Abstract
We have used Saccharomyces cerevisiae to identify toxicologically important proteins and pathways involved in arsenic-induced toxicity and carcinogenicity in humans. We performed a systemic screen of the complete set of 4,733 haploid S. cerevisiae single gene deletion mutants to identify those that have decreased or increased growth, relative to wild-type, after exposure to sodium arsenite (NaAsO2). IC50 values for all mutants were determined to further validate our results. Ultimately we identified 248 mutants sensitive to arsenite and 5 mutants resistant to arsenite exposure. We analyzed the proteins corresponding to arsenite-sensitive mutants and determined that they belonged to functional categories that include protein binding, phosphate metabolism, vacuolar/lysosomal transport, protein targeting, sorting, and translocation, cell growth/morphogenesis, cell polarity and filament formation. Furthermore, these data were mapped onto a protein interactome to identify arsenite toxicity-modulating networks. These networks are associated with the cytoskeleton, ubiquitination, histone acetylation and the MAPK signaling pathway. Our studies have potential implications for understanding toxicity and carcinogenesis in arsenic-induced human conditions, such as cancer and aging.
Keywords: Arsenite, Toxicity, Saccharomyces cerevisiae
Introduction
Arsenic (As) is a ubiquitously present metalloid and a human carcinogen that is associated with skin, bladder, lung, kidney and liver cancer [1; 2; 3]. It is also implicated in vascular diseases, neurological and neurobehavioral disorders, diabetes and as a teratogen [4; 5]. Paradoxically, arsenic trioxide is currently used in the treatment of acute promyelocytic leukemia (APL) [6; 7]. Inorganic arsenic is considered the most hazardous among all the arsenic species present in the environment. Inorganic arsenic exists in the environment in two major forms, arsenite [As (III)] or arsenate [As (V)]. In general, As (III) is more acutely toxic than As (V) [8]. Arsenic is a paradoxical non-mutagenic carcinogen, as there are arsenic-induced cancers observed in humans but there has been a lack of acceptable animal models. The mechanisms of arsenic-mediated toxicity and carcinogenesis are poorly understood, but it has been suggested that at least part of its toxicity is due to oxidative stress, which in turn causes protein denaturation, lipid damage and DNA strand breaks [9]. Arsenic activates signal transduction pathways including AP-1, NFκB and MAPK (see review [10]). In addition, arsenic is involved in epigenetic mechanisms, for example, the alteration of epigenetic marks such as H3K4 tri-methylation and H3K9 di-methylation [11].
The availability of a complete set of single gene deletion Saccharomyces cerevisiae strains has allowed us to functionally characterize the yeast genes that respond to cellular insults at the systemic level [12; 13]. The complete yeast nucleotide sequence contains ~6,300 genes [14; 15], but only the 4,733 nonessential yeast mutants can be examined because deletion of the essential genes is lethal. Since there is a high degree of homology among the eukaryotes, S. cerevisiae can be used as a model to identify genes that might be important in arsenic-induced carcinogenesis in other eukaryotes, including humans.
Cells have developed adaptive defense systems against environmental stress, such as detoxification, repair, removal of damaged molecules[16]. To better understand how cells respond to As (III) exposure, we screened the S. cerevisiae deletion strain set for sensitivity and resistance and identified the genes that have human homologues. In principle, genes whose deletion conferred sensitivity to arsenite would correspond to proteins involved in cellular recovery against arsenite-induced toxicity while genes whose deletion conferred resistance would correspond to proteins that arrest or reduce growth after arsenite exposure. We have analyzed the contribution of proteins that correspond to sensitive and resistant phenotypes in the framework of 12,232 protein-protein and protein-DNA interactions making up the known yeast interactome. Our results using this unbiased whole genomic approach reveal that genes whose deletion confers sensitivity to As (III) exposure correspond to proteins significantly enriched in various cellular functions, including vacuolar transport, cytoskeleton, acetylation and deacetylation processes, osmotic sensing and response, ubiquitination and proteosomal degradation, cell growth, regulation of carbon-compound and carbohydrate metabolism, protein binding, endocytosis, mitotic (M) phase, transport AT Pases, protein targeting, sorting and translocation, puine nucleotide/nucleoside/nucleobase anabolism, vacuole or lysosome function, homeostasis of protons, phosphate metabolism, stress response, budding, cell polarity and filament formation, cytoplasmic and nuclear protein degradation and MAPKKK cascade (Table 2). In contrast to 248 arsenite-toxicity sensitive proteins, only 5 arsenite-toxicity resistant proteins were identified (Mub1, Uth1, Fps1, Upf3, Ask10 and P15B12).
Table 2.
Categories of yeast deletion mutants sensitive to arsenite. The deletion mutants that showed arsenic-sensitive phenotype were categorized based on the biological functions using FunSpec. The category was ordered by p value. Some of these mutants are present in more than one category as they have several functions.
| Functional Category | p-value | In category from cluster | Arsenite Toxitciy Modulating |
Total in Category |
|---|---|---|---|---|
| vacuolar/lysosomal transport[20.09.13] | 3.31E-08 | VPS8 STP22 FEN1 CUP5 VPS25 VPS24 VPS51 SNF7 SRN2 PEP3 VPS36 VMA6 MVP1 VPS21 VMA4 VTS1 SNF8 VPS16 BRO1 |
19 | 153 |
| cytoskeleton/structural proteins[42.04] | 4.14E-07 | HSL7 BEM1 STE50 RES161 RPN4 RVS167 GIM4 PAC10 PFD1 SAC1 YKE2 TUB3 CLA4 SVL3 NIP100 |
15 | 113 |
| modification by acetylation, deacetylation [14.07.04] |
2.47E-06 | SGF29 MAK31 ADA2 SGF73 GCN5 ARD1 HDA1 PHO23 HFI1 MAK3 HDA3 |
11 | 69 |
| cell growth / morphogenesis [40.01] | 1.66E-05 | SLA1 HSL7 STE50 RVS161 FEN1 RPN4 HBT1 REG1 BMH2 SSD1 RVS167 HOC1 GRR1 CLA4 STI1 SVL3 NIP100 |
17 | 189 |
| regulation of C-compound and carbohydrate metabolism [01.05.25] |
4.11E-05 | REG1 PBS2 GRR1 VPS25 SNF7 HOG1 VPS36 PSY2 POP2 GAL11 SNF2 SNF8 SSN3 |
13 | 126 |
| protein binding [16.01] | 6.77E-05 | SLA1 PIN4 BEM1 STP22 RVS161 BMH2 MSN5 RVS167 GIM4 UBC8 PAC2 PAC10 RTT101 PBS2 PFD1 GRR1 VPS51 UBI4 SIC1 PFP3 YKE2 THP1 STI1 RBL2 NIP100 |
25 | 391 |
| endocytosis [20.09.18.09.01] | 0.0001976 | SLA1 EDE1 RVS161 DOA4 RVS167 CUP5 VPS21 SVL3 |
8 | 59 |
| osmosensing and response [34.11.03.13] | 0.0003469 | STE50 DOA4 PBS2 HOG1 NST1 SSK2 | 6 | 35 |
| M phase [10.03.01.01.11] | 0.0004621 | SPC72 DOC1 TUB3 CIK1 MCK1 CSE2 NIP100 | 7 | 51 |
| trapnsport ATPases [ 20.03.22] | 0.0005872 | PCA1 CUP5 SPF1 PMR1 PPA1 VMA6 VMA4 | 7 | 53 |
| protein targeting, sorting and translocation [14.04] |
0.000737 | VPS8 STP22 PEX19 MSN5 CUP5 KAP123 VPS25 VPS24 VPS51 SNF7 SRN2 PEP3 VPS36 MVP1 VPS21 VTS1 SNF8 VPS16 |
18 | 281 |
| purine nucleotide/nucleoside/nucleobase anabolism [01.03.01.03] |
0.00107 | ADE1 YSA1 ADE6 YNK1 BAS1 | 5 | 29 |
| vacuole or lysosome [42.25] | 0.001227 | VAC17 KCS1 DOA4 CUP5 PEP3 VPS16 | 6 | 44 |
| modification by ubiquitination deubiquitination [14.07.05] |
0.001451 | DOA4 UBC8 UBP3 RTT101 GRR1 UBI4 BRE5 UBP2 | 8 | 79 |
| homeostasis of protons [34.01.01.3] | 0.001743 | CUP5 PPA1 VPH2 MEH1 VMA6 VMA4 | 6 | 47 |
| proteasomal degradation (ubiquitin/proteasomal pathway) [14.13.01.01] |
0.0028 | RPN4 UBC8 BST1 DOC1 PRE9 RPL40A RTT101 GRR1 DOA1 BRO1 |
10 | 128 |
| vacuolar protein degradation [14.13.04.02] | 0.002932 | APE3 VID30 MEH1 | 3 | 11 |
| phosphate metabolism [01.04] | 0.00345 | YSA1 NPP1 RBK1 DUN1 KCS1 REG1 PRO1 DBF2 YTA4 KSP1 PBS2 YNK1 PEX1 SAC1 HOG1 PPZ1 CLA4 MCK1 PEX6 SSK2 SSN3 |
21 | 401 |
| stress response [32.01] | 0.005175 | NTH1 KCS1 SSD1 PBS2 GRR1 UBI4 TRM9 PPZ1 MCK1 SSK2 BRO1 |
11 | 162 |
| osmotic and salt stress response [32.01.03] |
0.005561 | STE50 REV161 RVS167 HOG1 NST1 GRX5 | 6 | 59 |
| budding, cell polarity and filament [43.01.03.05] |
0.005666 | SLA1 HSL7 BEM1 STE50 RVS161 FEN1 RPN4 BMH2 SSD1 RVS167 BEM2 HOC1 GRR1 CLA4 THR1 SVL3 NIP100 |
17 | 312 |
| cytoplasmic and nuclear protein degradation [14.13.01] |
0.006041 | DOA4 UBP3 YTA7 CPS1 UBI4 UBP2 | 6 | 60 |
| MAPKKK cascade [30.01.05.01.03] | 0.006082 | STE50 PBS2 HOG1 SSK2 | 4 | 27 |
Materials and Methods
Medium, Solution, and strains
All yeast strains were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose, 2% agar for plates) supplemented with 200 µg/ml G418. Sodium arsenite was purchased from sigma (St. Louis, MO). The complete set of 4,733 non-essential haploid S. cerevisiae single gene deletion mutants were obtained and described as before [17; 18].
High-Throughput screening
High-throughput genomic screening was performed using the complete set of 4,733 mutants, as described before [17; 18]. Briefly, 96-well master plates containing individual deletion strains were resuspended with 60 µl bursts of forced air from a Hydra liquid handling apparatus(Robbins Scientific, Sunnyvale, CA), and then 1 µl samples were spotted on YPD agar plates containing 0, 0.75, and 1 mM sodium arsenite. Inoculated plates were incubated for 60 h at 30 °C and the resulting plates were imaged using an AlphaImager (Alpha Innotech Corporation, San Leandro, CA). The mutants were scored as sensitive or resistant compared with the non-treated and the wild type strain (BY4741). The experiments were done in triplicate.
Determination of IC50
5 µl of log-phase yeast culture was transferred into 195 µl YPD medium containing sodium arsenite in 96 well plates. The concentrations of sodium arsenite were 0, 0.375, 0.75, 1, 1.25 and 2.5 mM for sensitive strains and 0, 1.5, 2, 2.5, 3.75 and 5 mM for resistant strains. The cultures were incubated at 30 °C for 20 h, and cell density was determined by measuring the absorbance at 590 nm by Perkin Elmer HTS 7000 Bio Assay Reader. The concentration responsible for half-maximal inhibition of growth (IC50) was calculated using GraphPad Prism 5 program.
