Abstract
Nrf2:INrf2(Keap1) are cellular sensors of chemical and radiation induced oxidative and electrophilic stress. Nrf2 is a nuclear transcription factor that controls the expression and coordinated induction of a battery of defensive genes encoding detoxifying enzymes and antioxidant proteins. This is a mechanism of critical importance for cellular protection and cell survival. Nrf2 is retained in the cytoplasm by an inhibitor INrf2. INrf2 functions as an adapter for Cul3/Rbx1 mediated degradation of Nrf2. In response to oxidative/electrophilic stress, Nrf2 is switched on and then off by distinct early and delayed mechanisms. Oxidative/electrophilic modification of INrf2cysteine151 and/or PKC phosphorylation of Nrf2serine40 results in the escape or release of Nrf2 from INrf2. Nrf2 is stabilized and translocates to the nucleus, forms heterodimers with unknown proteins, and binds antioxidant response element (ARE) that leads to coordinated activation of gene expression. It takes less than fifteen minutes from the time of exposure to switch on nuclear import of Nrf2. This is followed by activation of a delayed mechanism that controls switching off of Nrf2 activation of gene expression. GSK3β phosphorylates Fyn at unknown threonine residue(s) leading to nuclear localization of Fyn. Fyn phosphorylates Nrf2tyrosine568 resulting in nuclear export of Nrf2, binding with INrf2 and degradation of Nrf2. The switching on and off of Nrf2 protect cells against free radical damage, prevents apoptosis and promotes cell survival.
Introduction
Oxidative stress is induced by a vast range of factors including xenobiotics, drugs, heavy metals and ionizing radiation. Oxidative stress leads to the generation of Reactive Oxygen Species (ROS) and electrophiles. ROS and electrophiles generated can have a profound impact on survival, growth development and evolution of all living organisms [1,2] ROS include both free radicals, such as the superoxide anion and the hydroxyl radical, and oxidants such as hydrogen peroxide [3]. ROS and electrophiles can cause diseases such as cancer, cardiovascular complications, acute and chronic inflammation, and neurodegenerative diseases [1]. Therefore, it is obvious that cells must constantly labor to control levels of ROS, preventing them from accumulation.
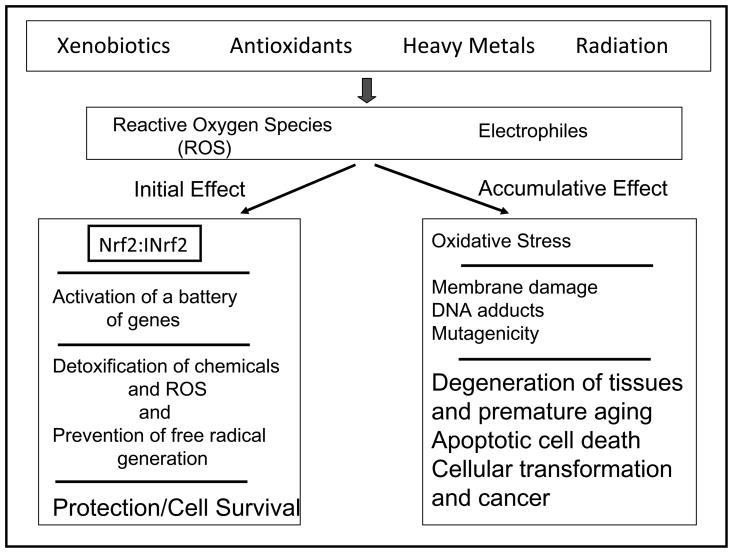
Much of what we know about the mechanisms of protection against oxidative stress has come from the study of prokaryotic cells [4,5]. Prokaryotic cells utilize transcription factors OxyR and SoxRS to sense the redox state of the cell, and during oxidative stress these factors induce the expression of nearly eighty defensive genes [5]. Eukaryotic cells have similar mechanisms to protect against oxidative stress [Fig. 1; ref. 3,6–9]. Initial effect of oxidative/electrophilic stress leads to activation of a battery of defensive gene expression that leads to detoxification of chemicals and ROS and prevention of free radical generation and cell survival [Fig. 1]. Of these genes, some are enzymes such as NAD(P)H:quinine oxidoreductase 1 (NQO1), NRH:quinone oxidoreductase 2 (NQO2), glutathione S-transferase Ya subunit (GST Ya Subunit), heme oxygenase 1 (HO-1), and γ-glutamylcysteine synthetase (γ-GCS), also known as glutamate cysteine ligase (GCL). Other genes have end products that regulate a wide variety of cellular activities including signal transduction, proliferation, and immunologic defense reactions. There is a wide variety of factors associated with the cellular response to oxidative stress. For example, NF-E2 related factor 2 (Nrf2), heat shock response activator protein 1, and NF-kappaB promote cell survival, where as activation of c-jun, N-terminal kinases (JNK), p38 kinase and TP53 may lead to cell cycle arrest and apoptosis [10]. The Nrf2 pathway is regarded as the most important in the cell to protect against oxidative stress. [3,6–9]. It is noteworthy that accumulation of ROS and/or electrophiles leads to oxidative/electrophile stress, membrane damage, DNA adducts formation and mutagenicity [Fig. 1]. These changes lead to degeneration of tissues and premature aging, apoptotic cell death, cellular transformation and cancer.
