Abstract
ADAR1 (adenosine deaminase acting on RNA) catalyses the deamination of adenosine to inosine on RNA substrates with double-stranded character. Here, we show that co-expression of ADAR1 in mammalian cells markedly increases plasmid-based gene expression in transfected cells. The enhanced expression was independent of the nature of the promoter (viral, cellular) used to drive gene expression, was independent of the protein reporter (luciferase, RRP) tested, and was independent of the human cell line examined (293T, HeLa). Exogenous protein levels were increased ~20 to ~50-fold when ADAR1 was co-expressed, whereas RNA transcript levels changed less than 2-fold. The activation of PKR protein kinase and the phosphorylation of translation initiation factor eIF-2α seen following plasmid DNA transfection were both greatly reduced in ADAR1-transfected cells. Stable knockdown of the PKR kinase increased reporter gene expression in the absence, but not the presence of ADAR1 co-expression. Both size forms of ADAR1, the p150 inducible form and the p110-like constitutive form, enhanced plasmid-based gene expression. Taken together, these results indicate that the ADAR1 deaminase increases exogenous gene expression at the translational level by decreasing PKR-dependent eIF-2α phosphorylation.
Keywords: ADAR1, PKR, Translational control, eIF-2α phosphorylation
INTRODUCTION
Among the proteins inducible by interferon (IFN) are two double-stranded (ds) RNA binding enzymes, ADAR1 and PKR.1 Adenosine deaminase acting on RNA (ADAR1) catalyzes the hydrolytic C-6 deamination of adenosine (A) in RNA substrates with ds-character to yield inosine (I).2–4 The resultant A-to-I substitutions destabilize dsRNA structures otherwise formed between complementary strands. Indeed, ADAR1 was first identified by its dsRNA-unwinding activity.2,5 Because I is recognized as G instead of A, A-to-I editing can alter several processes including mRNA translation by changing codons and hence protein amino acid sequence; pre-mRNA splicing by changing splice site recognition sequences; and RNA-structure dependent activities that entail binding of RNA by proteins.6,7 The single copy mammalian Adar1 gene encodes two size forms of ADAR1 protein: an IFN-inducible ~150 kDa form (p150) found in both the cytoplasm and nucleus and a constitutively expressed ~110 kDa form (p110), found predominantly if not exclusively in the nucleus.8,9 Alternative promoters, one of which is IFN-inducible, and alternative exon 1 splicing generate transcripts that encode p150 and p110 in mammalian cells.9,10 Both p150 and p110 are active deaminases, with the catalytic deaminase domain present in the C-terminal region and three copies of the canonical dsRNA binding motif in the central region of the proteins.8,11–13 The p150 form of ADAR1 also includes two copies of a Z-DNA binding motif (Zα, Zβ) with homology to the poxvirus E3L protein,8,14 whereas the N-terminally truncated p110 form (that initiates translation at the methionine corresponding to AUG-296 of p150 in human) lacks the Zα motif.10 A-to-I editing can be highly selective, as in the cases of the cellular pre-mRNAs for the glutamate and serotonin receptors, hepatitis D antigenome RNA, and herpes virus HHV8 kaposin mRNA.15–18 A-to-I editing also can occur at multiple sites, as observed in viral RNA genomes such as those of measles,19 hepatitis C20 and lymphocytic choriomeningitis viruses.21
Protein kinase regulated by RNA (PKR) is an IFN-inducible protein kinase that plays an important role in antiviral innate immunity, apoptosis, cell proliferation and stress signaling.1,4,22 The N-terminal region of PKR includes a repeated dsRNA binding motif through which RNA binding mediates the activation, or antagonism, of protein serine/threonine kinase activity. The C-terminal region of PKR contains the kinase catalytic domain.1 PKR is inactive as a monomer through interdomain autoinhibition.23,24 Binding of either dsRNA or structured single-stranded RNAs can mediate PKR autophosphorylation and autoactivation. A prototypical activator of PKR is dsRNA 30 to 50 bp or longer, which includes viral dsRNA produced during replication and synthetic dsRNA such as polyinosine-polycytidylic acid (polyI:polyC). The best-characterized substrate of PKR is protein synthesis initiation factor eIF-2α.25
A universal mechanism for the regulation of protein synthesis in eukaryotes involves eIF-2α phosphorylation that can occur in response to a diverse range of environmental stresses.26,27 The Ser-51-phosphorylated form of eIF-2α (P-eIF-2α) inhibits guanine nucleotide exchange of GDP with GTP bound to eIF-2 by the eIF-2B factor.28 The resulting reduction in eIF-2 activity leads to inhibition of translation initiation. Phosphorylation of eIF-2α is mediated by a family of protein kinases that are activated by different types of stress stimuli. Among them, in addition to the PKR kinase activated by RNA, are the heme-regulated inhibitor kinase (HRI) found in reticulocytes and activated by hemin deficiency and heavy metals;29,30 the general control non-derepressible kinase 2 (GCN2) activated by amino acid deprivation;31 and the endoplasmic reticulum related eIF-2α kinase (PERK) activated by protein misfolding in the endoplasmic reticulum.32 Several studies have demonstrated the importance of eIF-2α kinase pathways in disease pathologies, including viral infections and neurological disorders.4,22,33
Because A-to-I editing mediated by ADAR1 has the capacity to alter base pairing and destabilize dsRNA structures,2,4,5 and because some cellular proteins including PKR and 2′, 5′-oligoadenylate synthetase are regulated by dsRNA and inhibit gene expression,1,22 we tested the effect of ADAR1 on plasmid-based gene expression in transfected human cell lines. We found that ADAR1 overexpression decreased PKR activation and eIF-2α phosphorylation, and increased exogenous gene expression at the translational level.