Biological function analysis of arsenite toxicity modulating proteins
The deletion mutants that showed arsenic-sensitive phenotypes were categorized based on the biological functions using the program FunSpec (Functional Specification). The categories were downloaded from the MIPS Database and the GO Database. The p-values, calculated using the hypergeometric distribution, represent the probabilities that the intersection of a given list with any given functional category occurs by chance. Note that many genes are contained in many categories, especially in the MIPS database (which are hierarchical) and that this can create biases.
Interactome mapping analysis of arsenite toxicity modulating proteins
The deletion mutants were analyzed using the Cytoscope software for protein interaction networks as described [19]. S. cerevisiae protein–protein interaction information were obtained from the Database of Interacting Proteins [20]. In all we compiled 14,493 interactions between 5,433 proteins. The interactome is an extensive framework that can be used to identify protein networks activated by stress but it is a non saturated structure with regard to molecular interactions. None the less it provides a framework to analyze and associate discrete data points. Protein-protein interaction information was imported into Cytoscape for network visualization and subnetwork filtering. Subnetwork filtering was performed by tab selection of identified arsenite-toxicity modulation proteins and their associated protein-protein interactions. Analysis of the filtered interactome was set to identify sub networks => 4 nodes. It should be noted that interactome filtering does not use statistical validation to assign p-values to sub-networks. Instead, the filtering step identifies all connected As-toxicity modulating proteins in the interactome to provide a global view of how different functional activities are potentially coordinated.
Human homologues to yeast genes
Human homologues to the identified yeast genes were determined by BLAST using the tBLASTn program, which is available online from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) [21]. Note only the top scoring human homologue for each gene was used.
Results and discussion
Screening of single-gene deletion mutants of S. cerevisiae for arsenite sensitivity and resistance
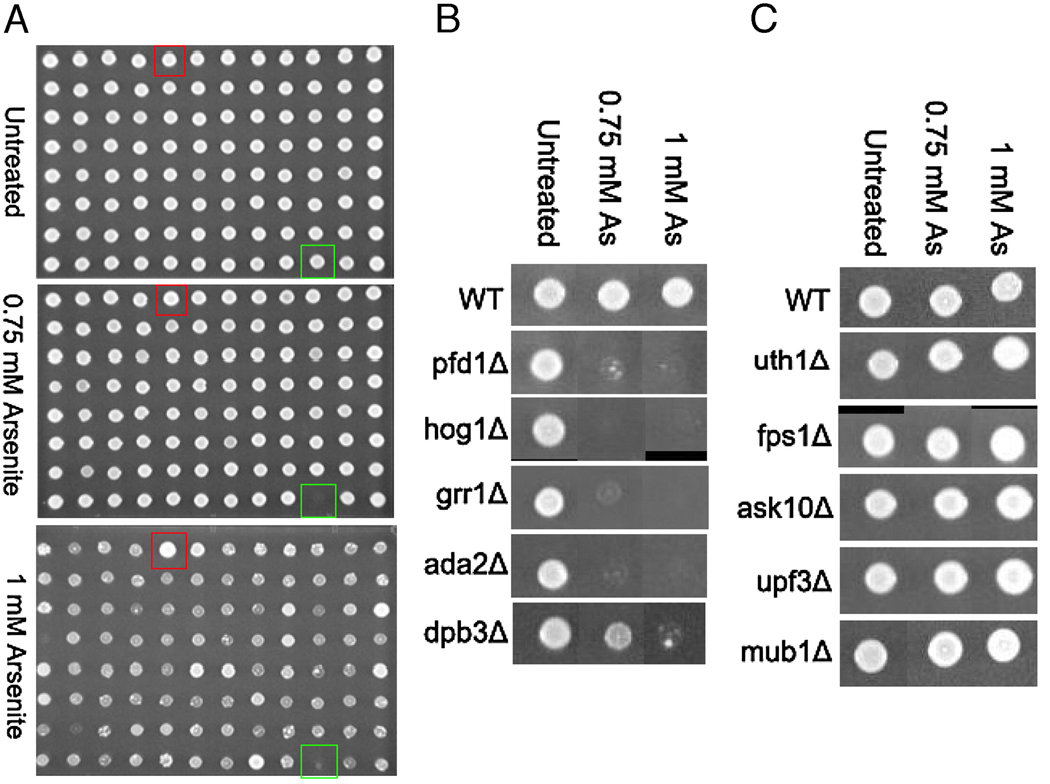
We screened a library of 4,733 S. cerevisiae gene deletion strains in triplicate to determine which proteins influence resistance or sensitivity after exposure to As (III). Strains from saturated cultures grown in 96-well format were robotically spotted onto agar plates with and without As (III) in the agar. Following 60 h incubation, plates were recorded by digital imaging of colony growth. By visual inspection of the imaged colonies, the strains were scored for sensitivity or resistance. The strains that had a significant decrease in the growth of colonies relative to wild type BY4741 were scored as sensitive, and the strains that displayed increased colony growth relative to wild-type were scored as resistant. Typical colony growth images were shown in Figure 1. In order to identify the possible pathways of arsenic response in humans, only the genes that have human homologues were listed (Table 1).
Figure 1.
High-throughput screen of arsenite. (A) 96 gene-deletion mutants were spotted onto agar plates containing 0.75 and 1.25 mM arsenite, incubated at 30 °C for 60 h, and imaged. Red squares denote the arsenite-resistant gene-deletion mutant fps1Δ. Green squares denote the arsenite-sensitive gene-deletion mutant hog1Δ. (B) Growth of wild type BY4741 (WT), pfd1Δ, hog1Δ, grr1Δ, ada2Δ and dpb3Δ. These mutants are the most sensitive gene-deletion mutants as determined by IC50. Images were cropped and recompiled together. (C) Growth of wild type BY4741 (WT), uth1Δ, fps1Δ, ask10Δ, upf3Δ and mub1Δ. These mutants are the most resistant gene-deletion mutants as determined by IC50. Images were cropped and recompiled together.
Table 1.
List of yeast deletion mutants that are sensitive or resistant to arsenite. All the sensitive and resistant mutants identified are presented in this table. The mutants are ordered from the most to the least sensitive. IC50 values are indicated in mM sodium arsenite.
| Yeast Protein name |
Yeast Function | IC50 | Human SwissProt number |
Human Function | |
|---|---|---|---|---|---|
| YJL179W | PFD1 | Protein with similarity to bovine prefoldin subunit 1 | 0.2444 | O60925 | Prefoldin subunit 1 |
| YLR113W | HOG1 | MAP kinase (MAPK) central to the high-osmolarity signal transduction pathway |
0.3059 | Q16539 | Mitogen-activated protein kinase 14 |
| YJR090C | GRR1 | F-box protein that targets G1 cyclins and Gic1p and other proteins for degradation by the SCF-Grr1p complex (Skp1p-Cdc53p-Cdc34p-Grr1p); also required for glucose repression and for glucose and cation transport |
0.3464 | AAH07557 | F-box/LRR-repeat protein 20 |
| YDR448W | ADA2 | Component of two nucleosomal histone | 0.375 | O75478 | Transcriptional adapter 2-alpha |
| YBR278W | DPB3 | DNA polymerase epsilon third subunit | 0.4023 | Q9NR33 | DNA polymerase epsilon subunit 4 |
| YCR027C | RHB1 | Putative Rheb-related GTPase involved in regulating canavanine resistance and arginine uptake; member of the Ras superfamily of G-proteins |
0.4121 | Q15382 | GTP-binding protein Rheb [Precursor] |
| YPL105C | SYH1 | Protein of unknown function that may interact with ribosomes, based on co-purification experiments; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
0.4152 | O75137 | PERQ amino acid-rich with GYF domain-containing protein 2 |
| YOL081W | IRA2 | GTPase-activating protein for Ras1p and Ras2p | 0.4156 | P21359 | Neurofibromin |
| YGL084C | GUP1 | Plasma membrane protein involved in remodeling GPI anchors; member of the MBOAT family of putative membrane-bound O-acyltransferases; proposed to be involved in glycerol transport |
0.4504 | Q9NVH9 | Protein-cysteine N- palmitoyltransferase HHAT |
| YNL229C | URE2 | Nitrogen catabolite repression transcriptional regulator that acts by inhibition of GLN3 transcription in good nitrogen source; altered form of Ure2p creates [URE3] prion |
0.4504 | P30711 | Glutathione S-transferase theta-1 |
| YEL003W | GIM4 | Prefoldin subunit 2, component of the Gim protein complex that promotes formation of functional alpha- and gamma-tubulin |
0.4787 | Q9UHV9 | Prefoldin subunit 2 |
| YBR298C | MAL31 | High affinity maltose/H+ symporter (maltose permease), member of the hexose transporter family of the major facilitator superfamily (MFS) |
0.4962 | P11168 | Solute carrier family 2, facilitated glucose transporter member 2 |
| YPL042C | SSN3 | Cyclin-dependent serine/threonine protein kinase of the RNA polymerase II holoenzyme complex and Kornberg's mediator (SRB) subcomplex |
0.4992 | P49336 | Cell division protein kinase 8 |
| YGR252W | GCN5 | Component of two nucleosomal histone acetyltransferase complexes |
0.4996 | Q92831 | Histone acetyltransferase PCAF |
| YJL128C | PBS2 | MAP kinase kinase (MEK) activated by high osmolarity through the Sln1p-Ypd1p-Ssk1p two-component osmosensor and the Sho1p osmosensor |
0.5 | P36507 | Dual specificity mitogen-activated protein kinase kinase 2 |
| YNL307C | MCK1 | Serine/threonine/tyrosine protein kinase, positive regulator of meiosis and spore formation |
0.5 | AAH27984 | Glycogen synthase kinase-3 alpha |
| YBL051C | PIN4 | Protein involved in G2/M phase progression and response to DNA damage, interacts with Rad53p; contains an RNA recognition motif, a nuclear localization signal, and several SQ/TQ cluster domains; hyperphosphorylated in response to DNA damage |
0.5056 | AAN05429 | Cleavage stimulation factor 64 kDa subunit, tau variant |
| YCL001W-A | Protein of unknown function | 0.5102 | |||
| YBR133C | HSL7 | Negative regulatory protein of the Swe1p protein kinase | 0.5163 | Q9UKH1 | Protein arginine N- methyltransferase 5 |
| YCR009C | RVS161 | Protein required for viability after N, C, or S starvation, for internalization step of endocytosis, and for cell fusion during mating; roles in endocytosis and in cell fusion are independent of one another |
0.5181 | Q9NQY0 | Bridging integrator 3 |
| YGL012W | ERG4 | Sterol C-24 (28) reductase | 0.5262 | Q14739 | Lamin-B receptor |
| YCL010C | SGF29 | SaGa associated Factor 29kDa; Probable 29kKDa Subunit of SAGA histone acetyltransferase complex |
0.527 | Q96ES7 | SAGA-associated factor 29 homolog |
| YNR031C | SSK2 | Map kinase kinase kinase (MAPKKK) of the high- osmolarity signal transduction pathway |
0.552 | BAA13204 | Mitogen-activated protein kinase kinase kinase 4 |
| YDR028C | REG1 | Regulatory subunit for protein phosphatase Glc7p, required for glucose repression |
0.5533 | Q9NZW4 | Dentin sialophosphoprotein [Precursor] |
| YBL007C | SLA1 | Protein involved in assembly of cortical actin cytoskeleton, has three SH3 domains |
0.5535 | Q9UIQ3 | Proto-oncogene tyrosine-protein kinase FGR |
| YER014C-A | BUD25 | Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild- type bipolar pattern |
0.5587 | ||
| YCR045C | Protein with similarity to protease B (Prb1p) and subtilisin family proteases |
0.5638 | Q8NBP7 | Proprotein convertase subtilisin/kexin type 9 [Precursor] |
|
| YER155C | BEM2 | GTPase-activating (GAP) protein; regulates Rho1p and has a role in bud emergence and cell cycle-related cytoskeletal reorganization |
0.5718 | AAH38976 | Rho GTPase-activating protein 15 |
| YCR026C | NPP1 | Nucleotide pyrophosphatase/phosphodiesterase family member; mediates extracellular nucleotide phosphate hydrolysis along with Npp2p and Pho5p; activity and expression enhanced during conditions of phosphate starvation |
0.5725 | Q9UJA9 | Ectonucleotide pyrophosphatase/phosphodiesterase e family member 5 [Precursor] |
| YER110C | KAP123 | Karyopherin-beta involved in nuclear import of ribosomal proteins |
0.5772 | Q8TEX9 | Importin-4 |
| YCL063W | VAC17 | Protein involved in vacuole inheritance; acts as a vacuole-specific receptor for myosin Myo2p |
0.5813 | AAH40354 | Caldesmon |