Fig. 1.
Chemical and radiation exposure and coordinated induction of defensive genes.
Antioxidant Response Element and Nrf2
Promoter analysis identified a cis-acting enhancer sequence designated as the antioxidant response element (ARE) that controls the basal and inducible expression of antioxidant genes in response to xenobiotics, antioxidants, heavy metals and UV light [11]. The ARE sequence is responsive to a broad range of structurally diverse chemicals apart from β-nafthoflavone and phenolic antioxidants [12]. Mutational analysis revealed GTGACA***GC to be the core sequence of the ARE [11,13–14]. This core sequence is present in all Nrf2 downstream genes that respond to antioxidants and xenobiotics [3, 6–9]. Nrf2 binds to the ARE and regulates ARE-mediated antioxidant enzyme genes expression and induction in response to a variety of stimuli including antioxidants, xenobiotics, metals, and UV irradiation [6, 15–21].
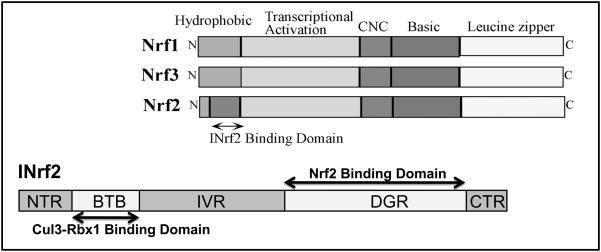
Nrf2 is ubiquitously expressed in a wide range of tissue and cell types [22–24] and belongs to a subset of basic leucine zipper genes (bZIP) sharing a conserved structural domain designated as a cap’n’collar domain which is highly conserved in Drosphila transcription factor CNC (Fig. 2; ref. 25]. The basic region, just upstream of the leucine zipper region, is responsible for DNA binding [3] and the acidic region is required for transcriptional activation. ARE-mediated transcriptional activation requires heterodimerization of Nrf2 with other bZIP proteins including Jun (c-Jun, Jun-D, and Jun-B) and small Maf (MafG, MafK, MafF) proteins [18–20, 26–27].
Fig. 2. Schematic Presentation of Various Domains of Nrf (Nrf1, Nrf2, Nrf3) and INrf2.
Nrf, NF-E2 Related Factor; INrf2, Inhibitor of Nrf2; NTR, N-Terminal Region; BTB, Broad complex, Tramtrack, Bric-a-brac; IVR, Intervening/linker Region; DGR, Kelch domain/diglycine repeats; CTR, C-Terminal Region.
Initial evidence demonstrating the role of Nrf2 in antioxidant-induction of detoxifying enzymes came from studies on the role of Nrf2 in ARE-mediated regulation of NQO1 gene expression [17]. Nrf2 was subsequently shown to be involved in the transcriptional activation of other ARE-responsive genes such as GST Ya, γ-GCS, HO-1, antioxidants, proteasomes, and drug transporters [3, 6–9, 28–33]. Overexpression of Nrf2 cDNA was shown to upregulate the expression and induction of the NQO1 gene in response to antioxidants and xenobiotics [17]. In addition, Nrf2-null mice exhibited a marked decrease in the expression and induction of NQO1, indicating that Nrf2 plays an essential role in the in vivo regulation of NQO1 in response to oxidative stress [26]. The importance of this transcription factor in upregulating ARE-mediated gene expression has been demonstrated by several in vivo and in vitro studies [reviewed in ref. 3]. The results indicate that Nrf2 is an important activator of phase II antioxidant genes [3, 8].
