PAGE gels. After electrophoresis, the proteins were transferred to PVDF membranes for Western blots. After probing for AcK382p53 residues, the blots were stripped and probed for total p53. Corresponding densitometry analysis is given. The blot shown is representative of two biological experiments.
NIC-mediated SIRT inhibition correlates with increased p53 binding to its consensus DNA binding sequence
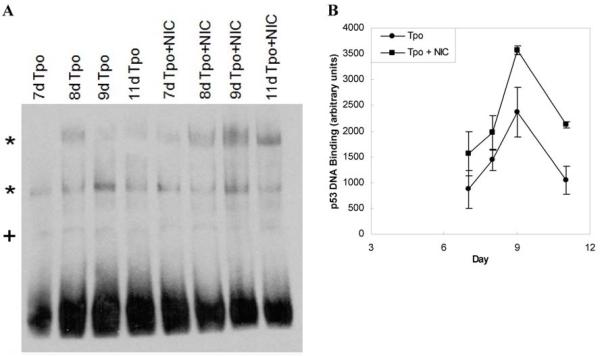
EMSA experiments were performed to determine whether NIC-mediated p53 acetylation increased p53 DNA-binding activity. As previously reported [20], two low-mobility DNA complexes were formed during these experiments and their levels increased during megakaryopoiesis (Fig. 6A). Both of the low-mobility DNA complexes were used in subsequent densitometry analyses because the binding affinity of both regions varied during the course of CHRF cell Mk differentiation and both were specific to the p53 consensus binding sequence oligonucleotides [20]. Consistent with the greater DNA-binding activity reported for acetylated p53 [56, 57], p53 DNA-binding activity was greater in cells cultured with NIC at every time point investigated (Fig. 6A,B). Although the differences were not statistically significant for the individual time points, nonparametric analysis of all of the EMSA data using the Kruskal-Wallis test confirmed that the consistently greater p53 DNA-binding activity observed for cells treated with NIC was significant (p = .033). Results at days 7, 9 and 11 from another biological experiment conducted using the TransBinding p53 ELISA kit (Panomics; Freemont, CA) also showed consistently greater p53 DNA-binding activity in the nuclear extracts of cells cultured with Tpo plus NIC compared to those cultured with Tpo alone (data not shown).
Figure 6.
NIC increases p53 DNA-binding activity. (A) mPB CD34+ cells were cultured using 100 ng/mL Tpo with or without 6.25 mM NIC beginning on day 5. At selected time points nuclear extracts were prepared for EMSA analysis. The gel shown is representative of 3 replicates from two biological experiments. Low mobility p53-DNA complexes (*) were formed using biotinylated p53 consensus binding sequence oligonucleotides. Non-specific binding is indicated by a (+) sign. (B) Densitometry of low mobility p53-DNA complexes is shown as the mean ± SEM of arbitrary density units for 2 replicates from one biological experiment.
Discussion
We previously showed that NIC increases Mk ploidy and proplatelet production without significantly altering Mk-specific gene expression in cultures of mPB CD34+ cells supplemented with Tpo [8]. Here, we show that NIC does not alter normal Mk ultrastructure (Fig. 1) and demonstrate the generality of NIC effects. NIC synergizes with Tpo to further enhance Mk maturation and is effective at increasing polyploidization under all of the Mk-promoting conditions we have investigated (Fig. 2). Therefore, NIC is likely to be effective under a wide range of other conditions that promote Mk maturation.