| YBR286W | APE3 | Aminopeptidase Y (yscIII, APY), major vacuolar aminopeptidase with preference for basic amino acids and proline |
0.582 | Q9UQQ1 | N-acetylated-alpha-linked acidic dipeptidase-like protein |
| YDR359C | EAF1 | Component of the NuA4 histone acetyltransferase complex; required for initiation of pre-meiotic DNA replication, probably due to its requirement for significant expression of IME1 |
0.5904 | Q96L91 | E1A-binding protein p400 |
| YCL032W | STE50 | Protein required for feedback control of pheromone- induced signal transduction |
0.5926 | O43419 | Intestinal mucin [Fragment] |
| YBR281C | DUG2 | Probable di- and tri-peptidase; forms a complex with Dug1p and Dug3p to degrade glutathione (GSH) and other peptides containing a gamma-glu-X bond in an alternative pathway to GSH degradation by gamma- glutamyl transpeptidase (Ecm38p) |
0.599 | Q96KN2 | Beta-Ala-His dipeptidase [Precursor] |
| YCR087C-A | LUG1 | Protein of unknown function | 0.6023 | ||
| YPL165C | SET6 | Protein of unknown function; deletion heterozygote is sensitive to compounds that target ergosterol biosynthesis, may be involved in compound availability |
0.6043 | Q9NRG4 | SET and MYND domain-containing protein 2 |
| YDR388W | RVS167 | Protein that affects actin distribution and bipolar budding, has an SH3 domain |
0.605 | Q96HL8 | SH3 domain-containing YSC84-like protein 1 |
| YER007W | PAC2 | Putative tubulin-specific chaperone, involved in formation of alpha-beta-tubulin heterodimer |
0.6177 | Q15813 | Tubulin-specific chaperone E |
| YCR036W | RBK1 | Ribokinase; member of a family of sugar kinases that includes Pfk2p |
0.6283 | Q9H477 | Ribokinase |
| YML121W | GTR1 | Cytoplasmic GTP binding protein and negative regulator of the Ran/Tc4 GTPase cycle; component of GSE complex, which is required for sorting of Gap1p; involved in phosphate transport and telomeric silencing; similar to human RagA and RagB |
0.6322 | AAH34726 | Ras-related GTP-binding protein B |
| YCL037C | SRO9 | Suppressor of ypt6 null and rho3 mutations | 0.6364 | Q9NW12 | La-related protein 2 |
| YLR079W | SIC1 | P40 inhibitor of Cdc28p-Clb protein kinase complex | 0.6452 | Q8NHA9 | Seven transmembrane helix receptor |
| YHR013C | ARD1 | Protein N-acetyltransferase subunit; mating functions are reduced in mutants due to derepression of silent mating type loci |
0.6467 | P41227 | N-terminal acetyltransferase complex ARD1 subunit homolog A |
| YJL136C | RPS21B | Ribosomal protein S21 (yeast S26; YS25; rat S21), to Rps21Ap |
0.6509 | Q8WVC2 | RPS21 protein |
| YAL047C | SPC72 | Component of spindle pole body that interacts with Stu2p |
0.6523 | ||
| YCL033C | Putative protein-methionine-R-oxide reductase; involved in response to oxidative stress; similar to mouse Sepx1p and fly SelRp; YCL033C is not an essential gene |
0.6639 | Q9Y3D2 | Methionine-R-sulfoxide reductase B2, mitochondrial [Precursor] |
|
| YPL059W | GRX5 | Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; mitochondrial matrix protein involved in the synthesis/assembly of iron- sulfur centers; monothiol glutaredoxin subfamily member along with Grx3p and Grx4p |
0.6678 | AAH23528 | Glutaredoxin-related protein 5 |
| YDR335W | MSN5 | Karyopherin involved in nuclear import and export; to be responsible for nuclear import of replication protein A and for export of several proteins including Swi6p, Far1p, and Pho4p; cargo dissociation involves binding to RanGTP |
0.6794 | Q9BZV5 | Exportin-5 |
| YOR290C | SNF2 | Component of SWI-SNF global transcription activator complex, acts to assist gene-specific activators through chromatin remodeling |
0.6797 | Q9HBD4 | SMARCA4 isoform 2 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4, isoform CRA_c) |
| YPL174C | NIP100 | Mitotic spindle positioning protein, dynactin complex protein associated with the spindle |
0.683 | P30622 | CAP-Gly domain-containing linker protein 1 |
| YKL006W | RPL14A | Ribosomal protein L14 (mammalian L14), nearly identical to Rpl14Bp |
0.6885 | P50914 | 60S ribosomal protein L14 |
| YGR078C | PAC10 | Protein required in the absence of Cin8p | 0.6981 | CAA76761 | Prefoldin subunit 3 |
| YCL060C | Protein of unknown function | 0.7028 | |||
| YBR231C | SWC5 | Protein of unknown function, component of the SWR1 complex, which exchanges histone variant H2AZ (Htz1p) for chromatin-bound histone H2A |
0.7067 | Q9UEE9 | Craniofacial development protein 1 |
| YEL027W | CUP5 | Protein of unknown function | 0.7128 | P27449 | Vacuolar ATP synthase 16 kDa proteolipid subunit |
| YEL031W | SPF1 | Putative Ca2+-transporting ATPases, member of the P- type ATPase superfamily |
0.7154 | Q9HD20 | Probable cation-transporting ATPase 13A1 |
| YPL084W | BRO1 | Protein that interacts with components of the PKC1- MAP kinase pathway |
0.7197 | Q9BX86 | Programmed cell death 6-interacting protein |
| YPL262W | FUM1 | Fumarate hydratase; mitochondrial and cytoplasmic fumarase, converts L-malate to fumarate as part of the cycle |
0.7284 | P07954 | Fumarate hydratase, mitochondrial [Precursor] |
| YOL001W | PHO80 | Cyclin that interacts with Pho85p protein kinase, regulates the phosphate pathway through phosphorylation of Pho4p |
0.7321 | Q9H4N0 | Uncharacterized protein C2orf24 |
| YOR360C | PDE2 | 3',5'-Cyclic-nucleotide phosphodiesterase, high-affinity | 0.7328 | Q13945 | 3',5'-cyclic AMP phosphodiesterase [Fragment] |
| YGL066W | SGF73 | Subunit of SAGA histone acetyltransferase complex; in formation of the preinitiation complex assembly at promoters; null mutant displays defects in premeiotic DNA synthesis |
0.7364 | Q9ULK2 | Ataxin-7-like protein 1 |
| YAR002aW | Protein of unknown function | 0.7652 | |||
| YNL087W | TCB2 | Bud-specific protein with a potential role in membrane trafficking; GFP-fusion protein migrates from the cell surface to intracellular vesicles near vacuole; contains 3 calcium and lipid binding domains; mRNA is targeted to the bud |
0.7657 | Q96LX0 | Multiple C2 and transmembrane domain-containing protein 1 |
| YKL213C | DOA1 | WD repeat protein required for ubiquitin-mediated protein degradation, forms complex with Cdc48p, plays a role in controlling cellular ubiquitin concentration; also promotes efficient NHEJ in postdiauxic/stationary phase |
0.77 | Q9UF53 | Phospholipase A-2-activating protein |
| YML124C | TUB3 | Tubulin alpha-3 chain, non-essential | 0.7732 | AAH06468 | Tubulin alpha-1A chain |
| YKL119C | VPH2 | Vacuolar H(+)-ATPase (V-ATPase) assembly protein in the endoplasmic reticulum |
0.7901 | O75336 | Liprin-beta-1 |
| YDR393W | SHE9 | Protein with similarity to Arabidopsis thaliana CIP1, lethality when overexpressed |
0.793 | P13533 | Myosin-6 |
| YOL012C | HTA3 | Histone-related protein that can suppress histone H4 point mutation |
0.8078 | AAH20936 | Histone H2A.Z |
| YDR069C | DOA4 | Ubiquitin-specific protease (ubiquitin C-terminal hydrolase), involved in recycling ubiquitin from protein substrates targeted to the proteasome and the vacuole |
0.8121 | P40818 | Ubiquitin carboxyl-terminal hydrolase |
| YBR085W | AAC3 | ADP/ATP transporter protein, member of the mitochondrial carrier (MCF) family |
0.8173 | AAH31912 | ADP/ATP translocase 3 |
| YBL047C | EDE1 | Key endocytic protein involved in a network of interactions with other endocytic proteins, binds membranes in a ubiquitin-dependent manner, may also bind ubiquitinated membrane-associated proteins |
0.8291 | Q9UBC2 | Epidermal growth factor receptor substrate 15-like 1 |
| YER151C | UBP3 | Ubiquitin-specific protease, ubiquitin C-terminal hydrolase |
0.834 | Q9BWG7 | Ubiquitin carboxyl-terminal hydrolase 10 |
| YJR073C | OPI3 | Phospholipid-N-methyltransferase; carries out second and third methylation steps of the phosphatidylcholine biosynthesis pathway |
0.8357 | Q9BW86 | Phosphatidylethanolamine N- methyltransferase |
| YCR020C-A | MAK31 | Protein required for structural stability of L-A double- stranded RNA- (dsRNA-) containing particles, has similarity to snRNA-associated proteins of the Sm family |
0.8388 | ||
| YNR010W | CSE2 | Component of RNA polymerase II mediator (SRB) subcomplex |
0.8433 | O75179 | Ankyrin repeat domain-containing protein 17 |
| YBR284W | Putative protein of unknown function | 0.8464 | Q96IA1 | AMP deaminase 2 | |
| YAL012W | CYS3 | Cystathionine gamma-lyase, generates cysteine from cystathionine |
0.8467 | P32929 | Cystathionine gamma-lyase |
| YPR179C | HDA3 | Subunit of a possibly tetrameric trichostatin A-sensitive class II histone deacetylase complex that contains an Hda1p homodimer and an Hda2p-Hda3p heterodimer; required for the activity of the complex; has similarity to Hda2p |
0.8494 | Q8N7Z2 | Golgin subfamily A member 6-like protein 1 |
| YNL201C | PSY2 | Putative subunit of an evolutionarily conserved protein phosphatase complex containing the catalytic subunit Pph3p and the regulatory subunit Psy4p; required for cisplatin and oxaliplatin resistance; localizes to nucleus |
0.8569 | BAC23106 | SMEK1 |
| YGR061C | ADE6 | 5'-phosphoribosylformyl glycinamidine synthetase, has glutamine amidotransferase domain and aminator domain |
0.8577 | Q9BX02 | |
| YNL107W | YAF9 | Subunit of both the NuA4 histone H4 acetyltransferase complex and the SWR1 complex, may function to antagonize silencing near telomeres; interacts directly with Swc4p, has homology to human leukemogenic protein AF9, contains a YEATS domain |
0.8635 | O95619 | YEATS domain-containing protein 4 |
| YCL008C | STP22 | Protein required for vacuolar targeting of temperature- sensitive plasma membrane proteins such as Ste2p and Can1p |
0.872 | Q9BUM5 | Tumor susceptibility gene 101 protein |
| YLR025W | SNF7 | Protein involved in glucose derepression and in protein sorting in pre-vacuolar endosome |
0.8731 | Q9H444 | Charged multivesicular body protein 4b |
| YBR200W | BEM1 | Protein required for cell polarization and bud formation, contains two SH3 domains |
0.8796 | Q9Y338 | Sorbin and SH3 domain-containing protein 1 |
| YJL080C | SCP160 | Protein involved in control of mitotic chromosome transmission, contains 14 KH motifs found in RNA- binding proteins such as Mer1p and mouse hnRNP X |
0.8926 | AAH01179 | Vigilin |