Negative Regulation of Nrf2 mediated by INrf2
A cytosolic inhibitor (INrf2), also known as Keap1 (Kelch-like ECH-associating protein 1), of Nrf2 was identified and reported [Fig. 2; ref. 34–35]. INrf2, existing as a dimer [36], retains Nrf2 in the cytoplasm. Analysis of the INrf2 amino acid sequence and domain structure-function analyses have revealed that INrf2 has a BTB(broad complex, tramtrack, bric-a-brac)/POZ (poxvirus, zinc finger) domain and a Kelch domain [34–35] also known as the DGR domain (Double glycine repeat) [37]. Keap1 has three additional domains/regions: the N-terminal region (NTR), the invervening region (IVR), and the C-terminal region (CTR) [8]. The BTB/POZ domain has been shown to be a protein-protein interaction domain. In the Drosophila Kelch protein, and in IPP, the Kelch domain binds to actin [38–39] allowing the scaffolding of INrf2 to the actin cytoskeleton which plays an important role in Nrf2 retention in the cytosol [40]. The main function of INrf2 is to serve as an adapter for the Cullin3/Ring Box 1 (Cul3/Rbx1) E3 ubiquitin ligase complex [41–43]. Cul3 serves as a scaffold protein that forms the E3 ligase complex with Rbx1 and recruits a cognate E2 enzyme [8]. INrf2 via its N-terminal BTB/POZ domain binds to Cul3 [44] and via its C-terminal Kelch domain binds to the substrate Nrf2 leading to the ubiquitination and degradation of Nrf2 through the 26S proteasome [45–49]. Under normal cellular conditions, the cytosolic INrf2/Cul3-Rbx1 complex is constantly degrading Nrf2. When a cell is exposed to oxidative stress Nrf2 dissociates from the INrf2 complex, stabilizes and translocates into the nucleus leading to activation of ARE-mediated gene expression [3, 6–9]. An alternative theory is that Nrf2 in response to oxidative stress escapes INrf2 degradation, stabilizes and translocates in the nucleus [49–50]. We suggested the theory of escape of Nrf2 from INrf2 [49] and similar suggestion was also made in another report [50]. However, the follow up studies in our laboratory could not support the escape theory. Escape theory is a possibility but has to be proven by experiments before it can be adapted. Therefore, we will use the release of Nrf2 from INrf2 in the rest of this review.
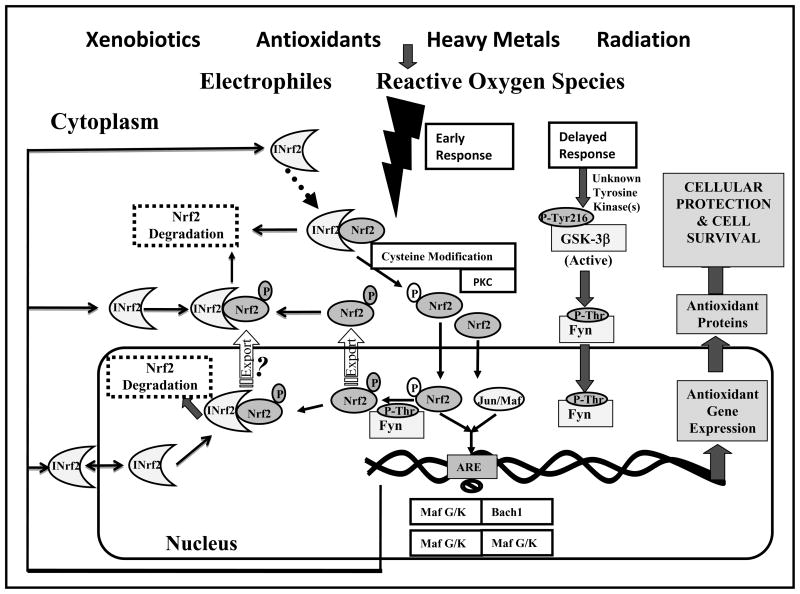
Numerous reports have suggested that any mechanism that modifies INrf2 and/or Nrf2 disrupting the Nrf2:INrf2 interaction will result in the upregulation of ARE-mediated gene expression. A model Nrf2:INrf2 signaling from antioxidant and xenobiotic to activation of ARE-mediated defensive gene expression is shown in Fig. 3. Since the metabolism of antioxidants and xenobiotics results in the generation of ROS and electrophiles [51], it is thought that these molecules might act as second messengers, activating ARE-mediated gene expression. Several protein kinases including PKC, ERK, MAPK, p38, and PERK [49, 52–56] are known to modify Nrf2 and activate its release from INrf2. Among these mechanisms, oxidative/electrophilic stress mediated phosphorylation of Nrf2 at serine40 by PKC is necessary for