Increases in Mk ploidy may be due to direct modulation by NIC, but may also be mediated indirectly by elevated levels of NAD(H). We observed a 5-fold increase in NAD(H) levels when 6.25 mM NIC was added to Mk cultures (Fig. 3). We also examined quinolinic acid, which is incorporated into NAD+ via the de novo synthesis pathway. QA at 10 mM increased the level of intracellular NAD(H) to a similar extent as 1 mM NIC, but had little effect on Mk ploidy. The de novo pathway is distributed throughout the cell [34], while the salvage pathway used for NIC incorporation has been shown to be primarily localized to the nucleus in yeast cells [35, 36]. Greater levels of nuclear NAD processing enzyme Nmnat1 in NIC-treated cells further supports the hypothesis that NAD(H) synthesis in these cells is enhanced in the nucleus. Increased nuclear levels of NADH, induced by hypoxic conditions, cause a conformational change in hCtBP proteins that reduces their ability to bind with transcriptional corepressors [58], such as hdm2, and together inhibit p53 activation. Thus, higher levels of nuclear NADH result in increased p53 activity and greater activation of its downstream targets [59]. However, we note that cambinol would not be expected to increase NAD+ synthesis and we did not observe any increase in NAD(H) levels when the megakaryocytic CHRF cell line was treated with cambinol (data not shown).
NIC has been extensively characterized as an inhibitor of sirtuins, which were identified as NAD+-dependent Class III histone deacetylases [39-41]. Sirtuins play important roles in transcriptional silencing, genetic stability, and cellular lifespan regulation [13, 14, 42]. Deacetylation by the nuclear protein SIRT1 [42] regulates the activity of proteins associated with DNA repair, apoptosis, and cell cycle regulation including NBS1, p53, FOXO3a, RelA/p65 (NFκB), and Ku70 [15, 54, 60-63]. In contrast, the tubulin deacetylase SIRT2 can be localized to either the nucleus or the cytoplasm [11, 43]. Recent studies suggest that SIRT2 regulation of α-tubulin acetylation plays an important role in modulating the extension of cell processes. Knockdown of SIRT2 increased hippocampal neurite outgrowth [64] and the arborization complexity of primary oligodendrocyte precursors [65]. The SIRT2-specific inhibitor AGK2 was 30% as effective as NIC in increasing Mk ploidy (Fig. 4C,D). In contrast, cambinol, which inhibits both SIRT1 and SIRT2, increased Mk ploidy almost as effectively as NIC (Fig. 4A,B). This suggests that NIC primarily increases Mk ploidy by inhibition of SIRT1 and/or SIRT2. Together with our previous finding that SIRT1 and SIRT2 mRNA levels increase during Mk maturation [8], our results suggest that SIRT1 and/or SIRT2 are involved in controlling the transition from endomitosis, and that inhibiting SIRT1/2 delays this process and increases Mk ploidy.
Functional inhibition of SIRT1 and/or SIRT2 in NIC-treated Mks was confirmed using two well-established targets of SIRT deacetylation: nucleosomes and p53 (Fig. 5). Histone deacetylation within nucleosomes generally leads to the repression of transcription since removal of acetyl groups from lysine residues leads to chromatin becoming more compact such that specific regions of DNA are less accessible for transcription factor binding [66]. Increased acetylation of residue K382 on p53 in Mks treated with NIC (Fig. 5A,B,E) was associated with increased binding of p53 to its consensus DNA binding sequence (Fig. 6). Acetylation of K382 on p53 can lead to cell cycle arrest and apoptosis [67, 68], as well as activation and stabilization of p53, which may prevent p53 ubiquitination and degradation [69]. Although Mk viability and apoptosis did not substantially change upon treatment with NIC (Fig. S2C,D,G,H), p53 has widespread effects through its regulation of other cell cycle, DNA repair, and lifespan-related targets including CDC2, GADD45, p21, and 14-3-3s [70, 71].
We previously demonstrated the importance of p53 in Mk polyploidization and apoptosis [20, 21, 72]. Gene expression analysis of mPB CD34+ cells cultured under Mk-promoting conditions revealed differential expression of several transcriptional targets of the p53 protein. Using RNAi to reduce p53 expression in CHRF cells, we showed that lower p53 activity leads to a greater fraction of polyploid cells, higher mean and maximal ploidy, accelerated DNA synthesis, and delayed apoptosis upon Mk differentiation [20]. Recent results in our laboratory indicate that posttranslational p53 modifications (phosphorylation and acetylation) take place in a Mk-specific manner and apparently play an important role in Mk maturation (data not shown). Consistent with this hypothesis are the findings reported here whereby NIC increased p53 acetylation and p53 DNA-binding activity. This suggests that the effect of NIC, as a SIRT1/2 inhibitor, is mediated at least in part by increased p53 acetylation. While it seems counterintuitive that increased p53 DNA-binding activity and p53 knockdown would both increase Mk ploidy, p53 has many different target genes, whose activity may be differentially regulated by SIRT1 and/or SIRT2. A genome-wide ChIP-on-chip analysis identified 1546 p53-binding sites after treatment with Actinomycin D [73].