| YOR124C | UBP2 | Ubiquitin-specific protease (ubiquitin C-terminal hydrolase), cleaves at the C-terminus of ubiquitin |
0.9048 | Q96RU2 | Ubiquitin carboxyl-terminal hydrolase 28 |
| YDR293C | SSD1 | Protein with a role in maintenance of cellular integrity, interacts with components of the TOR pathway; ssd1 mutant of a clinical S. cerevisiae strain displays elevated virulence |
0.9048 | Q9Y2L1 | Exosome complex exonuclease RRP44 |
| YML014W | TRM9 | tRNA methyltransferase, catalyzes esterification of modified uridine nucleotides in tRNA(Arg3) and tRNA(Glu), likely as part of a complex with Trm112p; deletion confers resistance to zymocin |
0.9082 | Q9P272 | Putative methyltransferase KIAA1456 |
| YDL101C | DUN1 | Protein kinase required for induction of Rnr3p and DNA repair genes after DNA damage |
0.9116 | O96017 | Serine/threonine-protein kinase Chk2 |
| YDL146W | LDB17 | Protein of unknown function | 0.912 | AAH16052 | NCK interacting protein with SH3 domain |
| YDR358W | GGA1 | Golgi-localized protein with homology to gamma- adaptin, interacts with and regulates Arf1p and Arf2p in a GTP-dependent manner in order to facilitate traffic through the late Golgi |
0.9303 | Q9NZ52 | ADP-ribosylation factor-binding protein GGA3 |
| YBR030W | RKM3 | Protein with weak similarity to Sin3p | 0.9322 | Q9H787 | SET domain-containing protein 6 |
| YLL039C | UBI4 | Ubiquitin, mature protein is cleaved from polyubiquitin (Ubi4p) or from fusions with ribosomal proteins Rps31p, Rpl40Ap, or Rpl40Bp |
0.9427 | AAH39193 | Ubiquitin |
| YLR417W | VPS36 | Protein involved in vacuolar sorting; mutant has a prominent novel pre-vacuolar organelle |
0.9471 | Q9H8Z5 | Vacuolar protein-sorting-associated protein 36 |
| YKR007W | MEH1 | Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; loss results in a defect in vacuolar acidification |
0.9476 | Q9NVC8 | Ubiquitin carboxyl-terminal hydrolase 36 |
| YBL064C | PRX1 | Mitochondrial peroxiredoxin (1-Cys Prx) with thioredoxin peroxidase activity, has a role in reduction of hydroperoxides; induced during respiratory growth and under conditions of oxidative stress; phosphorylated |
0.9489 | AAH35857 | Peroxiredoxin-6 |
| YLR447C | VMA6 | Vacuolar H(+)-ATPase (V-ATPase) 36 kDa subunit (subunit D) of membrane (V0) sector, required for V- ATPase assembly |
0.9493 | P12953 | Vacuolar proton pump subunit d 1 |
| YOR371C | GPB1 | Multistep regulator of cAMP-PKA signaling; inhibits PKA downstream of Gpa2p and Cyr1p, thereby increasing cAMP dependency; inhibits Ras activity through direct interactions with Ira1p/2p; regulated by G-alpha protein Gpa2p; homolog of Gpb2p |
0.9591 | P51610 | Host cell factor |
| YKL134C | 10/1/1999 | Mitochondrial intermediate peptidase | 0.9685 | Q96G65 | Mitochondrial intermediate peptidase [Precursor] |
| YDR300C | PRO1 | Glutamate 5-kinase, carries out first step in proline biosynthesis pathway |
0.9723 | P54886 | Delta-1-pyrroline-5-carboxylate synthetase |
| YCR034W | FEN1 | Protein involved in the elongation of fatty acids up to 24 carbons |
0.9727 | Q9NYP7 | Elongation of very long chain fatty acids protein 5 |
| YLR200W | YKE2 | Protein involved in microtubule biogenesis | 1.001 | AAH39033 | Prefoldin subunit 6 |
| YOR322C | LDB19 | Protein of unknown function involved in maintenance of proper telomere length; null mutant shows a reduced affinity for the alcian blue dye suggesting a decreased net negative charge of the cell surface |
1.005 | Q9UJF2 | Ras GTPase-activating protein nGAP |
| YOL039W | RPP2A | Acidic ribosomal protein P2A (L44; A2; YP2alpha; E. coli L12eIB; human P2alpha), plays a role in the elongation step |
1.007 | P05387 | 60S acidic ribosomal protein P2 |
| YPL158C | AIM44 | Protein of unknown function; GFP-fusion protein localizes to the bud neck; transcription is regulated by Swi5p; null mutant displays increased frequency of mitochondrial genome loss and reduced growth rate in minimal glycerol media |
1.009 | Q8NAM5 | Putative protein TPRXL |
| YKL041W | VPS24 | Protein involved in sorting of proteins in pre-vacuolar endosome |
1.012 | Q9Y3E7 | Charged multivesicular body protein 3 |
| YML057W | CMP2 | Calcineurin catalytic (A) subunit, protein serine/threonine phosphatase 2B (PP2B), member of the PPP family of protein phosphatases |
1.022 | Q8TAW9 | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform |
| YLR335W | NUP2 | Nuclear pore protein (nucleoporin) with XFXFG motifs; has functional overlap with other proteins of nuclear pore complex |
1.026 | P49792 | E3 SUMO-protein ligase RanBP2 |
| YGR101W | PCP1 | Mitochondrial serine protease required for the processing of various mitochondrial proteins and maintenance of mitochondrial DNA and morphology; belongs to the rhomboid-GlpG superfamily of intramembrane peptidases | 1.033 | Q96CQ4 | Presenilins-associated rhomboid-like protein, mitochondrial [Precursor] |
| YNL021W | HDA1 | Component of histone deacetylase A, 75 kDa subunit | 1.043 | Q9NSW6 | Putative uncharacterized protein DKFZp566E044 |
| YOR265W | RBL2 | Protein that rescues excess beta-tubulin lethality | 1.045 | O75347 | Tubulin-specific chaperone A |
| YDL020C | RPN4 | Subunit of the regulatory particle of the proteasome | 1.046 | AAH36038 | Zinc finger protein 25 |
| YHR082C | KSP1 | Serine/threonine kinase that suppresses prp20 mutant when oveproduced |
1.048 | Q9P0L2 | Serine/threonine-protein kinase MARK1 |
| YJL204C | RCY1 | F-box protein involved in recycling plasma membrane proteins internalized by endocytosis; localized to sites of polarized growth |
1.056 | O00471 | Exocyst complex component 5 |
| YHR026W | PPA1 | Proteolipid of the vacuolar H(+)-ATPase (V-ATPase) | 1.058 | Q99437 | Vacuolar ATP synthase 21 kDa proteolipid subunit |
| YKL212W | SAC1 | Phosphatidylinositol phosphate (PtdInsP) phosphatase involved in hydrolysis of PtdIns[4]P; transmembrane protein localizes to ER and Golgi; involved in protein trafficking and processing, secretion, and cell wall maintenance |
1.06 | O94935 | Phosphatidylinositide phosphatase SAC1 |
| YGL240W | DOC1 | Component of the anaphase-promoting complex (APC); required for Clb2p degradation and for the metaphase- anaphase transition |
1.06 | Q9Y5R0 | Anaphase-promoting complex subunit 10 |
| YKR031C | SPO14 | Phospholipase D, catalyzes the hydrolysis of phosphatidylcholine, producing choline and phosphatidic acid; involved in Sec14p-independent secretion; required for meiosis and spore formation; differently regulated in secretion and meiosis |
1.061 | O14939 | Phospholipase D2 |
| YFL025C | BST1 | GPI inositol deacylase of the ER that negatively regulates COPII vesicle formation, prevents production of vesicles with defective subunits, required for proper discrimination between resident ER proteins and Golgi- bound cargo molecules |
1.066 | Q9HA24 | GPI inositol-deacylase |
| YDL223C | HBT1 | Substrate of the Hub1p ubiquitin-like protein that localizes to the shmoo tip (mating projection); mutants are defective for mating projection formation, thereby implicating Hbt1p in polarized cell morphogenesis |
1.066 | Q9NZW4 | Dentin sialophosphoprotein, [Precursor] |
| YNL298W | CLA4 | Serine/threonine protein kinase required for cytokinesis | 1.068 | Q13153 | Serine/threonine-protein kinase PAK |
| YPL002C | SNF8 | Protein involved in glucose derepression | 1.076 | Q96H20 | Vacuolar-sorting protein SNF8 |
| YNL215W | IES2 | Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions; essential for growth under anaerobic conditions |
1.076 | Q16081 | Nexilin |
| YMR004W | MVP1 | Protein required for sorting proteins to the vacuole, interacts genetically with Vps1p |
1.083 | Q9Y5X2 | Sorting nexin-8 |
| YKR100C | SKG1 | Transmembrane protein with a role in cell wall polymer composition; localizes on the inner surface of the plasma membrane at the bud and in the daughter cell |
1.085 | Q9NW40 | Pre-mRNA-splicing factor 38B |
| YJL189W | RPL39 | Ribosomal protein L39 | 1.09 | P02404 | 60S ribosomal protein L39 |
| YAR015W | ADE1 | Phosphoribosylamidoimidazole-succinocarboxamide synthase; (SAICAR synthetase), catalyzes the seventh step in de novo purine biosynthesis pathway |
1.097 | P22234 | Multifunctional protein ADE2 |
| YNL097C | PHO23 | Protein involved in expression of PHO5 | 1.1 | Q9NXR8 | Inhibitor of growth protein 3 |
| YAL002W | VPS8 | Protein involved in vacuolar sorting | 1.102 | Q8N3P4 | Vacuolar protein sorting-associated protein 8 homolog |
| YML058C-A | Protein of unknown function | 1.11 | |||
| YJL047C | RTT101 | Protein of the cullin family, with similarity to Cdc53p | 1.11 | AAH09591 | Cullin-2 ( |
| YJR145C | RPS4A | Ribosomal protein S4 (yeast S7; YS6; rp5; rat and human S4), identical to Rps4Bp |
1.11 | P12750 | 40S ribosomal protein S4, X isoform |
| YPR030W | CSR2 | Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
1.111 | Q9NZW4 | Dentin sialophosphoprotein [Precursor] |
| YNR051C | BRE5 | Ubiquitin protease cofactor, forms deubiquitination complex with Ubp3p that coregulates anterograde and retrograde transport between the endoplasmic reticulum and Golgi compartments; null is sensitive to brefeldin A |
1.121 | Q9BX49 | Proteoglycan-4 [Precursor] |
| YGR180C | RNR4 | Ribonucleotide reductase small subunit | 1.123 | AAH30154 | Ribonucleoside-diphosphate reductase subunit M2 |
| YKL135C | APL2 | Beta-adaptin, large subunit of the clathrin-associated protein (AP) complex |
1.123 | Q96J19 | AP-2 complex subunit beta-1 |
| YJL036W | SNX4 | Sorting nexin, involved in retrieval of late-Golgi SNAREs from post-Golgi endosomes to the trans-Golgi network and in cytoplasm to vacuole transport; contains a PX phosphoinositide-binding domain; forms complexes with Snx41p and with Atg20p |
1.124 | O95219 | Sorting nexin-4 |
| YOL072W | THP1 | Nuclear pore-associated protein, forms a complex with Sac3p that is involved in transcription and in mRNA export from the nucleus; contains a PAM domain implicated in protein-protein binding |
1.133 | Q9NWH3 | PCI domain-containing protein 2 |
| YDL155W | CLB3 | G2/M-phase-specific cyclin | 1.137 | O95067 | G2/mitotic-specific cyclin-B2 |