Nrf2 release from INrf2, but is not required for Nrf2 accumulation in the nucleus [49, 52–53]. In addition to post-translational modification in Nrf2, several crucial residues in INrf2 have also been proposed to be important for activation of Nrf2. Studies based on the electrophile mediated modification, location and mutational analyses revealed that three cysteine residues, Cys151, Cys273 and Cys288 are crucial for INrf2 activity [50]. INrf2 itself undergoes ubiquitination by the Cul3 complex, via a proteasomal independent pathway, which was markedly increased in response to phase II inducers such as antioxidants [57]. It has been suggested that normally INrf2 targets Nrf2 for ubiquitin mediated degradation but electrophiles may trigger a switch of Cul3 dependent ubiquitination from Nrf2 to INrf2 resulting in ARE gene induction. The redox modulation of cysteines in INrf2 might be a mechanism redundant to the phosphorylation of Nrf2 by PKC, or that the two mechanisms work in concert. In addition to cysteine151 modification, phosphorylation of Nrf2 has also been shown to play a role in INrf2 retention and release of Nrf2. Serine104 of INrf2 is required for dimerization of INrf2, and mutations of serine104 led to the disruption of the INrf2 dimer leading to the release of Nrf2 [36]. Recently, Eggler at al. demonstrated that modifying specific cysteines of the electrophile-sensing human INrf2 protein is insufficient to disrupt binding to the Nrf2 domain Neh2 (58). Upon introduction of electrophiles, modification of INrf2C151 leads to a change in the conformation of the BTB domain by means of perturbing the homodimerization site, disrupting Neh2 ubiquitination, and causing ubiquitination of INrf2. Modification of INrf2 cysteines by electrophiles does not lead to disruption of the INrf2–Nrf2 complex. Rather, the switch of ubiquitination from Nrf2 to INrf2 leads to Nrf2 nuclear accumulation.
Fig. 3.
Nrf2 signaling in ARE-mediated coordinated activation of defensive genes.
More recently, our laboratory demonstrated that phosphorylation and de-phosphorylation of tyrosine141 in INrf2 regulates its stability and degradation, respectively [59]. The de-phosphorylation of tyrosine141 caused destabilization and degradation of INrf2 leading to the release of Nrf2. Furthermore, we showed that prothymosin-α mediates nuclear import of the INrf2/Cul3-Rbx1 complex [60]. The INrf2/Cul3-Rbx1 complex inside the nucleus exchanges prothymosin-α with Nrf2 resulting in degradation of Nrf2. These results led to the conclusion that prothymosin-α mediated nuclear import of INrf2/Cul3-Rbx1 complex leads to ubiquitination and degradation of nuclear Nrf2 presumably to regulate nuclear level of Nrf2 and rapidly switch off the activation of Nrf2 downstream gene expression. An auto-regulatory loop also exists within the Nrf2 pathway [61]. An ARE was identified in the INrf2 promoter that facilitates Nrf2 binding causing induction of the INrf2 gene. Nrf2 regulates INrf2 by controlling its transcription, and INrf2 controls Nrf2 by serving as an adaptor for degradation.
