Abstract
The retinoblastoma gene Rb is a prototype tumor suppressor which is conserved in Drosophila. Although much is known about the roles of Rb in cell proliferation and apoptosis, much less is known about how Rb regulates cell differentiation. Inactivation of Drosophila Rb (rbf) exhibited subtle differentiation defects similar to inactivation of Rb in mice, suggesting the existence of redundant mechanisms in the control of cell differentiation. To test this possibility and to characterize the role of Rbf in cell differentiation during retinal development, we carried out a genetic screen and identified a mutation in rhinoceros (rno), which leads to synergistic differentiation defects in conjunction with rbf inactivation. Characterization of an early differentiation defect, the multiple-R8 phenotype, revealed that this phenotype was caused by limiting amounts of Notch signaling due to reduced expression of the Notch ligand, Delta (Dl). Decreasing the gene dosage of Dl enhanced the multiple-R8 phenotype, while increasing the level of Dl suppressed this phenotype. Interestingly, removal of the transcriptional activation of dE2F1 partially restores Dl expression in rbf,rno mutant clones and suppresses the associated differentiation defects, indicating that this differentiation function of RBF is mediated by its regulation of dE2F1 activity.
Keywords: Rbf, Rno, Delta, R8 determination, differentiation
Introduction
The retinoblastoma protein pRb is a prototype tumor suppressor that is often mutated or inactivated in cancers (Classon and Harlow, 2002; Weinberg, 1995). pRb regulates a variety of normal cellular processes including cell proliferation, differentiation, as well as apoptosis (Du and Pogoriler, 2006; van den Heuvel and Dyson, 2008). Extensive studies have shown that pRb exerts its function by forming complexes with other proteins. The best-studied Rb interacting proteins are the E2F transcription factors, which are heterodimers composed of a subunit of the E2F family and a subunit of the DP family. In mammalian systems, there are eight E2F and three DP family members (DeGregori and Johnson, 2006). Although there is an abundance of evidence that E2F proteins mediate the role of Rb in cell proliferation and apoptosis regulation, it is not clear to what extent E2F mediate the differentiation function of Rb. In addition to E2F, pRb also binds to a large number of other proteins (Morris and Dyson, 2001), including a number of transcription factors involved in the differentiation of specific cell types such as MyoD, C/EBP β, and CBFA1. In vitro studies using cell culture systems have suggested that pRb directly interacted with and enhanced the activities of these transcription factors to promote differentiation (Chen et al., 1996; Gu et al., 1993; Novitch et al., 1999; Thomas et al., 2001). However, the significance of these cell culture-based observations has not been demonstrated in vivo in animal models. The study of Rb’s role in differentiation in vivo is complicated because differentiation defects observed in Rb mutant mice are relatively subtle and are generally associated with deregulation of the cell cycle and/or apoptosis (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992). In addition, the existence of a large family of E2F proteins in mammals also makes it difficult to examine the contribution of E2F proteins in mediating the differentiation function of Rb in vivo. Therefore, it is often difficult to determine whether the observed developmental defects in Rb mutants are simply consequences of deregulated cell cycle or apoptosis, and even more difficult to characterize the importance of E2F for various phenotypes of Rb mutants due to the presence of the large family of E2F proteins with overlapping functions.
The Rb/E2F pathway is highly conserved and much simpler in Drosophila. There is only one DP (dDP), two dE2F (dE2F1 and dE2F2), and two Rb family proteins (RBF and RBF2) in Drosophila (Du et al., 1996a; Dynlacht et al., 1994; Ohtani and Nevins, 1994; Sawado et al., 1998; Stevaux et al., 2002). The two Drosophila E2F proteins behave like the two different classes of mammalian E2Fs: dE2F1 mainly functions as a transcriptional activator (Du, 2000) similar to the activating E2Fs (E2F1-3), while dE2F2 mainly functions to mediate active repression similar to the repressive E2Fs (E2F4-5) in mammalian systems (Frolov et al., 2001). Similar to mammalian Rb, RBF can bind to both the activating E2F (dE2F1) as well as the repressive E2F (dE2F2), while RBF2 binds specifically to dE2F2, similar to the preferential binding of p107/p130 to the repressive E2F proteins in mammals (Stevaux et al., 2002). This simple and yet highly conserved Rb/E2F pathway, in conjunction with the available genetic and developmental tools, makes Drosophila an attractive model to study the role of Rb in vivo.
Similar to Rb knockout mice, rbf mutant flies exhibit deregulated cell proliferation and apoptosis but only subtle differentiation defects (Du, 2000; Du and Dyson, 1999). The subtle differentiation defects observed in Rb knockout mice or rbf mutant flies could be due to the existence of partially redundant mechanisms in the control of proper cell differentiation, similar to the role of Rb (lin-35) on multiple vulval induction in C. elegans (Lu and Horvitz, 1998). To test this possibility and to identify the genes that are required for the proper differentiation of rbf mutant cells in the developing eye, we have carried out a genetic screen to identify mutations that can lead to synergistic differentiation defects in conjunction with rbf inactivation.
The Drosophila developing eye has been extensively used as a model system to study cell proliferation and differentiation during development. Photoreceptor differentiation in the Drosophila developing eye initiates in the morphogenetic furrow (MF), which moves from the posterior of the eye disc to the anterior during the third larval instar. The first photoreceptor determined is R8, which can be identified by the expression of Senseless (Sens) (Frankfort et al., 2001). R8 determination is regulated by the expression of the bHLH protein Atonal (Ato), which is initially expressed in all the cells in the MF, and later Ato expression is upregulated and restricted to clusters of cells and finally to single R8 cells immediately posterior to the MF (Dokucu et al., 1996; Sun et al., 1998). Notch signaling is required both for the upregulation of Ato expression as well as for the restriction of Ato expression to single R8 cells (Baker and Yu, 1997; Baonza and Freeman, 2001; Li and Baker, 2001). Following R8 specification, EGFR signaling is required for the step wise recruitment of additional photoreceptor cells, cone cells, and other accessory cells to form the ommatidia (Freeman, 1996).
Here, we report the identification of a mutation that exhibited synergistic differentiation defects in conjunction with rbf inactivation. We found that this mutation was an allele of rhinoceros (rno) (Voas and Rebay, 2003). Characterization of rno and rbf single as well as rbf,rno double mutant clones in the developing eye revealed weak differentiation defects in each of the single mutant clones but much more dramatic defects in the double mutant clones. We further characterized an early differentiation defect, the multiple-R8 phenotype, and showed that this phenotype was caused by a limiting amount of Notch signaling due to reduced expression of the Notch ligand, Delta (Dl). Decreasing the gene dosage of Dl enhanced the multiple-R8 phenotype while increasing the level of Dl decreased the incidence of multiple-R8. Furthermore, we found that the differentiation effect of rbf is mediated by its regulation of dE2F1 activity, since removal of transcriptional activation by dE2F1 partially restores Dl expression in rbf,rno mutant clones and suppresses the associated differentiation defects.