| YJR075W | HOC1 | Subunit of the Anp1p-Hoc1p-Mnn11p-Mnn9p mannosyltransferase complex of the Golgi involved in cell wall integrity |
1.14 | Q9Y6P7 | snRNA-activating protein complex subunit 4 |
| YOR201C | PET56 | Ribose methyltransferase specific for G2270 in mitochondrial 21S rRNA |
1.145 | Q13395 | Probable methyltransferase TARBP1 |
| YKL143W | LTV1 | Protein required for viability at low temperature | 1.148 | Q96GA3 | Protein LTV1 homolog |
| YJL062W | LAS21 | Protein required for addition of a side chain to the glycosylphospatidylinositol (GPI) core structure |
1.151 | Q8NCC9 | GPI ethanolamine phosphate transferase 2 |
| YJR113C | RSM7 | Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S7 ribosomal protein |
1.152 | P46782 | 40S ribosomal protein S5 |
| YBR295W | PCA1 | P-type copper-transporting ATPase | 1.166 | P35670 | Copper-transporting ATPase 2 |
| YKL197C | PEX1 | AAA-peroxin that heterodimerizes with AAA-peroxin Pex6p and participates in the recycling of peroxisomal signal receptor Pex5p from the peroxisomal membrane to the cystosol; induced by oleic acid and upregulated during anaerobiosis |
1.176 | AAH35575 | Peroxisome biogenesis factor 1 |
| YDR099W | BMH2 | Homolog of mammalian 14-3-3 protein, has strong similarity to Bmh1p |
1.178 | AAH39025 | |
| YPL032C | SVL3 | Protein involved in vacuolar uptake of endocytosed vital dyes |
1.186 | Q8NAM5 | Putative protein TPRXL |
| YGR270W | YTA7 | Protein that localizes to chromatin and has a role in regulation of histone gene expression; has a bromodomain-like region that interacts with the N-terminal tail of histone H3, and an ATPase domain; potentially phosphorylated by Cdc28p |
1.196 | Q8N890 | ATPase family AAA domain- containing protein 2 |
| YDL081C | RPP1A | Acidic ribosomal protein P1A (A1; YP1alpha; E. coli L12eIIA; human and rat P1) |
1.208 | P05386 | 60S acidic ribosomal protein P1 |
| YLR268W | SEC22 | Synaptobrevin (v-SNARE) homolog involved in fusion of ER-to-Golgi transport vesicles; recognized by putative target t-SNARE (Sed5p) |
1.209 | O75396 | Vesicle-trafficking protein SEC22b |
| YPR051W | MAK3 | Protein N-acetyltransferase, acetylates N-terminus of L- A virus GAG protein |
1.212 | P41227 | N-terminal acetyltransferase complex ARD1 subunit homolog A |
| YGL167C | PMR1 | Ca2+-transporting P-type ATPase of Golgi membrane involved in Ca2+ import into Golgi |
1.212 | P98194 | Calcium-transporting ATPase type 2C member 1 |
| YNR006W | VPS27 | Protein involved in vacuolar sorting; mutants develop a prominent novel pre-vacuolar organelle |
1.213 | Q9NR36 | Hepatocyte growth factor-regulated tyrosine kinase substrate |
| YKL009W | MRT4 | Protein involved in mRNA turnover | 1.214 | Q9UKD2 | mRNA turnover protein 4 homolog |
| YPL079W | RPL21B | Ribosomal protein L21 (rat L21), nearly identical to Rpl21Ap |
1.229 | P46778 | 60S ribosomal protein L21 |
| YJR102C | VPS25 | Component of the ESCRT-II complex, which is involved in ubiquitin-dependent sorting of proteins into the endosome |
1.235 | Q9BRG1 | Vacuolar protein-sorting-associated protein 25 |
| YOR332W | VMA4 | Vacuolar H(+)-ATPase (V-ATPase) hydrophilic subunit (subunit E), 27 kDa subunit of V1 sector |
1.237 | AAH04443 | Vacuolar proton pump subunit E 1 |
| YNR071C | Putative protein of unknown function | 1.238 | Q96C23 | Aldose 1-epimerase | |
| YPL247C | Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; similar to the petunia WD repeat protein an11; overexpression causes a cell cycle delay or arrest |
1.242 | O15491 | WD repeat-containing protein 68 | |
| YMR052C-A | Protein of unknown function | 1.242 | |||
| YNL091W | NST1 | Protein with similarity to Uso1p and human NF2 neurofibromatosis type 2 gene product |
1.258 | Q8NAM5 | Putative protein TPRXL |
| YKL067W | YNK1 | Nucleoside diphosphate kinase, responsible for synthesis of all nucleoside triphosphates except ATP |
1.268 | P22392 | Nucleoside diphosphate kinase B |
| YBR111C | YSA1 | Nucleoside diphosphate-sugar hydrolase of the MutT (nudix) family |
1.27 | Q9UKK9 | ADP-sugar pyrophosphatase |
| YOR089C | VPS21 | GTP-binding protein of the rab family required for sorting of vacuolar proteins and involved in late stage of endocytosis |
1.272 | AAO15677 | RAB5A protein |
| YEL012W | UBC8 | Ubiquitin-conjugating enzyme that is able to ubiquitinate histones in vitro |
1.276 | P37286 | Ubiquitin-conjugating enzyme E2 H |
| YBL090W | MRP21 | Mitochondrial ribosomal protein of the small subunit | 1.282 | P82921 | 28S ribosomal protein S21, mitochondrial |
| YGL256W | ADH4 | Alcohol dehydrogenase IV | 1.283 | AAK44223 | Hydroxyacid-oxoacid transhydrogenase, mitochondrial precursor |
| YEL062W | NPR2 | Nitrogen permease regulator | 1.295 | Q9Y249 | Tumor suppressor candidate 4 |
| YJL112W | MDV1 | Peripheral protein of the cytosolic face of the mitochondrial outer membrane, required for mitochondrial fission; interacts with Fis1p and with the dynamin-related GTPase Dnm1p; contains WD repeats |
1.299 | Q96LE0 | F-box/WD repeat-containing protein 7 |
| YOR295W | UAF30 | Subunit of UAF (upstream activation factor), which is an RNA polymerase I specific transcription stimulatory factor composed of Uaf30p, Rrn5p, Rrn9p, Rrn10p, histones H3 and H4; deletion decreases cellular growth rate |
1.3 | Q96GM5 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 |
| YML081C-A | ATP18 | Subunit of the mitochondrial F1F0 ATP synthase, which is a large enzyme complex required for ATP synthesis; termed subunit I or subunit j; does not correspond to known ATP synthase subunits in other organisms |
1.31 | ||
| YML032C | RAD52 | Protein required for recombination and repair of X-ray damage, has a late function in meiotic recombination |
1.31 | P43351 | DNA repair protein RAD52 homolog |
| YMR116C | ASC1 | Abundant protein with effects on translational efficiency and cell size, has two WD (WD-40) repeats |
1.311 | AAH32006 | Guanine nucleotide-binding protein subunit beta-2-like 1 |
| YJR104C | SOD1 | Copper-zinc superoxide dismutase | 1.316 | AAL15444 | Superoxide dismutase |
| YJL121C | RPE1 | Ribulose-5-phosphate 3-epimerase, interconverts ribulose-5-phosphate and xylulose-5-phosphate |
1.317 | Q9BSB5 | Ribulose-phosphate 3-epimerase |
| YLR148W | PEP3 | Vacuolar peripheral membrane protein involved in vacuolar protein sorting and required for vacuole biogenesis |
1.317 | Q9P253 | Vacuolar protein sorting-associated protein 18 homolog |
| YOR258W | HNT3 | Member of the third branch of the histidine triad (HIT) superfamily of nucleotide-binding proteins; similar to Aprataxin, a Hint related protein that is mutated in individuals with ataxia with oculomotor apraxia |
1.322 | Q9NXM5 | |
| YMR198W | CIK1 | Coiled-coil protein of spindle pole body involved in spindle formation and the congression (nuclear migration) step of karyogamy |
1.322 | Q13439 | Golgin subfamily A member 4 |
| YKR020W | VPS51 | Component of the GARP (Golgi-associated retrograde protein) complex, Vps51p-Vps52p-Vps53p-Vps54p, which is required for the recycling of proteins from endosomes to the late Golgi; links the (VFT/GARP) complex to the SNARE Tlg1p |
1.324 | Q13999 | Kinectin |
| YJL102W | MEF2 | Mitochondrial translation elongation factor, promotes GTP-dependent translocation of nascent chain from A- site to P-site of ribosome |
1.33 | Q8N6D8 | G elongation factor, mitochondrial 2 |
| YJL063C | MRPL8 | Mitochondrial ribosomal protein of the large subunit (YmL8) |
1.338 | Q9C066 | 39S ribosomal protein L17, mitochondrial [Precursor] |
| YIL148W | RPL40A | Fusion protein comprised of ribosomal protein L40 (C- terminal half) and ubiquitin (N-terminal half) (rat L40), identical to Rpl40Bp |
1.338 | Q9BX98 | 60S ribosomal protein L40 |
| YPL045W | VPS16 | Vacuolar sorting protein; mutant has pleiotropic defects in vacuolar morphology and vacuolar protein targeting |
1.342 | Q9H269 | Vacuolar protein sorting-associated protein 16 homolog |
| YKL160W | ELF1 | Transcription elongation factor that contains a conserved zinc finger domain; implicated in the maintenance of proper chromatin structure in actively transcribed regions; deletion inhibits Brome mosaic virus (BMV) gene expression |
1.343 | Q96II4 | Transcription elongation factor 1 homolog |
| YPR024W | YME1 | Mitochondrial zinc-dependent protease of the AAA family of ATPases |
1.357 | AAH23507 | ATP-dependent metalloprotease YME1L1 |
| YGL136C | MRM2 | Mitochondrial 2' O-ribose methyltransferase, required for methylation of U(2791) in 21S rRNA; MRM2 deletion confers thermosensitive respiration and loss of mitochondrial DNA; has similarity to Spb1p and Trm7p, and to E. coli FtsJ/RrmJ |
1.357 | Q9UI43 | Putative ribosomal RNA methyltransferase 2 |
| YKR055W | RHO4 | Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely to be involved in the establishment of cell polarity |
1.36 | P06749 | Transforming protein RhoA precursor |
| YGR092W | DBF2 | Serine/threonine protein kinase related to Dbf20p, required for events in anaphase/telophase |
1.36 | Q9Y2H1 | Serine/threonine-protein kinase 38- like |
| YNL022C | Protein of unknown function | 1.377 | Q9NW70 | Putative methyltransferase NSUN5 | |
| YMR145C | NDE1 | Mitochondrial external NADH dehydrogenase, a type II NAD(P)H:quinone oxidoreductase that catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p provide cytosolic NADH to the mitochondrial respiratory chain |
1.378 | AAH23601 | Apoptosis-inducing factor 2 |
| YKL211C | TRP3 | Bifunctional enzyme exhibiting both indole-3-glycerol- phosphate synthase and anthranilate synthase activities, forms multifunctional hetero-oligomeric anthranilate synthase:indole-3-glycerol phosphate synthase enzyme complex with Trp2p |
1.379 | AAH12178 | GMP synthase |
| YML016C | PPZ1 | Protein serine/threonine phosphatase required for normal osmoregulation, member of the PPP family of protein phosphatases and related to PP1 phosphatases |
1.383 | P36873 | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit |
| YKR099W | BAS1 | Transcription factor; involved in regulation of basal and induced activity of histidine and adenine biosynthesis genes |
1.388 | Q9Y6P7 | snRNA-activating protein complex subunit 4 |