Other Regulatory Mediators of Nrf2
Bach1 (BTB and CNC homology 1, basic leucine zipper transcription factor 1) is a transcription repressor [62] that is ubiquitously expressed in tissues [63–64] and distantly related to Nrf2 [8]. In the absence of cellular stress, Bach1 heterodimers with small Maf proteins [65] that bind to the (ARE) [66] repressing gene expression. In the presence of oxidative stress, Bach1 releases from the ARE and is replaced by Nrf2. Bach1 competes with Nrf2 for binding to the ARE leading to suppression of Nrf2 downstream genes [66].
Nuclear import of Nrf2, from time of exposure to stabilization, takes roughly two hours [67]. This is followed by activation of a delayed mechanism involving Glycogen synthase kinase 3 beta (GSK3β) that controls switching off of Nrf2 activation of gene expression (Fig. 3). GSK3β is a multifunctional serine/threonine kinase, which plays a major role in various signaling pathways [68]. GSK3β phosphorylates Fyn, a tyrosine kinase, at unknown threonine residue(s) leading to nuclear localization of Fyn [69]. Fyn phosphorylates Nrf2 tyrosine 568 resulting in nuclear export of Nrf2, binding with INrf2 and degradation of Nrf2 [70].
The negative regulation of Nrf2 by Bach1 and GSK3β/Fyn are important in repressing Nrf2 downstream genes that were induced in response to oxidative/electrophilic stress. The tight control of Nrf2 is vital for the cells against free radical damage, prevention of apoptosis and cell survival [3, 6–9, 70].
Nrf2 in Cytoprotection, Cancer and Drug Resistance
Nrf2 is a major protective mechanism against xenobiotics capable of damaging DNA and initiating carcinogenesis [71]. Inducers of Nrf2 function as blocking agents that prevents carcinogens from reaching target sites, inhibits parent molecules undergoing metabolic activation, or subsequently preventing carcinogenic species from interacting with crucial cellular macromolecules, such as DNA, RNA, and proteins [72]. A plausible mechanism by which blocking agents impart their chemopreventive activity is the induction of detoxification and antioxidant enzymes [73]. Oltipraz, 3H-1,2,-dithiole-3-thione (D3T), Sulforaphane, and Curcumin can be considered potential chemopreventive agents because these compounds have all been shown to induce Nrf2 [74–81].
Studies have shown a role of Nrf2 in protection against cadmium and manganese toxicity [82]. Nrf2 also plays an important role in reduction of methyl mercury toxicity [83]. Methylmercury activates Nrf2 and the activation of Nrf2 is essential for reduction of methylmercury by facilitating its excretion into extracellular space. In vitro and in vivo studies have shown a role of Nrf2 in neuroprotection and protection against Parkinson’s disease [84–86]. Disruption of Nrf2 impairs the resolution of hyperoxia-induced acute lung injury and inflammation in mice [87]. Nrf2-knockout mice were more prone to tumor growth when exposed to carcinogens such as benzo[a]pyrene, diesel exhaust, and N-nitrosobutyl (4-hydroxybutyl) amine [88–90]. INrf2/Nrf2 signaling is also shown to regulate oxidative stress tolerance and lifespan in Drosophila [91].
A role of Nrf2 in drug resistance is suggested based on its property to induce detoxifying and antioxidant enzymes (92–97). The loss of INrf2 (Keap1) function is shown to lead to nuclear accumulation of Nrf2, activation of metabolizing enzymes and drug resistance (95). Studies have reported mutations resulting in dysfunctional INrf2 in lung, breast and bladder cancers (96–100). A recent study reported that somatic mutations also occur in the coding region of Nrf2, especially in cancer patients with a history of smoking or suffering from squamous cell carcinoma (101). These mutations abrogate its interaction with INrf2 and nuclear accumulation of Nrf2. This gives advantage to cancer cell survival and undue protection from anti-cancer treatments. However, the understanding of the mechanism of Nrf2 induced drug resistance remains in its infancy. In addition, the studies on Nrf2 regulated downstream pathways that contribute to drug resistance remain limited.