Results
Mutation of rno synergizes with loss of rbf in inducing adult eye defects
We have used a RBF genomic rescue construct on 3L (RBF-G3) to carry out a genetic screen on this arm of the chromosome to identify genes that can modulate the consequences of rbf inactivation (see Materials and Methods for details). From this screen, we identified a mutation, 1k, which exhibited a much more dramatic adult eye defect in conjunction with rbf mutation than the defects caused by either mutation alone. As shown in Fig. 1, rbf and 1k single mutant clones exhibited only weak defects and the exterior morphology in adult eyes were largely normal (white patches in Fig. 1A,B). In contrast, rbf,1k double mutant clones exhibited very severe defects characterized by shiny eye surfaces and irregular ommatidia shape and disorganization of the adult eye structure (Fig. 1C, white patches). Therefore, 1k mutation synergizes with loss of rbf in inducing severe eye developmental defects.
Figure 1.
Abnormal adult eye morphology of rbf,rno mutant clones. Images of adult eyes carrying mosaic clones of different genotypes as indicated are shown. Mutant tissues correspond to the white patches. Clones that are singly mutant for rbf (A), 1k (B), or rno3 (E) did not show striking defects in morphology of the adult eye. In contrast, double mutant clones of rbf,1k (C) or rbf,rno3 (F) had significant defects. Blocking apoptosis by mutation of dronc in rbf,dronc (G), rbf,1k,dronc (H), and rbf,rno3,dronc (I) increased clone sizes but did not alter the morphological defects (compare A with G, C with H, and F with I).
Recombination mapping of the 1k mutation showed that it lies close to the tip of chromosome 3L. Further deficiency mapping and complementation tests identified that 1k is an allele of rno based on following observations: 1) 1k mutation failed to complement the lethality of two previously identified null alleles of rno, rno1 and rno3; 2) both rno1 and rno3 alleles showed the same interaction with loss of rbf as the 1k mutation in inducing the severe shiny eye differentiation defects; and 3) single mutant clones of the rno alleles exhibited only slight defects similar to those of the 1k mutant clones (Fig. 1E, F and data not shown). These results indicated that 1k is an allele of rno (referred to as rnoK), which encodes a large PHD domain-containing protein that showed genetic interaction with the EGFR signaling pathway (Voas and Rebay, 2003).
Synergistic differentiation defects in larval and pupal eye discs in the absence of rbf and rno
At high magnification, the shiny rbf,rno double mutant clones in adult eyes were largely devoid of bristles. This no-bristle phenotype was also observed in pupal eye discs stained with Phalloidin. As shown in Fig. 2A–C, no bristles were observed in rbf,rno double mutant clones (marked by absence of GFP) even though they were observed in the surrounding normal tissues (Fig. 2C). In contrast, bristles were observed in both rbf and rno single mutant clones (Fig. 2A and B).
Figure 2.
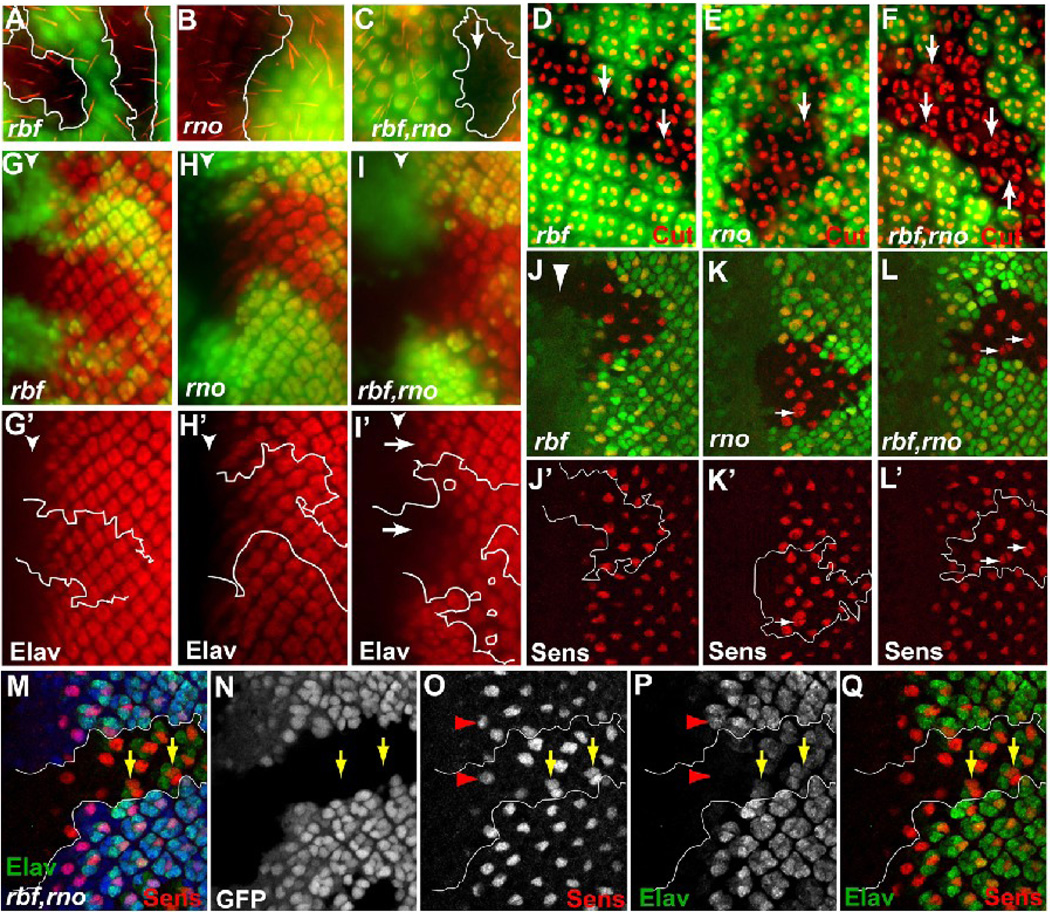
Eye differentiation phenotypes of rbf,rno mutant clones at the larval and pupal stages. For these and all subsequent images of the larval eye disc, anterior is to the left, mutant tissues are marked by the absence of GFP. (A–F) Pupal eye discs containing indicated mutant clones were stained with Phalloidin to visualize bristles (A–C) or anti Cut antibody to visualize developing cone cells (D–F). Arrows in (D–F) point to ommatidia with different numbers of cone cells. (G–L’) 3rd instar larval eye discs containing indicated mutant clones were stained with anti Elav antibody to visualize developing photoreceptors (G–I’) and anti Sens antibody to visualize developing R8 cells (J–L’). Arrows in (I’) show delayed photoreceptor differentiation in rbf,rno clones relative to neighboring wild-type tissue. Arrows in K–L’ point to occasional “multiple-R8” phenotype in rno clones (arrows in K and K’) and more frequent multiple-R8 phenotype in rbf,rno (arrows in L and L’). (M–Q) Sens and Elav co-labeling in rbf,rno clones showed that multiple-R8s were present within single Elav clusters (yellow arrows in M–Q).