It is likely that mechanistic understanding of the modulation of p53 activity and its targets on Mk differentiation, as well as those of other SIRT target proteins, will lead to additional means for enhancing Mk polyploidization. One promising p53 target with increased transcription as a result of increased p53 acetylation is p21WAF1 [54]. Levels of p21WAF1 were ca. 3-fold higher in a Sir2-deficient cell line than with WT Sir2 [54]. Using RT-PCR, we found that p21 expression was significantly reduced in p53 knockdown CHRF cells [20]. Expression of p53 and p21WAF1 was correlated with increasing nuclei size and DNA content in a human lung carcinoma cell line [74]. In addition, p21WAF1 expression increased in an erythroleukemia cell line induced to undergo polyploidization [75]. We are investigating p21 expression in Mks with inhibited or knocked down SIRT1 and/or SIRT2 in comparison to primary human Mks cultured with Tpo alone to determine whether p21 expression correlates with increased Mk ploidy.
Supplementary Material
Figure S1. NIC and cambinol synergize to slightly increase Mk ploidy. (A-B) mPB CD34+ cells were cultured with 100 ng/mL Tpo. On day 5 cultures were supplemented with 6.25 mM NIC, 10 μM cambinol, or 3.125 mM NIC plus 5 μM cambinol (n = 4). (C-D) Cells from a subset of the experiments shown in A-B were also treated with 6.25 mM NIC plus 10 μM cambinol (n = 2). Flow cytometry was used to determine (A,C) the percentage of CD41+ cells with DNA content ≥ 8N (high-ploidy Mks) or (B,D) the high-ploidy Mk distribution on day 11. Data shown represent the mean ± SEM.
Acknowledgements
We are grateful to Genentech for Tpo donation and to Dr. Antonio Bedalov (Fred Hutchinson Cancer Research Center) for donation of cambinol and helpful discussions. We thank Aaron Kuhl for initial development of the NAD(H) assay. We thank Dr. Mohamed Eldibany of the NorthShore University HealthSystem for help with analysis of EM images and Lennell Reynolds of the NU Cell Imaging facility for EM sample preparation. Supported by NIH Grant R01HL48276, a grant from the NorthShore University HealthSystem Pathology Department, and the Robert H. Lurie Comprehensive Cancer Center of Northwestern University Malkin Family Scholarship. S.P. was supported in part by NIH Biotechnology Predoctoral Training Grant T32 GM 008449. P.A.A. was supported in part by funds through the Delaware Biotechnology Institute and an Onassis Foundation fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosure
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
References
- [1].Elting LS, Rubenstein EB, Martin CG, et al. Incidence, cost, and outcomes of bleeding and chemotherapy dose modification among solid tumor patients with chemotherapy-induced thrombocytopenia. J Clin Oncol. 2001;19:1137–1146. doi: 10.1200/JCO.2001.19.4.1137. [DOI] [PubMed] [Google Scholar]
- [2].Vadhan-Raj S, Patel S, Bueso-Ramos C, et al. Importance of Predosing of Recombinant Human Thrombopoietin to Reduce Chemotherapy-Induced Early Thrombocytopenia. J Clin Oncol. 2003;21:3158–3167. doi: 10.1200/JCO.2003.08.003. [DOI] [PubMed] [Google Scholar]
- [3].Ryu KH, Chun S, Carbonierre S, et al. Apoptosis and megakaryocytic differentiation during ex vivo expansion of human cord blood CD34+ cells using thrombopoietin. Br J Haematol. 2001;113:470–478. doi: 10.1046/j.1365-2141.2001.02762.x. [DOI] [PubMed] [Google Scholar]
- [4].Falcieri E, Bassini A, Pierpaoli S, et al. Ultrastructural characterization of maturation, platelet release, and senescence of human cultured megakaryocytes. Anat Rec. 2000;258:90–99. doi: 10.1002/(SICI)1097-0185(20000101)258:1<90::AID-AR10>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [5].Li J, Kuter DJ. The end is just the beginning: megakaryocyte apoptosis and platelet release. Int J Hematol. 2001;74:365–374. doi: 10.1007/BF02982078. [DOI] [PubMed] [Google Scholar]