Future Perspectives
Nrf2 creates a new paradigm in cytoprotection, cancer prevention and drug resistance. Considerable progress has been made to better understand all mechanisms involved within the intracellular pathways regulating Nrf2 and its downstream genes. Preliminary studies demonstrate that deactivation of Nrf2 is as important as activation of Nrf2. Further studies are needed to better understand the negative regulation of Nrf2. Also better understanding of the negative regulation of Nrf2 could help design a new class of effective chemopreventive compounds not only targeting Nrf2 activation, but also targeting the negative regulators of Nrf2.
Acknowledgments
This work was supported by NIGMS grant RO1 GM047466 and NIEHS grant RO1 ES012265.
Abbreviations
- Nrf2
NF-E2 related factor 2
- INrf2
Inhibitor of Nrf2 also known as Keap1
- ROS
Reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Breimer LH. Molecular Mechanisms of oxygen radical carcinogenesis and mutagenesis: the role of DNA base damage. Mol Carcinog. 1990;3:188–197. doi: 10.1002/mc.2940030405. [DOI] [PubMed] [Google Scholar]
- 2.Meneghini R. Iron homeostasis, oxidative stress, and DNA damage. Free Radic Biol Med. 1997;23:783–792. doi: 10.1016/s0891-5849(97)00016-6. [DOI] [PubMed] [Google Scholar]
- 3.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 4.Bauer CE, Elsen S, Bird TH. Mechanisms for redox control of gene expression. Annu Rev Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 5.Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharm. 2000;59:1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 6.Dhakshinamoorthy S, Long DJ, II, Jaiswal AK. Antioxidant regulation of genes encoding enzymes that detoxify xenobiotics and carcinogens. Current Topics in Cellular Regulation. 2000;36:201–206. doi: 10.1016/s0070-2137(01)80009-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Copple IM, Goldring CE, Kitteringham NR, Park BK. The Nrf2-Keap1 defense pathway: role in protection against drug-induced toxicity. Toxicology. 2008;246:24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 10.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 4. Oxford University Press; 2007. [Google Scholar]
- 11.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Actiavtion by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 12.Rushmore TH, King RG, Paulson KE, Pickett CB. Regulation of glutathione S-transferase Ya subunit gene expression: identification of a unique xenobiotics-responsive element controlling inducible expression by planar aromatic compounds. Proc Natl Acad Sci USA. 1990;87:3826–3830. doi: 10.1073/pnas.87.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie T, Belinsky M, Xu Y, Jaiswal AK. ARE and TRE-mediated regulation of gene expression: response to xenobiotics and antioxidants. J Biol Chem. 1995;270:6894–6900. doi: 10.1074/jbc.270.12.6894. [DOI] [PubMed] [Google Scholar]
- 14.Rushmore TH, Pickett CB. Glutathione S-transferases, structures, regulation, and therapeutic implications. J Biol Chem. 1993;268:11475–11478. [PubMed] [Google Scholar]
- 15.Jaiswal AK. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic Biol Med. 2000;29:254–252. doi: 10.1016/s0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- 16.Bloom D, Dhakshinamoorthy S, Wang W, Celli CM, Jaiswal AK. Role of NF-E2 related factors in oxidative stress. In: Storey KB, Storey JM, editors. Cell and Molecular responses to stress vol 2 Protein adaptation and signal transduction. Amsterdam; Elsevier: 2001. pp. 229–238. [Google Scholar]
- 17.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alam J, Stewart D, Touchard C, Boinapally S, Choi MK, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 19.Wild AC, Moinova HR, Mulcahy RT. Regulation of γ-glutamylcysteine synthetase subunit gene expression by transcription factor Nrf2. J Biol Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element: activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 21.Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orikin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic leucine zipper protein. Nature. 1993;339:722–727. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 22.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 23.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JY, Han XL, Kan YW. Isolation of cDNA encoding human NF-E2 protein. Proc Natl Acad Sci USA. 1993;90:11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohler J, Mahaffey JW, Deutsch E, Vani K. Control of Drosphila head segment identity by the bZIP homeotic gene cnc. Development. 1995;121:237–247. doi: 10.1242/dev.121.1.237. [DOI] [PubMed] [Google Scholar]
- 26.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh K, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small maf heterodimers mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 27.Venugopal R, Jaiswal AK. Nrf2 and Nrf2 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 28.Kwak MK, Cho JM, Huang B, Shin S, Kensler TW. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neruoblastoma cells. Free Radic Biol Med. 2007;43:809–917. doi: 10.1016/j.freeradbiomed.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 30.Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–962. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- 31.Slitt AL, Cherrington NJ, Dieter MZ, Aleksunes LM, Scheffer GL, Huang W, Moore DD, Klaassen CD. trans-Stilbene oxide induces expression of genes involved in metabolism and transport in mouse liver via CAR and Nrf2 transcription factors. Mol Pharmacol. 2006;69:1554–1563. doi: 10.1124/mol.105.014571. [DOI] [PubMed] [Google Scholar]