| YAL043C-A | Protein of unknown function | 1.395 | |||
| YDL065C | PEX19 | Farnesylated protein required for peroxisome biogenesis (peroxin) |
1.395 | Q9HAT7 | Ninein |
| YOR027W | STI1 | Stress-induced protein required for optimal growth at high and low temperature, has tetratricopeptide (TPR) repeats |
1.397 | P31948 | Stress-induced-phosphoprotein 1 |
| YPL254W | HFI1 | Component of the ADA complex, interacts functionally with histone H2Ai |
1.398 | Q96FJ7 | Protein ADRM1 |
| YAL042W | ERV46 | Protein localized to COPII-coated vesicles, forms a complex with Erv41p; involved in the membrane fusion stage of transport |
1.403 | Q9Y282 | Endoplasmic reticulum-Golgi intermediate compartment protein 3 |
| YOR359W | VTS1 | Post-transcriptional gene regulator, RNA-binding protein containing a SAM domain; shows genetic interactions with Vti1p, which is a v-SNARE involved in cis-Golgi membrane traffic |
1.406 | Q9UPU9 | Sterile alpha motif domain- containing protein 4A |
| YLR119W | SRN2 | Suppressor of rna1-1 mutant | 1.407 | Q96KQ3 | Apoptosis-stimulating of p53 protein 2 |
| YML026C | RPS18B | Ribosomal protein S18 (E. coli S13; rat S18), identical to Rps18Ap |
1.413 | P25232 | 40S ribosomal protein S18 |
| YFL001W | DEG1 | Pseudouridine synthase that catalyzes the formation of pseudouridine-38 and -39 in cytoplasmic and mitochondrial tRNAs |
1.415 | Q96NB4 | tRNA pseudouridine synthase 3 |
| YOR123C | LEO1 | Protein of unknown function, extremely hydrophilic | 1.42 | Q8WVC0 | RNA polymerase-associated protein LEO1 |
| YKL197C | PEX1 | Peroxisomal biogenesis protein (peroxin); member of the AAA family of ATPases |
1.424 | AAH35575 | Peroxisome biogenesis factor 1 |
| YML033W | Protein of unknown function | 1.424 | |||
| YNL299W | TRF5 | Protein functionally similar to DNA topoisomerase I | 1.424 | Q9Y6C1 | DNA polymerase sigma |
| YGL227W | VID30 | Protein involved in proteasome-dependent catabolite degradation of fructose-1,6-bisphosphatase (FBPase); binds FBPase; shifts the balance of nitrogen metabolism toward glutamate production; localizes to the nucleus and the cytoplasm |
1.44 | Q9P264 | Ran-binding protein 10 |
| YLR068W | FYV7 | Essential protein required for maturation of 18S rRNA; required for survival upon exposure to K1 killer toxin |
1.453 | Q05682 | Caldesmon |
| YMR039C | SUB1 | Transcriptional coactivator; may be involved in the release of TFIIB from the transcription complex during RNA polymerase II transcription initiation |
1.464 | Q96L29 | Activated RNA polymerase II transcriptional coactivator p15 |
| YMR234W | RNH1 | Ribonuclease H, endonuclease that degrades RNA in RNA-DNA hybrids |
1.47 | O60930 | Ribonuclease H1 |
| YOL051W | GAL11 | Component of RNA polymerase II holoenzyme and Kornberg's mediator complex with positive and negative effects on transcription of individual genes |
1.479 | Q8NAM5 | Putative protein TPRXL |
| YDR017C | KCS1 | Potential transcription factor of the basic leucine zipper (bZIP) type, suppressor of temperature-sensitive growth and hyperrecombination in pkc1-4 |
1.482 | Q96PC2 | Inositol hexaphosphate kinase 3 |
| YNR052C | POP2 | Component of the CCR4 complex required for glucose derepression |
1.482 | AAH17366 | CCR4-NOT transcription complex subunit 8 |
| YMR060C | TOM37 | Component of mitochondrial outer membrane receptor complex, needed only at high temperature, has tetratricopeptide (TPR) repeats |
1.494 | Q13505 | Metaxin-1 |
| YDR001C | NTH1 | Neutral trehalase | 1.494 | O43280 | Trehalase [Precursor] |
| YNL329C | PEX6 | Peroxisomal biogenesis protein (peroxin) of the AAA family of ATPases |
1.497 | Q8WYQ2 | Peroxisome assembly factor 2 |
| YJL172W | CPS1 | Gly-X carboxypeptidase yscS, involved in nitrogen metabolism |
1.507 | Q96DM4 | Probable carboxypeptidase PM20D1 [Precursor] |
| YHR004C | NEM1 | Protein required for nuclear morphology | 1.51 | Q96GQ9 | Serine/threonine-protein phosphatase dullard |
| YJL029C | VPS53 | Component of the GARP (Golgi-associated retrograde protein) complex, Vps51p-Vps52p-Vps53p-Vps54p, which is required for the recycling of proteins from endosomes to the late Golgi; required for vacuolar protein sorting |
1.554 | Q9BY02 | Vacuolar protein sorting-associated protein 53 homolog |
| YMR219W | ESC1 | Protein of unknown function | 1.586 | Q9NZW4 | Dentin sialophosphoprotein [Precursor] |
| YDR162C | NBP2 | Nap1p-binding protein, has an SH3 domain | 1.612 | Q9P234 | Putative E3 ubiquitin-protein ligase SH3RF1 |
| YGR135W | PRE9 | Proteasome subunit alpha3_sc | 1.619 | Q8TBD1 | Proteasome subunit alpha type-4 |
| YNR032W | PPG1 | Protein serine/threonine phosphatase involved in glycogen accumulation, member of the PPP family of protein phosphatases and related to PP2A phosphatases |
1.718 | P33172 | Serine/threonine-protein phosphatase 4 catalytic subunit |
| YHR066W | SSF1 | Protein with a potential role in mating | 1.731 | ||
| YGR085C | RPL11B | Ribosomal protein L11 (yeast L16; YL22; rp39B; E. coli L5; rat L11), nearly identical to Rpl11Ap |
1.736 | Q9Y674 | 60S ribosomal protein L11 |
| YNL248C | RPA49 | RNA polymerase I third largest subunit | 1.753 | Q96L20 | DNA-directed RNA polymerase I subunit RPA49 |
| YNL302C | RPS19B | Ribosomal protein S19 (rp55; YS16B; rat S19), nearly identical to Rps19Ap |
1.757 | Q8WVX7 | Ribosomal protein S19 [Fragment] |
| YGL228W | SHE10 | Protein that causes lethality when overexpressed | 1.808 | O60437 | Periplakin |
| YDL096C | OPI6 | Protein of unknown function | 1.921 | ||
| YLR061W | RPL22A | Ribosomal protein L22, similar to Rpl22Bp | 1.932 | P35268 | 60S ribosomal protein L22 |
| YDR227W | SIR4 | Coiled-coil protein involved in maintenance of silencing of HMR, HML, and telomeres |
1.936 | BAA74868 | Neurofilament heavy polypeptide |
| YHR178W | STB5 | Protein with similarity to transcription factors | 1.996 | Q8NAM5 | Putative protein TPRXL |
| YKL191W | DPH2 | Protein required, along with Dph1p, Kti11p, Jjj3p, and Dph5p, for synthesis of diphthamide, which is a modified histidine residue of translation elongation factor 2 (Eft1p or Eft2p); may act in a complex with Dph1p and Kti11p |
2 | Q9BQC3 | Diphthamide biosynthesis protein 2 |
| YLR388W | RPS29A | Ribosomal protein S29 (yeast S36; YS29; rat S29), similar to Rps29Bp |
2.098 | AAH35313 | 40S ribosomal protein S29 |
| YGL203C | KEX1 | Carboxypeptidase specific for terminal arg or lys, involved in processing precursors of alpha-factor and K1 and K2 killer toxins |
2.115 | Q9BR08 | Lysosomal protective protein [Precursor] |
| YCR031C | RPS14A | Ribosomal protein S14A (rp59, E. coli S11, rat and human S14) involved in crytopleurine resistance, nearly identical to Rps14Bp |
2.199 | P06366 | 40S ribosomal protein S14 |
| YKL190W | CNB1 | Calcineurin regulatory (B) subunit | 2.274 | AAH27913 | Calcineurin subunit B type 1 |
| YNL246W | VPS75 | NAP family histone chaperone; binds to histones and Rtt109p, stimulating histone acetyltransferase activity; possesses nucleosome assembly activity in vitro; proposed role in vacuolar protein sorting and in double-strand break repair |
2.5 | Q9UJ03 | |
| YLR024C | UBR2 | Cytoplasmic ubiquitin-protein ligase (E3); required for ubiquitylation of Rpn4p; mediates formation of a Mub1p- Ubr2p-Rad6p complex |
2.5 | AAL32101 | E3 ubiquitin-protein ligase UBR2 |
| YMR304W | UBP15 | Putative ubiquitin-specific protease, ubiquitin C-terminal hydrolase |
2.5 | Q93009 | Ubiquitin carboxyl-terminal hydrolase 7 |
| YAL023C | PMT2 | Mannosyltransferase; (dolichyl phosphate-D-mannose | 2.932 | Q9P1W0 | Protein O-mannosyl-transferase 2 |
| YML106W | URA5 | Orotate phosphoribosyltransferase 1; fifth step in pyrimidine biosynthesis pathway |
3.233 | BAB93468 | Uridine 5'-monophosphate synthase |
| YOL008W | COQ10 | Coenzyme Q (ubiquinone) binding protein, functions in the delivery of Q6 to its proper location for electron transport during respiration; START domain protein with homologs in bacteria and eukaryotes |
3.487 | Q9BUP4 | Protein COQ10 A, mitochondrial [Precursor] |
| YOR039W | CKB2 | Casein kinase II Protein kinase CK2), regulatory (beta- prime) subunit |
3.597 | AAH35349 | |
| YOR084W | LPX1 | Oleic acid-inducible, peroxisomal matrix localized lipase; transcriptionally activated by Yrm1p along with genes involved in multidrug resistance; peroxisomal import is dependent on the PTS1 receptor, Pex5p and on self- interaction |
3.599 | Q9NVT5 | Protein phosphatase methylesterase 1 |
| YMR214W | SCJ1 | Homolog of E. coli DnaJ, functions in the endoplasmic reticulum by interaction with Kar2p |
3.943 | O60884 | DnaJ homolog subfamily A member 2 |
| YMR100W | MUB1 | Zinc finger protein, involved in the regulation of bud site selection |
4.642 | CAC16691 | Zinc finger MYND domain- containing protein 19 |
| YKR042W | UTH1 | Mitochondrial outer membrane and cell wall localized SUN family member required for mitochondrial autophagy; involved in the oxidative stress response, life span during starvation, mitochondrial biogenesis, and cell death |
above 5 | BAB79693 | Receptor activator of nuclear factor kappa B ligand 3 |
| YLL043W | FPS1 | Glycerol channel protein, member of the major intrinsic protein (MIP) family of transmembrane channel proteins |
above 5 | O43315 | Aquaporin-9 |
| YGR072W | UPF3 | Protein involved with Nam7p and Nmd2p in decay of mRNA containing nonsense codons |
above 5 | Q9H1J0 | Regulator of nonsense transcripts 3B |
| YGR097W | ASK10 | Potential transcription factor involved in Skn7p-mediated two-component regulatory system |
above 5 | O75404 | Pre-mRNA-processing factor 40 homolog A |
To identify genes whose deletion renders yeast most sensitive or resistant to arsenite toxicity, we determined the IC50 of the indentified mutants as well as the wild type strain. The degree of sensitivity or resistance of each gene was ordered based on the IC50 value (Table 1). The IC50 of the wild type strain for sodium arsenite is 4.47 mM. While most mutants are sensitive to As (III), a few of them (Mub1, Uth1, Fps1, Upf3, Ask10 and P15B12) are resistant to As (III) when compared to the IC50 of the wild type (Table 1).