- [6].Mattia G, Vulcano F, Milazzo L, et al. Different ploidy levels of megakaryocytes generated from peripheral or cord blood CD34+ cells are correlated with different levels of platelet release. Blood. 2002;99:888–897. doi: 10.1182/blood.v99.3.888. [DOI] [PubMed] [Google Scholar]
- [7].Trowbridge EA, Martin JF, Slater DN, et al. The origin of platelet count and volume. Clin Phys Physiol Meas. 1984;5:145–170. doi: 10.1088/0143-0815/5/3/007. [DOI] [PubMed] [Google Scholar]
- [8].Giammona LM, Fuhrken PG, Papoutsakis ET, Miller WM. Nicotinamide (vitamin B3) increases the polyploidisation and proplatelet formation of cultured primary human megakaryocytes. Br J Haematol. 2006;135:554–566. doi: 10.1111/j.1365-2141.2006.06341.x. [DOI] [PubMed] [Google Scholar]
- [9].Yang J, Klaidman LK, Adams JD. Medicinal chemistry of nicotinamide in the treatment of ischemia and reperfusion. Mini Rev Med Chem. 2002;2:125–134. doi: 10.2174/1389557024605483. [DOI] [PubMed] [Google Scholar]
- [10].Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- [11].North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Molecular cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- [12].Denu JM. The Sir 2 family of protein deacetylases. Current opinion in chemical biology. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [13].Grubisha O, Smith BC, Denu JM. Small molecule regulation of Sir2 protein deacetylases. Febs J. 2005;272:4607–4616. doi: 10.1111/j.1742-4658.2005.04862.x. [DOI] [PubMed] [Google Scholar]
- [14].Blander G, Guarente L. The Sir2 family of protein deacetylases. Annual Review of Biochemistry. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- [15].Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annual review of biochemistry. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- [16].Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- [17].Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD(+) salvage pathway. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McLure KG, Takagi M, Kastan MB. NAD+ modulates p53 DNA binding specificity and function. Mol Cell Biol. 2004;24:9958–9967. doi: 10.1128/MCB.24.22.9958-9967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. Biochem J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fuhrken PG, Apostolidis PA, Lindsey S, Miller WM, Papoutsakis ET. Tumor suppressor protein p53 regulates megakaryocytic polyploidization and apoptosis. J Biol Chem. 2008;283:15589–15600. doi: 10.1074/jbc.M801923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fuhrken PG, Chen C, Miller WM, Papoutsakis ET. Comparative, genome-scale transcriptional analysis of CHRF-288-11 and primary human megakaryocytic cell cultures provides novel insights into lineage-specific differentiation. Exp Hematol. 2007;35:476–489. doi: 10.1016/j.exphem.2006.10.017. [DOI] [PubMed] [Google Scholar]
- [22].Wagner TC, Scott MD. Single Extraction Method for the Spectrophotometric Quantification of Oxidized and Reduced Pyridine Nucleotides in Erythrocytes. Analytical Biochemistry. 1994;222:417–426. doi: 10.1006/abio.1994.1511. [DOI] [PubMed] [Google Scholar]
- [23].Bernofsky C, Swan M. An improved cycling assay for nicotinamide adenine dinucleotide. Analytical biochemistry. 1973;53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- [24].Cramer EM, Norol F, Guichard J, et al. Ultrastructure of platelet formation by human megakaryocytes cultured with the Mpl ligand. Blood. 1997;89:2336–2346. [PubMed] [Google Scholar]
- [25].Guerriero R, Mattia G, Testa U, et al. Stromal cell-derived factor 1alpha increases polyploidization of megakaryocytes generated by human hematopoietic progenitor cells. Blood. 2001;97:2587–2595. doi: 10.1182/blood.v97.9.2587. [DOI] [PubMed] [Google Scholar]
- [26].Bruno S, Gunetti M, Gammaitoni L, et al. In vitro and in vivo megakaryocyte differentiation of fresh and ex-vivo expanded cord blood cells: rapid and transient megakaryocyte reconstitution. Haematologica. 2003;88:379–387. [PubMed] [Google Scholar]