- 32.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- 33.Maher JM, Aleksunes LM, Dieter MZ, Tanaka Y, Peters J, Manautou JE, Klaassen CD. Nrf2 and PPAR{alpha}-Mediated Regulation of Hepatic Mrp transporters after Exposure to Perfluorooctanoic Acid and Perfluorodecanoic Acid. Toxicol Sci. 2008;106:319–328. doi: 10.1093/toxsci/kfn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to that amino-terminal Neh2 domain. Genes Dev. 1999;13:76–96. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhakshinamoorthy S, Jaiswal AK. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2001;20:3906–3917. doi: 10.1038/sj.onc.1204506. [DOI] [PubMed] [Google Scholar]
- 36.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 37.Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/Kelch proteins in animals. BMC Bioinformatics. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA-and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 39.Kim IF, Mohammadi E, Huang RCC. Isolation and characterization of IPP, a novel human gene coding an actin-binding KELCH-like protein. Gene. 1999;228:73–83. doi: 10.1016/s0378-1119(99)00006-2. [DOI] [PubMed] [Google Scholar]
- 40.Kang M, Kobayashi A, Wakabayashi N, Kim S, Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as a key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci USA. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functionas as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin3-Rox1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekhar KR, Yan XX, Freeman ML. Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene. 2002;21:6829–6834. doi: 10.1038/sj.onc.1205905. [DOI] [PubMed] [Google Scholar]
- 46.Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin–proteasome pathway and stabilization by cadmium. J Biol Chem. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2- mediated transcriptional activation of the antioxidant response element: degradation of Nrf2 by the 26S proteasome. J Biol Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 48.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M. Keap1 regulates both cytoplasmic–nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 49.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser(40) by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Long MJ, Santamaria AB, Talalay P. Role of cytochrome P1-450 in the induction of NAD(P)H:quinone reductase in a murine hepatoma cell line and its mutants. Carcinogenesis. 1987;8:1549–1553. doi: 10.1093/carcin/8.10.1549. [DOI] [PubMed] [Google Scholar]
- 52.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–74. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 53.Buckley BJ, Marshall ZM, Whorton AR. Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochem Biophys Res Commun. 2003;307:973–979. doi: 10.1016/s0006-291x(03)01308-1. [DOI] [PubMed] [Google Scholar]
- 54.Yu R, Mandlekar S, Weber MJ, Der CJ, Wu J, Tony-Kong AN. Role of mitogen-activated protein kinase pathway in the induction of phaseII detoxifying enzymes by chemicals. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 55.Zipper LM, Mulcahy RT. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 2000;278:484–492. doi: 10.1006/bbrc.2000.3830. [DOI] [PubMed] [Google Scholar]
- 56.Cullinan SB, Zhang D, Hannik M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 58.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophilie-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad SCi USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain AK, Mahajan S, Jaiswal AK. Phosphorylation and dephosphorylation of tyrosine 141 regulate stability and degradation of INrf2: A novel mechanism in Nrf2 activation. J Biol Chem. 2008;283:17712–17720. doi: 10.1074/jbc.M709854200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Niture SK, Jaiswal AK. Prothymosin-a mediates nuclear import of INrf2/Cul3-Rbx1 complex to degrade nuclear Nrf2. J Biol Chem. 2009;284:13856–13868. doi: 10.1074/jbc.M808084200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Lee O, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 62.Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H, Yamamoto M. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J Biol Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 63.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–95. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;11:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280:16891–900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 67.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 68.Kannoji A, Phukan S, Sudher Babu V, Balaji VN. GSK3beta: a master switch and a promising target. Expert Opin Ther Targets. 2008;11:1443–1455. doi: 10.1517/14728222.12.11.1443. [DOI] [PubMed] [Google Scholar]