Biological categories of arsenite-toxicity modulating proteins
Several studies have used S. cerevisiae as a tool to identify the molecules and cellular pathways linking arsenic induced toxicity and carcinogenicity. Nucleic acid metabolism, oxidative phosphorylation, protein synthesis and vacuolar acidification were involved in either arsenite sensitivity or resistance as determined by screening single gene knockout strains of S. cerevisiae in mitochondrial biogenesis and function [22]. The strains whose deletion confers sensitivity to arsenic trioxide were found to be significantly enriched in the biological processes of osmoregulation, stress-related transcription regulation, cytoskeletal assembly and maintenance, signal transduction, DNA repair, oxidative stress, glutathione synthesis, secretory pathways and vacuole function, and general defense mechanisms [23]. Here we analyzed the effect of arsenite on yeast single-gene deletion mutants. Our studies with sodium arsenite found many genes in common with those of studies done by others using arsenic trioxide, and yet additional genes whose deletion leads to sensitivity or resistance exclusively to arsenite were also identified in this study.
Cytoskeleton and structure proteins
The strain most sensitive to sodium arsenite exposure, with an IC50 of 0.24 mM, lacks Pfd1 (Table 1). Pfd1 is subunit 1 of prefoldin, involved in the biogenesis of actin and of alpha- and gamma-tubulin, which are, in turn, important for cytoskeleton stability. A strain that lacks Gim4, which is prefoldin subunit 2 and a component of the Gim protein complex that promotes formation of functional alpha- and gamma-tubulin [24], was the second most sensitive strain to sodium arsenite exposure in the category of cytoskeleton assembly and maintenance. Thirteen other mutants that were sensitive to As (III), including Hsl7, Bem1, Ste50, Rvs161, Rpn4, Rvs167, Pac10, Sac1, Yke2, Tub3, Cla4, Svl3 and Nip100, correspond to cytoskeleton or structural proteins (Table 2). Cytoskeleton formation is important in establishing cell shape, providing mechanical strength, regulation of cell motility, chromosome separation in mitosis and meiosis, and intracellular transport of vesicles and protein complexes. Microtubules are one of the components of the cytoskeleton and are polymers of α- and β-tubulin dimers. Sodium arsenite directly interacts with the sulfhydryl-containing cysteine residues of tubulin, disrupting tubulin organization and microtubule assembly, and is proposed to induce aneuploidy in arsenite-treated human lymphocytes [25]. Thus, the requirement of Pdf1 and Gim4 in synthesizing tubulins, as well as other proteins that maintain cell structure, should be important in protecting cells from arsenite-induced damage to the cytoskeleton.
Acetylation and deacetylation
Histone acetylation is associated with activation of gene expression and it also seems to be affected by arsenic. We have found that eleven of the S. cerevisiae strains sensitive to arsenite lack proteins involved in the acetylation or deacetylation process (Table 2), and these include Sgf29, Mak31, Ada2, Sgf73, Gcn5, Ard1, Hda1, Pho23, Hfi1, Mak3 and Hda3. Ada2, Gcn5 and Sgf29 are part of Spt-Ada-Gcn5 acetyltransferase(SAGA) complex which contains more than 20 subunits [26]. The IC50 of Ada2, Gcn5 and Sgf29 deletion mutants are 0.375, 0.5 and 0.53, respectively. SAGA preferentially acetylates multiple lysine residues on the N-terminal tails of histone H3 and H2B [27], including acetylation of K9, K14, K18 and K23 of H3 [28]. The component protein Gcn5 (general control nonderepressible 5) has histone acetyltransferase activity [29] and Ada2 potentiates Gcn5 acetyltransferase activity [30]. SAGA regulates transcription of approximately 10% of the genome, most of which are upregulated in response to environmental stresses, including heat, oxidation, acidity, DNA damage, carbon or nitrogen starvation, and excess unfolded proteins [31].
Osmotic stress response and MAPK pathway
Six of the S. cerevisiae strains sensitive to As (III) were missing genes whose corresponding proteins were involved in osmoregulation. These included Ste50, Doa4, PbsS2, Hog1, Nst1 and Ssk2. In yeast, cells respond to osmotic stress through a high-osmolarity glycerol (HOG1) pathway to maintain optimal cell volume and viability [32]. In humans, the mitogen-activated protein kinase (MAPK) super-family consists of three major sets of kinases: the extracellular-receptor kinases (ERKs), the c-Jun N-terminal kinases/stress-activated protein kinases (JNK/SAPK), and the p38 MAPK. Hog1 is homologous to the p38 MAPK [32] and it activates its targets, including several transcription factors, which in turn activate genes devoted to osmoadaptation [33; 34; 35]. Osmotic stress activates Hog1 through the MAPKKK Ssk2 and the MAPKK Pbs2. Notably, HOG1 is the second most sensitive mutant with an IC50 of 0.31 mM (Table 1). Strains lacking Pbs2 and Ssk2 are also very sensitive to As (III), with IC50 of 0.5 mM and 0.55 mM, respectively (Table 1). In mammalian cells, the MAPK p38 pathway is activated by As (III) [36]. Similarly, tolerance of fission yeast Schizosaccharomyces pombe to As (III) involves the MAPK Spc1, a homologue of mammalian p38 MAPK [37].
Vacuolar transport
Vacuoles function to compartmentalize materials that may be harmful to cells. Glutathione-conjugated arsenic can be sequestered by Ycf1 in the vacuole, which contributes to cellular tolerance of arsenic [38]. The proteins whose deletions confer sensitivity to As(III) in the category of vacuolar transport are Yps8, Stp22, Fen1, Cup5, Vps25, Vps24, Vps51, Snf7, Srn2, Pep3, Vps36, Vma6, Mvp1, Vps21, Vma4, Vts1, Snf8, Vps16 and Bro1(Table 2).
Ubiquitination and proteosomal degradation
Removal of damaged molecules is a defense mechanism that maintains cellular and genetic integrity in response to environmental insults. Proteins are generally degraded by the ubiquitin (Ub)-mediated protein degradation pathway. Ub is conjugated to proteins by ubiquitin ligases. This tagging process leads to their recognition by the 26S proteasome, and ubiquitinated proteins are targeted to the 26S proteasome for degradation. Eight of the sensitive mutants lack genes whose corresponding proteins are involved in ubiquitination and deubiquitination, including Doa4, Ubc8, Ubp3, Rtt101, Grr1, Ubi4, Bre5 and Ubp2, and ten of the sensitive mutants lack proteins involved in proteosomal degradation, namely Rpn4, Ubc8, Bst1, Doc1, Pre9, Rpl40A, Rtt101, Grr1, Doa1 and Bro1(Table 2).
In the category of ubiquitination and proteosomal degradation, Grr1 displays a significant sensitivity to As (III) with an IC50 of 0.35 mM (Table 1). Similarly, Grr1 null yeast cells exhibit an elongated sausage-shape, and are sensitive to osmotic stress caused by ethylene glycol [39]. Grr1 is an F-box protein and is part of the SCF ubiquitin ligase complexes [40]. SCF consists of four proteins, Skp1, Cdc53/cullin, Rbx1/Roc1 and an F-box protein. The F-box protein functions as a substrate adaptor and mediates substrate specificity. Although it’s known that Grr1 is involved in glucose repression and that it targets the G1 cyclins Cln1 and Cln2 for degradation [39; 41; 42], the mechanism of As (III) induced-sensitivity of Grr1 mutants is unclear.
Arsenite-resistance modulating proteins
For a complete understanding of the toxicity induced by arsenite, it is important to study the function and regulation of uptake or secretory pathways. As(III) is transported into the cells through the aquaglyceroporin Fps1 [43]. Deletion of Fps1 decreases As (III) influx into the cell and allows glycerol accumulation when cells are treated with As (III). Fps1 deletion mutants are resistant to As (III) toxicity (Table 2). Interestingly, the activity of Fps1 is modulated by Hog1 [44]. Hog1 inactivates Fps1 by phosphorylation on T231 within the N-terminal domain of Fps1 [44].
Another of the most resistant strains lacks the yeast aging gene Uth1. It is a member of the family of yeast genes termed the “SUN family”. It is the first indentified gene providing a link between oxidative stress response, aging and mitochondria [45]. It has been shown to interfere with mitochondria biogenesis and it is involved in the autophagic degradation of mitochondria [46; 47]. It is also required for Bax-induced cell death in yeast [48]. Since arsenic induces oxidative stress, it’s very likely that Uth1 is important in mediating arsenic-induced toxicity through oxidative stress.
Computational interactome mapping of genomic screening data
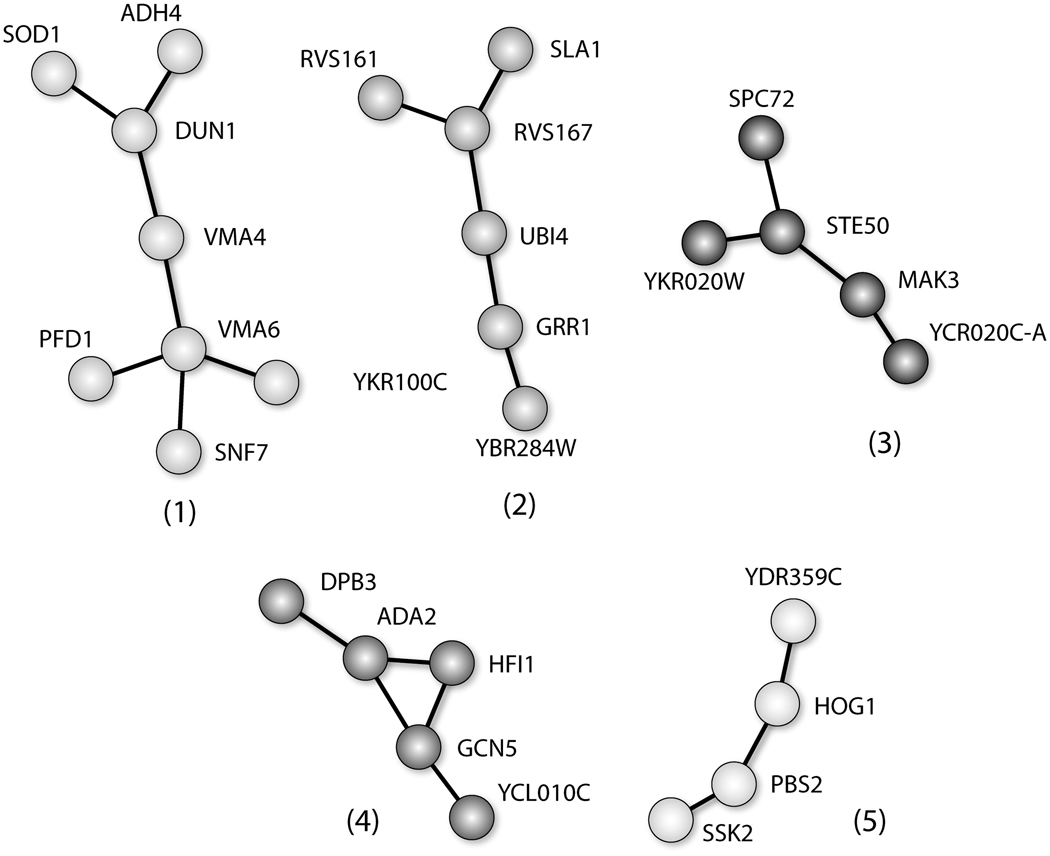
The genes whose inactivation led to arsenic sensitivity were analyzed for various cellular interactions. Using the Cytoscape software, protein-protein or protein-DNA interactions were analyzed. Toxicity modulating subnetworks consisting of greater than 3 connected nodes, corresponding to sensitive strains, are shown in Figure 2.