- [27].Norol F, Vitrat N, Cramer E, et al. Effects of cytokines on platelet production from blood and marrow CD34+ cells. Blood. 1998;91:830–843. [PubMed] [Google Scholar]
- [28].Lindsley JE, Rutter J. Nutrient sensing and metabolic decisions. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:543–559. doi: 10.1016/j.cbpc.2004.06.014. [DOI] [PubMed] [Google Scholar]
- [29].LaIuppa JA, Papoutsakis ET, Miller WM. Evaluation of cytokines for expansion of the megakaryocyte and granulocyte lineages. Stem Cells. 1997;15:198–206. doi: 10.1002/stem.150198. [DOI] [PubMed] [Google Scholar]
- [30].Evans J, Wang TC, Heyes MP, Markey SP. LC/MS analysis of NAD biosynthesis using stable isotope pyridine precursors. Analytical biochemistry. 2002;306:197–203. doi: 10.1006/abio.2002.5715. [DOI] [PubMed] [Google Scholar]
- [31].Micheli V, Simmonds HA, Sestini S, Ricci C. Importance of nicotinamide as an NAD precursor in the human erythrocyte. Archives of biochemistry and biophysics. 1990;283:40–45. doi: 10.1016/0003-9861(90)90609-3. [DOI] [PubMed] [Google Scholar]
- [32].Grant RS, Kapoor V. Murine glial cells regenerate NAD, after peroxide-induced depletion, using either nicotinic acid, nicotinamide, or quinolinic acid as substrates. Journal of neurochemistry. 1998;70:1759–1763. doi: 10.1046/j.1471-4159.1998.70041759.x. [DOI] [PubMed] [Google Scholar]
- [33].O’Dorisio MS, Barker KL. Effects of estradiol on the biosynthesis of pyridine nucleotide coenzymes in the rat uterus. Biology of reproduction. 1976;15:504–510. doi: 10.1095/biolreprod15.4.504. [DOI] [PubMed] [Google Scholar]
- [34].Katoh A, Hashimoto T. Molecular biology of pyridine nucleotide and nicotine biosynthesis. Front Biosci. 2004;9:1577–1586. doi: 10.2741/1350. [DOI] [PubMed] [Google Scholar]
- [35].Anderson RM, Bitterman KJ, Wood JG, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- [36].Gallo CM, Smith DL, Jr., Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- [38].Schweiger M, Hennig K, Lerner F, et al. Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS letters. 2001;492:95–100. doi: 10.1016/s0014-5793(01)02180-9. [DOI] [PubMed] [Google Scholar]
- [39].Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Molecular cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- [40].Sanders BD, Zhao K, Slama JT, Marmorstein R. Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes. Molecular cell. 2007;25:463–472. doi: 10.1016/j.molcel.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- [42].Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- [43].Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007;6:1011–1018. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- [44].Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Heltweg B, Gatbonton T, Schuler AD, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer research. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- [46].Jin YH, Kim YJ, Kim DW, et al. Sirt2 interacts with 14-3-3 beta/gamma and down-regulates the activity of p53. Biochem Biophys Res Commun. 2008;368:690–695. doi: 10.1016/j.bbrc.2008.01.114. [DOI] [PubMed] [Google Scholar]
- [47].Ota H, Akishita M, Eto M, Iijima K, Kaneki M, Ouchi Y. Sirt1 modulates premature senescence-like phenotype in human endothelial cells. Journal of molecular and cellular cardiology. 2007;43:571–579. doi: 10.1016/j.yjmcc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- [48].Ota H, Tokunaga E, Chang K, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- [49].Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- [50].Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci U S A. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Molecular cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- [52].Posakony J, Hirao M, Bedalov A. Identification and characterization of Sir2 inhibitors through phenotypic assays in yeast. Combinatorial chemistry & high throughput screening. 2004;7:661–668. doi: 10.2174/1386207043328346. [DOI] [PubMed] [Google Scholar]