The shiny exterior of the adult eye of rbf,rno mutant clones could be related to cone cell defects. We used an antibody against Cut, which labels the cone cells in the developing eye discs, to compare the cone cell developmental defects in rbf or rno single mutant clones and in rbf,rno double mutant clones. At 48 hours after puparium formation (APF), Cut-positive cone cells can be observed in rbf and rno single mutant clones as well as in rbf,rno double mutant clones. While WT ommatidia contain four cone cells per cluster, rbf mutant clusters contain between three to five cone cells per cluster (Fig. 2D, arrows). On the other hand, rno mutant ommatidia often exhibited five cone cells per cluster (Fig. 2E, arrow), consistent with the previous report (Voas and Rebay, 2003). Interestingly, rbf,rno double mutant ommatidia exhibited much more variable number of cone cells per cluster, with some clusters containing almost twice the number of normal cone cells (Fig. 2F, arrows). In addition, the ommatidia arrangements were much more disorganized in rbf,rno double mutant clones (Fig. 2F).
As cone cells are recruited into the ommatidia clusters after the photoreceptor cells, it is possible that the observed cone cell defects were consequences of defects in photoreceptor differentiation. Therefore we examined the photoreceptor differentiation using Elav and Senseless, which label all developing photoreceptor cells and R8, respectively. Interestingly, staining with Sens and Elav revealed that two or more R8s (multiple-R8) were often found to be in the same cluster in the rbf,rno double mutant clones (Fig. 2L–Q arrows). The incidences of multiple-R8 in rbf,rno1 and rbf,rno3 are 19.3±3.6% and 18.4±5.9%, respectively (Fig. 4A). Occasionally multiple-R8 was observed at the border of the mutant clones and involving one WT and one mutant cell, however multiple-R8 from two WT cells was never observed (over 500 positions counted). Careful examination of the rbf and rno single mutant clones revealed a low level of multiple-R8 in rno single mutant clones (Fig. 2K, incidences are 3.2±2.5% and 3.5±2.5% for rno1 and rno3 single mutant clones, respectively) but no multiple-R8 in rbf single mutant clones (Fig. 2J, J’). The incidence of multiple-R8s are significantly different between rno1 and rbf,rno1 (P<0.006) and between rno3 and rbf,rno3 (P<0.0001). In contrast, no significant difference in the multiple-R8 incidence was observed between rno1 and rno3 (P=0.80) or between rbf,rno1 and rbf,rno3 (P=0.76). Therefore, rno1 and rno3 exhibited the same multiple-R8 phenotype and showed similar synergistic differentiation defects in conjunction with loss of rbf. Interestingly, although R8 photoreceptor differentiation was not obviously delayed as identified by Sens staining (Fig. 2O, Q), the differentiation of the remaining photoreceptors were significantly delayed in rbf,rno double mutant clones as judged by Elav staining (Fig. 2M–Q, I, I’, an average of three rows of delay, N>10 clones). On the other hand, photoreceptor cell differentiation as judged by Elav staining was not detectably delayed in rbf single mutant clones, but was slightly delayed in rno single mutant clones (Fig. 2G–H’, an average of one row of delay, N>10 clones).
Figure 4.
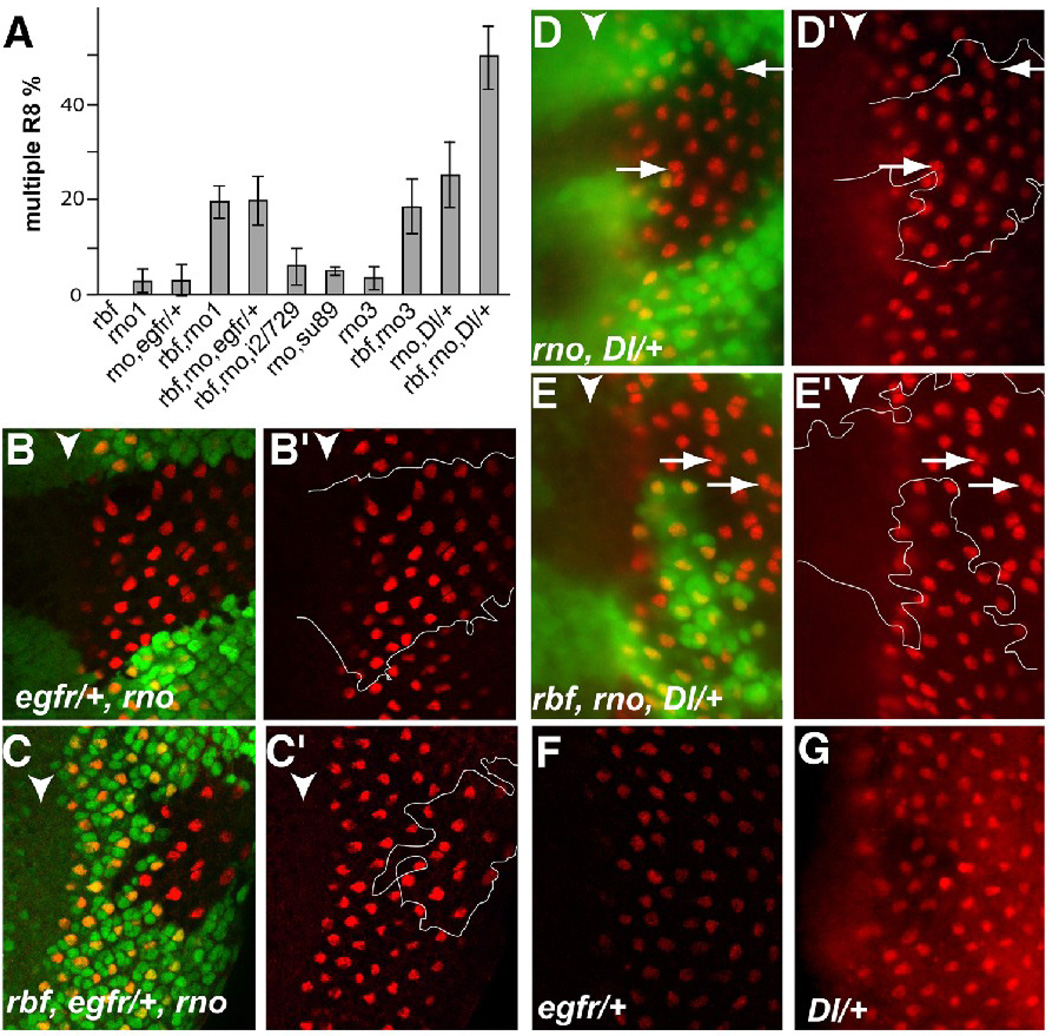
Reduction of Dl gene dosage enhanced the multiple-R8 phenotype of rbf,rno. (A) A diagram summarizes the incidences of multiple-R8 phenotypes in different genetic background as indicated. i2/729 and su89 indicate de2f1i2/de2f1rm729 and de2f1su89 mutant backgrounds, respectively. (B–G) Sens staining was used to characterize the multiple-R8 phenotype. Gene dosage reduction of egfr did not affect multiple-R8 incidence in rno (B, B’) or rbf,rno clones (C,C’). In contrast, gene dosage reduction of the Notch signaling ligand Dl led to a significant increase in multiple-R8 phenotype in both rno and rbf,rno clones (D, D’ and E, E’ respectively). Heterozygosity of egfr (F) or Dl (G) does not induce multiple-R8 phenotype.