Figure 2.
Cellular interaction analysis of the mutants whose deletion confers sensitivity to arsenite exposure. Cytoscape software was used to analyze protein-protein interaction analysis.
Figure 2 illustrates the engagement of various cellular processes aiding the recovery of S. cerevisiae from arsenite exposure; the processes embraced by each subnetwork are indicated in Table 2. All of the proteins in each network confer recovery of S. cerevisiae from arsenite exposure. Subnetwork (1) contains cytoskeleton/structural maintaining proteins (Pfd1), as well as vacuolar transport proteins (Snf7, Vma6, Vma4). Subnetwork (2) contains a group of proteins involved in ubiquitination (Grr1 and Ubi4). Subnetwork (2) also contains proteins involved in budding, cell polarity and filament formation during endocytosis (Rvs167, Rvs161 and Sla1) as well as a protein of unknown function (YBR284W). Subnetwork (3) contains Ste50, which encodes for a protein that is involved in mating response, invasive/filamentous growth, and osmotolerance. Subnetwork (4) contains components of ADA and SAGA histone acetyltransferase complexes (Ada2, Gcn5 and Sgf29). Histone acetylation is a modification mark of active gene transcription. Histone acetyltransferase complexes may provide resistance by participating in transcriptional activation of genes whose products aid recovery. Subnetwork (5) is dominated by proteins involved in the high osmolarity MAPK signaling pathway, Hog1 (MAPK), Pbs2 (MEK) and Ssk2 (MAPKKK).
Conclusion
Several studies have been done to screen the S. cerevisiae gene deletion strains to assess the role of nonessential proteins in modulating toxicity upon exposure to arsenic compounds. Haugen et al. [49] identified two metabolic networks, L-threonine and L-homoserine synthesis/degradation and the sikimate pathway, that are important for sodium arsenite tolerance. Jin et al. [50] have shown that the mutants engaged in S. cerevisiae toxicity to sodium arsenite functioning in processes of stress-related transcription regulation, tubulin folding, signal transduction, secretory pathway, and response to stimulus. Dilda et al., [23] identified the sensitive mutant involved in the processes to include the high osmolarity glycerol stress signaling pathway, storage carbohydrate metabolism, DNA repair, oxidative stress defense, ergosterol biosynthesis, actin function, vacuolar acidification, secretory pathway function and NADPH biosynthesis. The focus of our study was to elucidate previously unidentified mechanisms and cellular pathways important for regulating the toxicity of arsenic in human cells, and restrict our studies to those sensitive and resistant strains whose gene deletion product has a human homologue. In this study, we have identified 248 arsenite-sensitive and 5 arsenite-resistant mutants by performing a genome-wide screen of genes in yeast. Functional categorization and interactome mapping suggests that cells develop multiple pathways to defend against arsenic-induced toxicity. In addition to the previously identified genes and pathways that confer sensitivity to arsenic, we have identified pathways of acetylation and deacetylation processes, cell growth/morphogenesis, endocytosis, M phase, protein targeting, sorting and translocation, purine nucleotide/nucleoside/nucleobase anabolism, homoestasis of protons, budding, cell polarity and filatment formation. This knowledge can be utilized to determine and understand the molecular and biological mechanisms by which arsenic induces toxicity. Future studies will determine if the identified genes control the activity of arsenic uptake or efflux, by measuring the concentration of arsenic in the mutant yeast cells after arsenic exposure, and if the human homologue of the yeast protein whose absence renders the cells either more sensitive or resistant to arsenite and therefore may have a direct role in the toxicity of arsenic compounds to human cells.
Acknowledgements
This work was supported by grant numbers ES014454, ES005512, ES000260 from the National Institutes of Environmental Health Sciences, grant number CA16087 from the National Cancer Institute (to M.C.), and grants T32 ES07324-08 and T32 NIEHS 007267-16 (to T.P.E.).
References
- 1.Simeonova PP, Luster MI. Mechanisms of arsenic carcinogenicity: genetic or epigenetic mechanisms? J Environ Pathol Toxicol Oncol. 2000;19:281–286. [PubMed] [Google Scholar]
- 2.Morales KH, Ryan L, Kuo TL, Wu MM, Chen CJ. Risk of internal cancers from arsenic in drinking water. Environ Health Perspect. 2000;108:655–661. doi: 10.1289/ehp.00108655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinmaus C, Moore L, Hopenhayn-Rich C, Biggs ML, Smith AH. Arsenic in drinking water and bladder cancer. Cancer Invest. 2000;18:174–182. doi: 10.3109/07357900009038249. [DOI] [PubMed] [Google Scholar]
- 4.Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure--a critical review. Toxicol Pathol. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- 5.Smith AH, Hopenhayn-Rich C, Bates MN, Goeden HM, Hertz-Picciotto I, Duggan HM, Wood R, Kosnett MJ, Smith MT. Cancer risks from arsenic in drinking water. Environ Health Perspect. 1992;97:259–267. doi: 10.1289/ehp.9297259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ, Corso D, DeBlasio A, Gabrilove J, Scheinberg DA, Pandolfi PP, Warrell RP., Jr Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 7.Soignet SL, Frankel SR, Douer D, Tallman MS, Kantarjian H, Calleja E, Stone RM, Kalaycio M, Scheinberg DA, Steinherz P, Sievers EL, Coutre S, Dahlberg S, Ellison R, Warrell RP., Jr United States multicenter study of arsenic trioxide in relapsed acute promyelocytic leukemia. J Clin Oncol. 2001;19:3852–3860. doi: 10.1200/JCO.2001.19.18.3852. [DOI] [PubMed] [Google Scholar]
- 8.Barrett JC, Lamb PW, Wang TC, Lee TC. Mechanisms of arsenic-induced cell transformation. Biol Trace Elem Res. 1989;21:421–429. doi: 10.1007/BF02917284. [DOI] [PubMed] [Google Scholar]
- 9.Hei TK, Liu SX, Waldren C. Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc Natl Acad Sci U S A. 1998;95:8103–8107. doi: 10.1073/pnas.95.14.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C, Ke Q, Costa M, Shi X. Molecular mechanisms of arsenic carcinogenesis. Mol Cell Biochem. 2004;255:57–66. doi: 10.1023/b:mcbi.0000007261.04684.78. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X, Sun H, Ellen TP, Chen H, Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008;29:1831–1836. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 13.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 14.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274(546):563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 15.Johnston M. The yeast genome: on the road to the Golden Age. Curr Opin Genet Dev. 2000;10:617–623. doi: 10.1016/s0959-437x(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 16.Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 17.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 18.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Rooney JP, George AD, Patil A, Begley U, Bessette E, Zappala MR, Huang X, Conklin DS, Cunningham RP, Begley TJ. Systems based mapping demonstrates that recovery from alkylation damage requires DNA repair, RNA processing, and translation associated networks. Genomics. 2009;93:42–51. doi: 10.1016/j.ygeno.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Vujcic M, Shroff M, Singh KK. Genetic determinants of mitochondrial response to arsenic in yeast Saccharomyces cerevisiae. Cancer Res. 2007;67:9740–9749. doi: 10.1158/0008-5472.CAN-07-1962. [DOI] [PubMed] [Google Scholar]
- 23.Dilda PJ, Perrone GG, Philp A, Lock RB, Dawes IW, Hogg PJ. Insight into the selectivity of arsenic trioxide for acute promyelocytic leukemia cells by characterizing Saccharomyces cerevisiae deletion strains that are sensitive or resistant to the metalloid. Int J Biochem Cell Biol. 2008;40:1016–1029. doi: 10.1016/j.biocel.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez P, Eastmond DA, Laclette JP, Ostrosky-Wegman P. Disruption of microtubule assembly and spindle formation as a mechanism for the induction of aneuploid cells by sodium arsenite and vanadium pentoxide. Mutat Res. 1997;386:291–298. doi: 10.1016/s1383-5742(97)00018-5. [DOI] [PubMed] [Google Scholar]
- 26.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 28.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 29.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 30.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 31.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 32.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 33.Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- 34.Proft M, Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell. 2004;118:351–361. doi: 10.1016/j.cell.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 35.de Nadal E, Casadome L, Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavigelli M, Li WW, Lin A, Su B, Yoshioka K, Karin M. The tumor promoter arsenite stimulates AP-1 activity by inhibiting a JNK phosphatase. EMBO J. 1996;15:6269–6279. [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Gabriel MA, Russell P. Distinct signaling pathways respond to arsenite and reactive oxygen species in Schizosaccharomyces pombe. Eukaryot Cell. 2005;4:1396–1402. doi: 10.1128/EC.4.8.1396-1402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh M, Shen J, Rosen BP. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flick JS, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 41.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 42.Bailey RB, Woodword A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- 43.Wysocki R, Chery CC, Wawrzycka D, Van Hulle M, Cornelis R, Thevelein JM, Tamas MJ. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol Microbiol. 2001;40:1391–1401. doi: 10.1046/j.1365-2958.2001.02485.x. [DOI] [PubMed] [Google Scholar]
- 44.Thorsen M, Di Y, Tangemo C, Morillas M, Ahmadpour D, Van der Does C, Wagner A, Johansson E, Boman J, Posas F, Wysocki R, Tamas MJ. The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol Biol Cell. 2006;17:4400–4010. doi: 10.1091/mbc.E06-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camougrand N, Rigoulet M. Aging and oxidative stress: studies of some genes involved both in aging and in response to oxidative stress. Respir Physiol. 2001;128:393–401. doi: 10.1016/s0034-5687(01)00314-0. [DOI] [PubMed] [Google Scholar]
- 46.Camougrand NM, Mouassite M, Velours GM, Guerin MG. The "SUN" family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;375:154–160. doi: 10.1006/abbi.1999.1655. [DOI] [PubMed] [Google Scholar]
- 47.Camougrand N, Kissova I, Velours G, Manon S. Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res. 2004;5:133–140. doi: 10.1016/j.femsyr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 48.Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule JJ, Manon S. The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol Microbiol. 2003;47:495–506. doi: 10.1046/j.1365-2958.2003.03311.x. [DOI] [PubMed] [Google Scholar]
- 49.Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin YH, Dunlap PE, McBride SJ, Al-Refai H, Bushel PR, Freedman JH. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 2008;4:e1000053. doi: 10.1371/journal.pgen.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]