- [53].Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- [54].Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- [55].Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- [56].Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- [57].Roy S, Tenniswood M. Site-specific acetylation of p53 directs selective transcription complex assembly. J Biol Chem. 2007;282:4765–4771. doi: 10.1074/jbc.M609588200. [DOI] [PubMed] [Google Scholar]
- [58].Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- [59].Mirnezami AH, Campbell SJ, Darley M, Primrose JN, Johnson PW, Blaydes JP. Hdm2 recruits a hypoxia-sensitive corepressor to negatively regulate p53-dependent transcription. Curr Biol. 2003;13:1234–1239. doi: 10.1016/s0960-9822(03)00454-8. [DOI] [PubMed] [Google Scholar]
- [60].Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends in pharmacological sciences. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- [61].Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- [62].Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO journal. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Molecular cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pandithage R, Lilischkis R, Harting K, et al. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. The Journal of cell biology. 2008;180:915–929. doi: 10.1083/jcb.200707126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li W, Zhang B, Tang J, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27:2606–2616. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fouladi M. Histone deacetylase inhibitors in cancer therapy. Cancer investigation. 2006;24:521–527. doi: 10.1080/07357900600814979. [DOI] [PubMed] [Google Scholar]
- [67].Roy S, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005;12:482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- [68].Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ito A, Lai CH, Zhao X, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. The EMBO journal. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- [71].Tokino T, Nakamura Y. The role of p53-target genes in human cancer. Critical reviews in oncology/hematology. 2000;33:1–6. doi: 10.1016/s1040-8428(99)00051-7. [DOI] [PubMed] [Google Scholar]
- [72].Chen C, Fuhrken PG, Huang LT, et al. A systems-biology analysis of isogenic megakaryocytic and granulocytic cultures identifies new molecular components of megakaryocytic apoptosis. BMC Genomics. 2007;8:384. doi: 10.1186/1471-2164-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Smeenk L, van Heeringen SJ, Koeppel M, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic acids research. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Grove LE, Ghosh RN. Quantitative characterization of mitosis-blocked tetraploid cells using high content analysis. Assay and drug development technologies. 2006;4:421–442. doi: 10.1089/adt.2006.4.421. [DOI] [PubMed] [Google Scholar]
- [75].Matsumura I, Tanaka H, Kawasaki A, et al. Increased D-type cyclin expression together with decreased cdc2 activity confers megakaryocytic differentiation of a human thrombopoietin-dependent hematopoietic cell line. J Biol Chem. 2000;275:5553–5559. doi: 10.1074/jbc.275.8.5553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. NIC and cambinol synergize to slightly increase Mk ploidy. (A-B) mPB CD34+ cells were cultured with 100 ng/mL Tpo. On day 5 cultures were supplemented with 6.25 mM NIC, 10 μM cambinol, or 3.125 mM NIC plus 5 μM cambinol (n = 4). (C-D) Cells from a subset of the experiments shown in A-B were also treated with 6.25 mM NIC plus 10 μM cambinol (n = 2). Flow cytometry was used to determine (A,C) the percentage of CD41+ cells with DNA content ≥ 8N (high-ploidy Mks) or (B,D) the high-ploidy Mk distribution on day 11. Data shown represent the mean ± SEM.