In conclusion, the rbf,rno mutant cells exhibited multiple synergistic differentiation defects, including the multiple-R8 phenotype, delayed photoreceptor differentiation, and defective cone cell differentiation.
The rbf,rno double mutant phenotypes are unlikely caused by a modulation of the apoptosis or proliferation of rbf mutant cells
Mutation of rbf in the developing eye leads to ectopic cell proliferation in the posterior and increased apoptosis near the MF (Du, 2000; Firth and Baker, 2005; Moon et al., 2006). It is possible that the observed severe differentiation defects in rbf,rno double mutant clones are caused by a modulation of the effect of apoptosis or ectopic proliferation induced by rbf mutation. Consistent with previous reports, rbf mutant clones led to significant amounts of apoptosis near the furrow as shown by increased staining by activated caspase-3 (C3) staining (Fig. 3A). In contrast, no significant C3 staining was observed in rbf mutant clones in the far anterior or in the far posterior eye disc (Fig. 3A). C3 staining of eye discs with rno or rbf,rno mutant clones showed that mutation of rno alone did not increase apoptosis (Fig. 3B) and that rbf,rno double mutant clones exhibited the same pattern of apoptosis as rbf single mutant clones, with significantly increased apoptosis near the furrow (Fig. 3A,C). Furthermore, while mutation of dronc in conjunction with rbf and rno significantly decreased the level of apoptosis (Fig. 3D) and led to the development of adult eyes with larger mutant clones (white patches in Fig. 1G–I), mutation of dronc did not suppress the adult eye phenotypes of rbf,rno mutant clones (Fig. 1H–I) or the multiple R8 phenotype. The level of multiple R8 in rbf,rno,dronc clones were also determined to be 14±4%, which is not significantly different from the level of multiple R8 in rbf,rno double mutant clones (P>0.1). Therefore, the observed rbf,rno mutant phenotypes are unlikely to result from an effect of rno mutation on the apoptosis induced by rbf.
Figure 3.
Cell proliferation and apoptosis assay of rbf and rno mutant clones. (A–D) Analysis of apoptosis via activated caspase-3 (C3) staining. rbf clones (A) showed increased levels of apoptosis, particularly in the region near the morphogenetic furrow (marked by white arrowhead). rno clones (B) showed no increased apoptosis. Clones of cells that are mutant for both rbf and rno (C) have similar pattern of apoptosis as those observed for rbf clones. Mutation of dronc significantly decreased the level of apoptosis of in rbf,rno,dronc mutant clones (D). (E–G) Analysis of cell proliferation by EdU incorporation. The second mitotic wave (SMW) is marked by the white arrows. Yellow arrows point to EdU-positive cells posterior to the SMW, a region of the eye disc where cells are normally non-proliferating. rbf clones (E) contain EdU-positive cells posterior to the SMW while rno clones (F) did not. Clones of cells that are mutant for both rbf and rno (G) also exhibited ectopic EdU incorporation in the posterior of the eye disc. Anterior is to the left.
Cell proliferation in the developing eye discs were analyzed by the Click-it Edu cell proliferation assay system (Salic and Mitchison, 2008). As shown in Fig. 3E–G, EdU incorporation assay showed that rbf,rno double mutant clones exhibited increased EdU incorporation in the posterior similar to that observed in rbf single mutant clones (Fig. 3E, G). S phase cells/unit area in the posterior rbf and rbf,rno clones are 6.8±4.6 and 7.9±4.9, respectively. Student’s t-tests showed that S phase cells numbers are not significantly different between the rbf and rbf,rno clones (P=0.5). In contrast, rno mutation alone did not have a significant effect on EdU incorporation in the developing eye disc (Fig. 3F). The number of S phase cells/unit area in rno clones in the posterior eye disc is 1.1±0.7, which is the same as the number of S phase cells in WT areas from the same eye discs (1.1±0.7) but significantly different from those in rbf or rbf,rno clones in the posterior eye discs (P≤0.01).
Therefore, the observed synergistic differentiation defects in rbf,rno mutant clones are unlikely to result from a modulation of the cell proliferation or apoptosis effect of rbf in the absence of rno. This is consistent with the observation that the rbf,rno mutant phenotypes are distinct from phenotypes resulting from increased or decreased cell proliferation (de Nooij and Hariharan, 1995; Du et al., 1996b). Since Drosophila eye development initiates with R8 determination, which in turn recruits additional photoreceptors, cone cells, and other accessory cells, we focused on the characterization of the multiple-R8 phenotype observed in rno and rbf,rno mutant clones in the remainder of this study to determine how rbf regulates differentiation.
Reducing the dosage of Dl significantly enhanced the multiple-R8 phenotype of rno and rbf,rno mutant clones
Multiple-R8 phenotypes have been observed in eye discs with mutation or overexpression that affect the Notch or the EGFR signaling. For example, mutations in scabrous or echinoid, or expression of activated Ras, activated Raf or Pointed-P1, all lead to multiple-R8 phenotypes (Rawlins et al., 2003; Spencer and Cagan, 2003; Spencer et al., 1998). In addition, overexpression of Ato or mutation of rough also led to multiple-R8s (Dokucu et al., 1996). To determine if the multiple-R8 phenotype of rbf,rno clones was sensitive to changes in the level of Notch or EGFR signaling, we carried out experiments to determine the genetic interactions between rno or rbf,rno and Dl or egfr.