- 69.Jain AK, Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem. 2007;282:16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]
- 70.Jain AK, Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J Biol Chem. 2006;281:12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 71.Giudice A, Montella M. Activation of the Nrf2-ARE signaling pathway: a promising strategy in cancer prevention. Bioessays. 2006;28:169–181. doi: 10.1002/bies.20359. [DOI] [PubMed] [Google Scholar]
- 72.Surh Y-J. Cancer chemoprevention with dietary phytochemicals. Nature Rev Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 73.Lee J-S, Surh Y-J. Nrf2 as a novel molecular target for chemoprevention. Cancer Letters. 2005;224:171–184. doi: 10.1016/j.canlet.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 74.Kang KW, Cho IJ, Lee CH, Kim SG. Essential role of phosphatidylinositol 3-kinase-dependent CCAAT/enhancer binding protein beta activation in the induction of glutathione S-transferase by oltipraz. J Natl Cancer Inst. 2003;95:53–66. doi: 10.1093/jnci/95.1.53. [DOI] [PubMed] [Google Scholar]
- 75.Kensler TW, Qian GS, Chen JG, Groopman D. Translational strategies for cancer prevention in liver. Nature Rev Cancer. 2003;3:321–329. doi: 10.1038/nrc1076. [DOI] [PubMed] [Google Scholar]
- 76.Pietsch EC, Chan JY, Torti FM, Torti V. Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones. J Biol Chem. 2003;278:2361–2369. doi: 10.1074/jbc.M210664200. [DOI] [PubMed] [Google Scholar]
- 77.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1–Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 78.Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, Yamamoto M, Kensler TW. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat Res. 2001;480/481:305–315. doi: 10.1016/s0027-5107(01)00190-7. [DOI] [PubMed] [Google Scholar]
- 79.Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the Nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1- dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 81.Balogun E, Hoque M, Gong P, Killeen E, Green CJ, Foresti R, Alam J, Motterlini R. Curcumin activates the haemoxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Casalino E, Calzaretti G, Landriscina M, Sblano C, Fabiano A, Landriscina C. The Nrf2 transcription factor contributes to the induction of alpha-class GST isoenzymes in liver of acute cadmium or maganese intoxicated rats: comparison with the toxic effect on NAD(P)H:quinone oxidoreductase. Toxicology. 2007;237:24–34. doi: 10.1016/j.tox.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 83.Toyama T, Sumi D, Shinkai Y, Yasutake A, Taguchi K, Tong KI, Yamamoto M, Kumagai; Y. Cytoprotective role of Nrf2/Keap1 system in methylmercury toxicity. Biochem Biophys Res Commun. 2007;363:645–650. doi: 10.1016/j.bbrc.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 84.Rajo AI, Rada P, Egea J, Rosa AO, Lopez MG, Cuadrado A. Functional interference between glycogen synthase kinase-3 beta and the transcription factor Nrf2 in protection against kainate-induced hippocampal cell death. Mol Cell Neurosci. 2008;39:125–132. doi: 10.1016/j.mcn.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 85.Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, Chen PC. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann N Y Acad Sci. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen PC, Vargas MR, Pank AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: critical role for the astrocyte. Proc Natl Acad Sci USA. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reddy NM, Kleeberger SR, Kensler TW, Yamamoto M, Hassoum PM, Reddy SP. Disruption of Nrf2 impairs the resolution of hyperoxia-induced acute lung injury and inflammation in mice. J Immunol. 2009;182:7264–7271. doi: 10.4049/jimmunol.0804248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharm. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 89.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme induccers is lost in Nrf2 transcription factor deficient mice. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, Shimazui T, Akaza H, Yamamoto M. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 91.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vollrath V, Wielandt AM, Iruretagoyena M, Chianale M. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 94.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 95.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng YX, Wondrack GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provided by its point mutations in lung cancer. Mol Cell. 2006;21:689–734. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 97.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional Keap1-Nrf2 interaction in non-small-cell lung cancer. PLos Med. 2006;3:1865–1876. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 99.Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun. 2007;362:816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 100.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 101.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in Nrf2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]