As shown in Fig. 4, reducing the gene dosage of egfr did not significantly affect the incidence of multiple-R8s either in rno1 single mutant clones or in rbf,rno1 double mutant clones (Fig. 4B–C’). The incidence of multiple-R8s in rno1 mutant clones with reduced EGFR gene dosage was 2.8±3.3%, not significantly different from that in rno1 mutant clones with normal EGFR gene dosage (Fig. 4A, P=0.83). Similarly, the multiple-R8 incidence in rbf, rno1 mutant clones with reduced EGFR gene dosage was 19.6±5.6%, which was not significantly different from that in rbf, rno1 mutant clones with normal EGFR gene dosage (Fig. 4A, P=0.92). In contrast, previous studies showed that reducing the gene dosage of egfr significantly reduced the incidence of multiple-R8s that resulted from the echinoid mutation (Rawlins et al., 2003). Therefore, the multiple-R8 phenotype in the rbf,rno mutant clones was not sensitive to changes in the gene dosage of egfr even though gene dosage reduction of egfr was sufficient to suppress the multiple-R8 phenotype of the echinoid mutation. These observations suggest that distinct mechanisms are involved in the development of multiple-R8 phenotypes between rbf,rno and echinoid.
Since Notch signaling has also been linked with the multiple-R8 phenotype, we tested the effect of altering the signaling of the Notch pathway on the multiple-R8 phenotype in rbf,rno clones. Removing one copy of Dl increased the incidence of multiple-R8s in rno1 mutant clones from 3.2±2.5% to 26.3±7.2% (P<0.0001, Fig. 4A, D, D’). Similarly, reducing the gene dosage of Dl also significantly increased the incidence of multiple-R8s in rbf,rno mutant clones from 18.4±5.9% to 49.2±6.4% (Fig. 4A, E, E’, P<0.0001). It should be pointed out that reducing the gene dosage of either egfr or Dl alone did not lead to the multiple-R8s (Fig. 4F and G). Therefore, the multiple-R8 phenotypes observed in rno and rbf,rno clones are very sensitive to changes in Dl gene dosage.
Reduced expression of Dl in rno and rbf,rno mutant clones
Dl expression is first detected in the morphogenetic furrow in the developing eye disc and later elevated Dl levels are observed in the developing photoreceptor clusters (Baker and Yu, 1998; Baker and Zitron, 1995; Parks et al., 1995). To determine if Dl expression was affected in rno and rbf,rno mutant clones, we first examined the level of Dl protein in rno and rbf,rno mutant clones. As shown in Fig. 5, removing rbf alone had no effect on the level of Dl protein while removing rno alone led to a slight reduction (Fig. 5A–B’). On the other hand, removing both rno and rbf led to a strong reduction in the level of Dl protein (Fig. 5C, C’), indicating that loss of both rbf and rno caused a synergistic decrease in the level of Dl protein.
Figure 5.
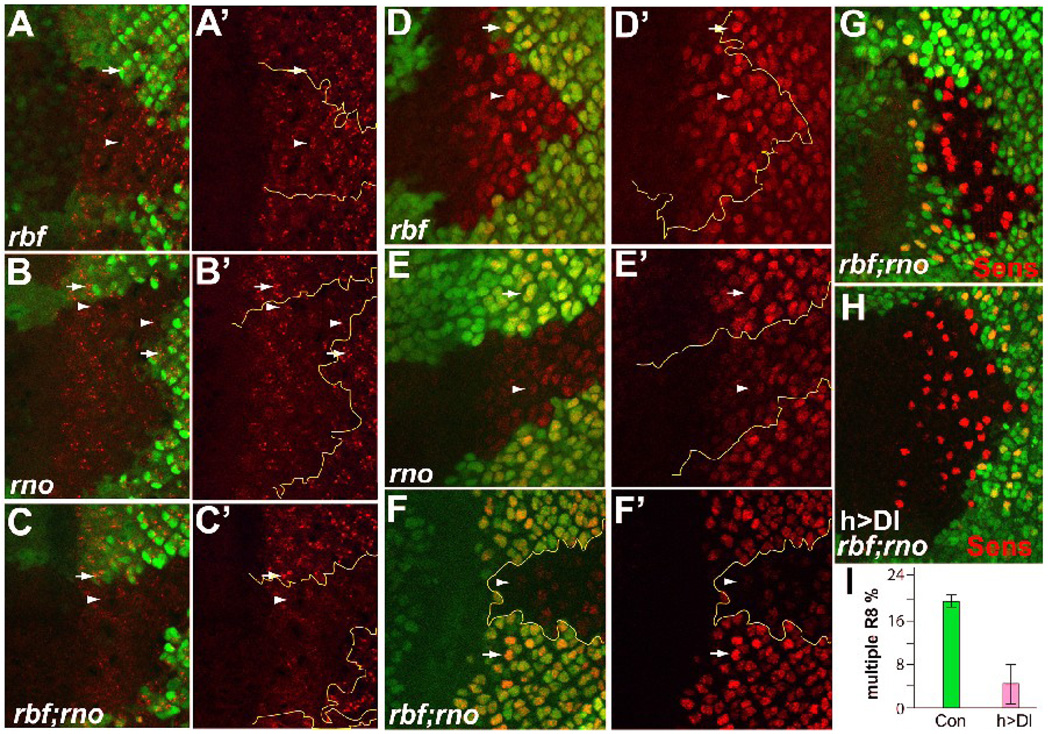
Expression of Dl was reduced in rbf,rno mutant clones. (A–F’) Antibody staining to visualize the level of Dl protein (A–C’) and Dl-lacZ reporter expression (D–F’) in rbf mutant clones (A, A’ and D, D’), rno clones (B and B’ and E, E’), and rbf,rno clones (C, C’ and F, F’). (G–I) Expression of Dl in rbf,rno clones suppressed the multiple-R8 phenotype. The high level of multiple-R8s observed in rbf,rno mutant clones (G) was dramatically suppressed by expression of Dl under the control of hh10-GAL4 driver (H). The quantification of the multiple-R8 incidence is shown in (I). Dl protein, Dl reporter expression, and Sens are shown in red; mutant clones are marked by absence of GFP and are outlined. White arrows and arrowheads point to Dl protein or Dl reporter expression in WT and mutant clones, respectively.
To determine whether the decrease in Dl protein in rno and rbf,rno clones was due to transcriptional control, the activity of a Dl-lacZ reporter expression in rno, or rbf,rno double mutant clones was examined. Similar to the observed effect of loss of rbf and rno on Dl protein levels, Dl expression was not affected by removal of rbf alone, while a reduced level of Dl expression was observed in rno single mutant clones, and a strong reduction was observed in rbf,rno double mutant clones (Fig. 5D–F’). We conclude from these results that the increased multiple-R8 phenotype of rbf,rno double mutant clones correlates with reduced Dl expression.
Expression of Dl suppressed the multiple-R8 phenotype of rbf,rno mutant clones
The above results showed that rbf,rno mutant clones have significantly reduced levels of Dl expression and that the multiple-R8 phenotype is sensitive to a reduction of Dl gene dosage. To determine if a decreased level of Dl is a cause of the observed multiple-R8 phenotype in rbf,rno double mutant clones, we decided to determine the effect of increasing the level of Dl in rbf,rno double mutant clones on the multiple-R8 phenotype. Since R8 is the first photoreceptor determined, we selected a Gal4 driver that can drive Dl expression before R8 determination and does not affect R8 differentiation. The hH10-Gal4 driver was selected because expression of Dl using this driver does not significantly alter the number of R8s specified as described earlier (Li and Baker, 2004). As hH10-Gal4 drives Dl expression immediately anterior to the MF, which will likely persist in the MF and in the posterior region close to the MF, we compared the incidence of multiple-R8s in rbf,rno mutant clones within the first 10 rows of photoreceptor differentiation with or without Dl expression. As shown in Fig. 5G–I, expression of Dl in rbf,rno mutant clones using the hH10-Gal4 driver significantly suppressed the multiple-R8 phenotype, reducing the incidence of multiple-R8s from 17.7±1.0% to 4.0±3.3% (Fig. 5I, P=0.002). These results demonstrate that reduced Dl level in rbf,rno mutant clones is a cause for the multiple-R8 phenotype.
Removal of the activation function of dE2F1 suppressed the multiple-R8 phenotype and partially restored Dl level in rbf,rno double mutant clones
The observed requirement of RBF for proper R8 differentiation and Dl expression resembles the reported role of Rb in the differentiation of mammalian cells. Several reports have suggested that the role of Rb in terminal differentiation is mediated by its direct interactions with the differentiation-promoting transcription factors to synergistically activate transcription. Such models would predict E2F-independent roles of Rb in regulating differentiation. However, due to the presence of large number of E2F proteins with related functions, such E2F-independent roles of Rb in differentiation have not been rigorously tested in vivo.
The streamlined version of dE2F proteins in Drosophila prompted us to examine the involvement of deregulated E2F activity in the absence of rbf in the proper differentiation of R8 neurons in rbf,rno double mutant cells. Specifically, we generated rbf,rno double mutant clones in the de2f1i2/rm729 background. de2f1i2 is an allele of de2f1 that has a stop codon mutation after the DNA binding and dimerization domain and de2f1rm729 is a P-element allele that behaves like a null (Duronio et al., 1995; Royzman et al., 1997). Importantly, transheterozygotes of de2f1i2/rm729 can rescue the lethality of rbf null mutants, suggesting that the essential function of rbf is to inhibit the transcription activation function of dE2F1 (Du, 2000). As shown in Fig. 6, rbf,rno1 double mutant clones in the de2f1i2/rm729 background did not show the shiny morphology in the exterior adult eye (Fig. 6A,B). In addition, examination of the multiple-R8 phenotype by Sens staining showed that the incidence of multiple-R8s in rbf,rno1 clones was also significantly suppressed (Fig. 6D, incidence reduced to 6.0±4.0% in de2f1i2/rm729 background as compared to 19.3±3.6% in WT background, P=0.007). In fact, the multiple-R8 incidence in rbf,rno1,de2f1i2/rm729 background was similar to that in rno1 single mutant clones (P=0.29). Since our results suggested that reduced levels of Dl are responsible for the observed multiple-R8 phenotype in rbf,rno mutant clones, we examined the level of Dl protein in rbf,rno1 mutant clones in de2f1i2/rm729 background. As shown in Fig. 6, the level of Dl was partially restored in rbf,rno1 mutant clones in the absence of dE2F1 transcription activation (Fig. 6F–F”). We conclude from these results that the observed differentiation role of RBF is mediated by its ability to regulate the activity of dE2F1.
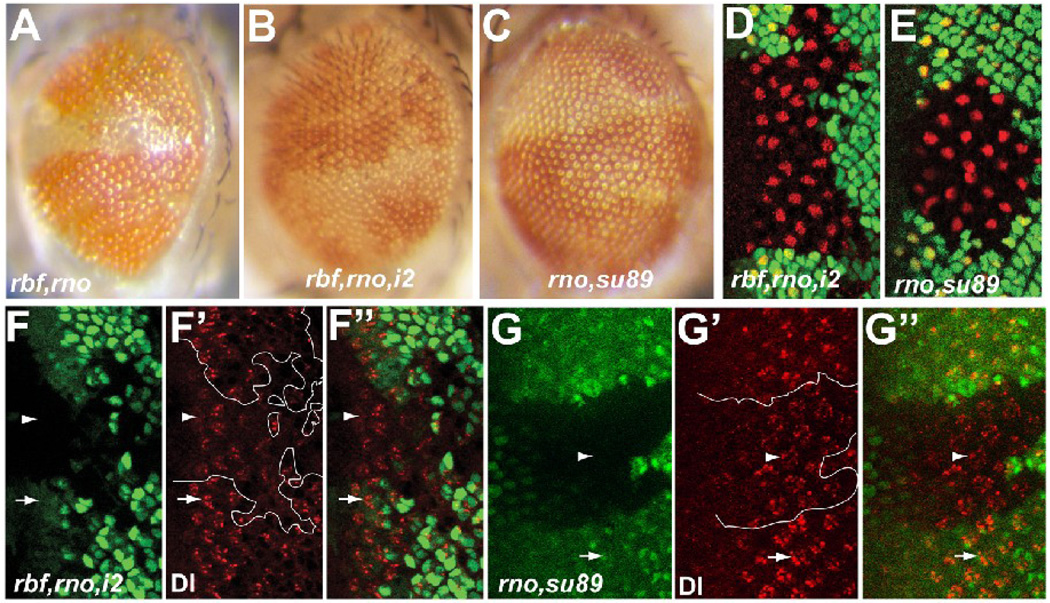
Figure 6.
Removal of the transcription activation domain of dE2F1 suppressed differentiation defects in rbf,rno mutant clones. (A–C) Adult eye images of rbf,rno clones in WT (A), rbf,rno clones in de2f1i2/rm729 (B), and rno clones de2f1su89 (C) background. (D–E) Anti Sens staining (shown in red) revealed low incidence of multiple-R8 phenotypes in rbf,rno clones in the de2f1i2/rm729 background (D) and in rno clones in the de2f1su89 background (E). (F–G”) Anti Dl antibody staining of Dl protein (red) in rbf,rno clones in the de2f1i2/rm729 (F–F”) and rno clones in de2f1su89 (G–G”) backgrounds are shown. Mutant clones were marked by absence of GFP and are outlined in F’ and G’. White arrows and arrowheads point to Dl protein expression in WT and mutant clones, respectively.
To further determine if the dE2F1su89, an allele of dE2F1 that can not bind RBF but can still activate transcription, can mimic the effect of rbf mutation in synergizing with rno to induce differentiation defects, we generated rno mutant clones in dE2F1su89 mutant background. As shown in Fig. 6C, dE2F1su89 mutation did not significantly enhance the adult phenotype of rno mutant clones (compare 6C and 1E). Consistent with this, a low level of multiple-R8 and slightly reduced Dl protein staining was observed in rno,dE2F1su89 mutants (Fig. 6E and 6G–G”, multiple-R8 incidence is 5.0±0.8%). This multiple-R8 incidence is not significantly different from that of rno single mutant clones (P>0.2). Taken together, these results indicated that while dE2F1 is required for the observed differentiation function of RBF, it is likely that this role of RBF is not simply mediated by its binding to dE2F1. However, it is formally possible that there is still residual interaction between RBF and dE2F1su89 that is sufficient to suppress the transactivation capability of dE2F1 in rno mutant clones to provide normal level of Dl expression and R8 differentiation.
Discussion
Studies of how Rb regulates differentiation in fly and mouse models are hampered in part by the fact that the differentiation defects observed by inactivation of Rb in mice or flies are quite subtle and are associated with deregulated cell proliferation and increased apoptosis (Clarke et al., 1992; Du, 2000; Du and Dyson, 1999; Jacks et al., 1992; Lee et al., 1992). On the other hand, inactivation of Rb in C. elegans has revealed its roles mostly in development and differentiation (Lu and Horvitz, 1998). We hypothesized that the weak differentiation defects observed in Rb knockout mice or flies were due to the presence of partially redundant pathways involved in normal differentiation, similar to the effect of Rb/lin-35 inactivation in inducing the synthetic multivulva phenotype in C. elegans (Lu and Horvitz, 1998). Consistent with this idea, we found that mutation of rno and rbf exhibited synergistic differentiation defects during eye development. These observations indicate that Rb regulates development and differentiation in conjunction with other partially redundant mechanisms in both Drosophila and C. elegans. Similarly, it is likely that Rb regulates differentiation in mammalian systems in conjunction with other partially redundant mechanisms. The strong differentiation defects of rbf,rno double mutant clones should provide a nice model to further characterize the role of RBF in differentiation.
Rno is a large PHD domain containing protein shown to antagonize Ras signaling in eye development (Voas and Rebay, 2003). PHD proteins are generally nuclear proteins that function in the control of chromatin or transcription (Bienz, 2006). Based on its domain structure, it is likely that Rno functions in the nucleus to modulate target gene expression. Failure to properly regulate the expression of Rno target gene expression likely mediates the observed rno differentiation defects. Consistent with this, we found that Dl expression was decreased in rno mutant clones and was further reduced in rbf,rno double mutant clones. Furthermore decreasing the level of Dl expression significantly enhanced the multiple-R8 phenotype of rbf,rno mutants while increasing the level of Dl suppressed it. These results suggest that Dl is a key target of RBF and Rno in preventing the multiple-R8 differentiation defects.
Based on the previous reports in mammalian systems that Rb mediates cell differentiation by direct interaction with transcription factors involved in differentiation, we had predicted that the role of RBF in regulating Dl expression and R8 differentiation would be independent of dE2F proteins. Unexpectedly, removal of the activation domain of dE2F1 using the de2f1i2 allele significantly suppressed the differentiation defects of rbf,rno double mutant clones, including the multiple-R8 phenotype and the shiny exterior eye morphology. Furthermore, an increased level of Dl was observed in rbf,rno double mutant clones in de2f1i2 background. Therefore, the role of RBF to promote proper differentiation in the developing eye is, at least in part, mediated by its ability to regulate the transcriptional activation function of dE2F1. Interestingly, a recent report showed that Rb inactivation blocks TCF/β-catenin-dependent transcription, due to an effect of E2F1 in inducing the post-translational degradation of β-catenin (Morris et al., 2008). Although we do not know exactly how RBF/dE2F1 regulates Dl expression, our results suggest that deregulated dE2F1 activity interferes with the expression of Dl. Dl expression in the developing eye is regulated by EGFR signaling through a complex containing the F-box/ WD40 protein Ebi (Tsuda et al., 2002). Interestingly, the phenotype derived from overexpression of dE2F1 and dDP were enhanced by reducing the gene dosage of ebi (Boulton et al., 2000). Therefore, Ebi or other factors in the regulation of Dl expression could be inactivated by deregulated dE2F1 activity. Further studies will be needed to establish the precise mechanisms by which RBF and dE2F1 regulate Dl expression.
Given the similarity of the role of Rb in regulating cell differentiation between Drosophila and C. elegans and given the high level of functional conservation between the Rb/E2F proteins between flies and mammals, it is tempting to speculate that the role of E2F in mediating the differentiation function of Rb is also conserved. In support of this idea, a recent study suggested that deregulated E2F-2 activity in differentiating erythroblasts mediated the erythroid maturation defects of Rb null red cells in mice (Dirlam et al., 2007). Further studies will be needed to determine the generality of deregulated E2F activities in mediating the differentiation function of Rb in mammals.
Materials and methods
Drosophila stocks
The following fly stocks were used in this study: rbf15aΔ, which is derived from excision of the P element insertion of the rbf15a allele (Du and Dyson, 1999), rno1 and rno3 (Voas and Rebay, 2003), dE2F1i2 (Royzman et al., 1997), dE2F1rm729 (Duronio et al., 1995), egfrf2 (BL-2768), and the Delta-lacZ05151 enhancer trap line (BL-11651). Stocks with numbers in parentheses indicated above were obtained from the Bloomington Stock Center.
Drosophila genetics
Flies were cultured at 25°C on standard cornmeal-yeast-agar medium. Ethyl methanesulfonate (EMS)-induced Rbf modifier screening was carried out on 3L. 3-day-old w; p{ry+, neoFRT80B} ry506 (BL#1988) males flies were fed overnight with 5mM EMS. These male flies were mated to rbf15aΔ,w, eyFLP; p{w+, Rbf-G3} p{w+, Ubi-GFP} p{ry+, neoFRT80B} virgin females and cultured at 25°C. The p{w+, Rbf-G3} is a Rbf genomic rescue transgene integrated in an unknown site on the 3L-chromosome. F1 male flies that exhibit significant developmental defects in mutant clones were isolated and were subsequently crossed with w; TM3,Ser/TM6b,Tb virgin females to establish balanced stocks. Balanced males were crossed with rbf15aΔ,w, eyFLP; p{w+, Rbf-G3} p{w+, Ubi-GFP} p{ry+, neoFRT80B} or w, eyFLP; p{w+, Ubi-GFP} p{ry+, neoFRT80B} virgin females for retest and rbf dependence test of the observed differentiation defect, respectively. 1k (later found to be an allele of rno) is a mutation isolated from the screen that showed dramatic differentiation defects in conjunction with rbf mutation. The genotypes of larvae analyzed in the studies were:
rbf15aΔ w, eyFLP (or HsFLP)/Y; egfrf2/+; Rbf-G3, Ubi-GFP, FRT80B/rno, FRT80B
w, eyFLP (or HsFLP)/Y; egfrf2/+; Ubi-GFP, FRT80B/rno, FRT80B
rbf15aΔ,w, eyFLP (or HsFLP)/Y; Rbf-G3, Ubi-GFP, FRT80B / rno, FRT80B, Dl
rbf15aΔ,w, eyFLP (or HsFLP)/Y; Rbf-G3, Ubi-GFP, FRT80B / rno ,dronc FRT80B
w, eyFLP (or HsFLP)/Y; Ubi-GFP, FRT80B/ rno, FRT80B,Dl
rbf15aΔ,w, eyFLP(or HsFLP)/Y;UAS-Dl/+; Rbf-G3,Ubi-GFP,FRT80B /rno, hH10,FRT80B
rbf15aΔ,w, eyFLP(or HsFLP)/Y;Rbf-G3,Ubi-GFP, FRT80B,de2f1729/rno,FRT80B, de2f1i2
w, eyFLP(or HsFLP)/Y;Rbf-G3,Ubi-GFP, FRT80B,de2f1su89/rno,FRT80B, de2f1su89
Edu Cell proliferation assay
EdU (Invitrogen) incorporation assay was performed according to the manufacturer’s instructions. EdU labeling was done at 10uM for 30 minutes and detected using the AlexaFluor 594 azide. To minimize GFP quenching, the fluorophore reaction was performed for 5 minutes. S phase cells/unit area in WT or mutant clones located posterior to the SMW were determined. The S phase cells in mutant clones were calculated by subtracting the background level of EdU incorporation in the wild type (GFP-positive) cells posterior to the SMW.
Quantification and statistics
To quantify multiple-R8 phenotypes, eye discs containing clones of indicated genotypes were induced via hsFLP and stained with Sens to visualize R8 cells. The number of R8 positions that have single or multiple-R8 were determined. Two or more Sens-positive cells that were touching were scored as one multiple-R8 position while a separated Sens-positive cell was scored as one single R8 position. A minimum of 150 total R8 positions were counted for each genotype, with an average of about 50 positions per replicate experiment. Student’s t-test was used to determine the statistical significance.
Immunohistochemistry
Unless otherwise indicated, all steps were completed at room temperature. Pupal retinas or larval imaginal discs were dissected in 1x PBS, fixed in 4% formaldehyde in 1x PBS for 30 minutes, and incubated in primary antibody diluted in 1x PBS plus 0.3% Triton-X100 (PBSTx) with 10% normal goat serum overnight at 4°C. Primary antibodies used were mouse anti-Cut (1:10; DSHB), guinea pig-anti-Sens (1:500; provided by H. Bellen), mouse anti-Delta (1:10; DSHB). Following incubation with primary antibodies, samples were washed three times (10 minutes each) in PBSTx, and incubated in secondary antibodies from Jackson ImmunoResearch (1:200 to 1:400 dilution). Phalloidin-TRITC labeled (Sigma) was used at 1:100 dilution. The Cut and Delta antibodies, developed by Gerald M. Rubin and Spyros Artavanis-Tsakonas respectively, were obtained from the Developmental Studies Hybridoma Bank (DSHB), developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
Acknowledgements
We would like to thank Drs. Ilaria Rebay and Hugo Bellen, the Bloomington Stock Center, and the Developmental Studies Hybridoma bank at the University of Iowa for generously supplying fly stocks and reagents. We also thank Jie Zhang for technical assistance and Dr. Jennifer Searle for reading this manuscript. This work was supported by a grant from the National Institute of Health (GM 074197) and by a grant from the America Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker NE, Yu SY. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol. 1997;7:122–132. doi: 10.1016/s0960-9822(06)00056-x. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech Dev. 1998;74:3–14. doi: 10.1016/s0925-4773(98)00054-9. [DOI] [PubMed] [Google Scholar]
- Baker NE, Zitron AE. Drosophila eye development: Notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech Dev. 1995;49:173–189. doi: 10.1016/0925-4773(94)00314-d. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Notch signalling and the initiation of neural development in the Drosophila eye. Development. 2001;128:3889–3898. doi: 10.1242/dev.128.20.3889. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Brook A, Staehling-Hampton K, Heitzler P, Dyson N. A role for Ebi in neuronal cell cycle control. Embo J. 2000;19:5376–5386. doi: 10.1093/emboj/19.20.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-L, Riley DJ, Chen Y, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes and Development. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- Clarke A, Maandag E, van Roon M, van der Lugt N, van der Valk M, Hooper M, Berns A, Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Hariharan IK. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- Dirlam A, Spike BT, Macleod KF. Deregulated E2f-2 underlies cell cycle and maturation defects in retinoblastoma null erythroblasts. Mol Cell Biol. 2007;27:8713–8728. doi: 10.1128/MCB.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Du W. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development. 2000;127:367–379. doi: 10.1242/dev.127.2.367. [DOI] [PubMed] [Google Scholar]
- Du W, Dyson N. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 1999;18:916–925. doi: 10.1093/emboj/18.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25:5190–5200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996a;10:1206–1218. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- Du W, Xie JE, Dyson N. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. Embo J. 1996b;15:3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH, Xie J-E, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Brook A, Dembski MS, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc. Natl. Acad. Sci. USA. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ. Functional antagonism between E2F family members. Genes Dev. 2001;15:2146–2160. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Schneider JW, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis [see comments] Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol. 2001;11:330–338. doi: 10.1016/s0960-9822(01)00093-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev Biol. 2004;4:5. doi: 10.1186/1471-213X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol. 2006;26:7601–7615. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Spicer DB, Kim PS, Cheung WL, Lassar AB. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Nevins JR. Functional properties of a Drosophila homolog of the E2F1 gene. Mol. Cell. Biol. 1994;14:1603–1612. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Turner FR, Muskavitch MA. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev. 1995;50:201–216. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, White NM, Jarman AP. Echinoid limits R8 photoreceptor specification by inhibiting inappropriate EGF receptor signalling within R8 equivalence groups. Development. 2003;130:3715–3724. doi: 10.1242/dev.00602. [DOI] [PubMed] [Google Scholar]
- Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes & Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Yamaguchi M, Nishimoto Y, Ohno K, Sakaguchi K, Matsukage A. dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem Biophys Res Commun. 1998;251:409–415. doi: 10.1006/bbrc.1998.9407. [DOI] [PubMed] [Google Scholar]
- Spencer SA, Cagan RL. Echinoid is essential for regulation of Egfr signaling and R8 formation during Drosophila eye development. Development. 2003;130:3725–3733. doi: 10.1242/dev.00605. [DOI] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. Embo J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. The novel plant homeodomain protein rhinoceros antagonizes Ras signaling in the Drosophila eye. Genetics. 2003;165:1993–2006. doi: 10.1093/genetics/165.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]