RESULTS
ADAR1 increases plasmid-based gene expression
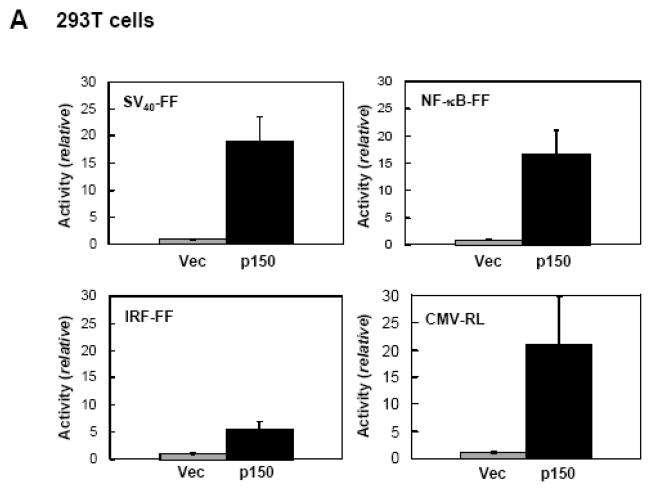
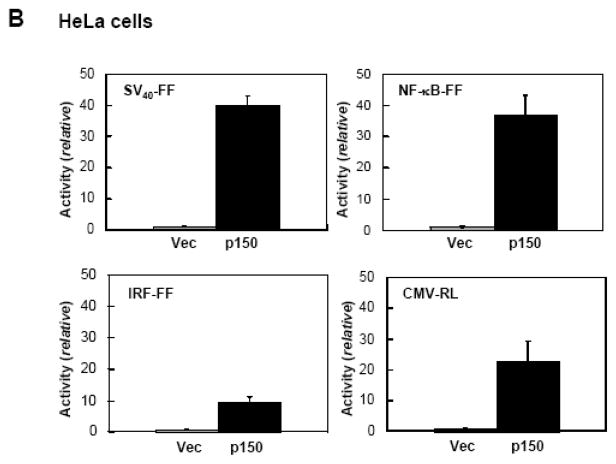
Because dsRNA is an established effector of gene regulation27,34 and ADAR activity was originally discovered as a dsRNA unwinding activity,2,5 we tested the effect of ADAR1 on gene expression in transfected cells. As shown in Figure 1A, when 293T cells were transfected with pGL3 control plasmid encoding a firefly luciferase reporter driven by the SV40 promoter, cotransfection with pCTAP ADAR1 p150 strongly up-regulated luciferase activity compared to cotransfection with the empty pCTAP vector. This enhancing effect of ADAR1 p150 on luciferase expression was independent of the promoter used to drive reporter expression, and the nature of the luciferase reporter. ADAR1 p150 co-expression also increased expression of an NF-κB promoter driven firefly luciferase reporter, an IRF promoter driven firefly luciferase reporter, and a CMV promoter driven Renilla luciferase reporter (Fig. 1A). The enhancing effect of ADAR1 p150 was independent of the cell-type transfected. Results similar to those seen in 293T cells (Fig. 1A) were obtained with HeLa cells (Fig. 1B). Western blot analysis confirmed that ADAR1 p150 was overexpressed in 293T and HeLa cells after cotransfection (data not shown).
Figure 1. ADAR1 enhances plasmid-based gene expression.
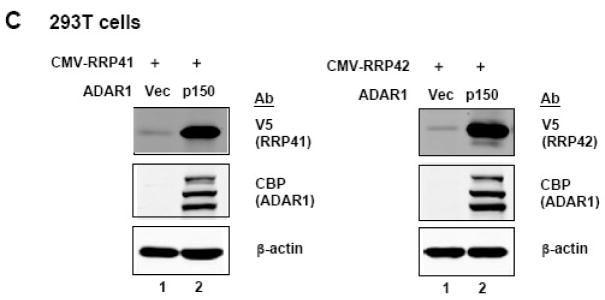
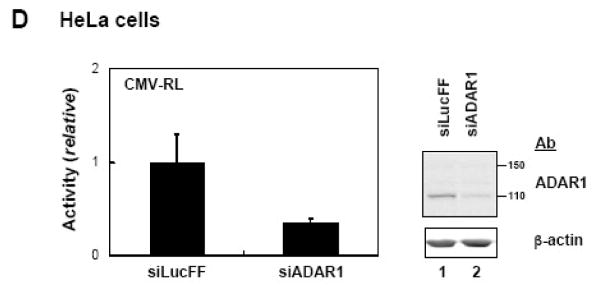
A, 293T cells were transfected with the indicated luciferase reporter construct and either empty pCTAP vector (Vec) or pCTAP p150 encoding the full-length long form of ADAR1 (p150). Cells were harvested 24 h after transfection and analyzed for luciferase activity. Reporter plasmids expressing firefly (FF) or Renilla (RL) luciferase were driven by an SV40, CMV, IRF or NF-κB-dependent promoter. Standard deviations were determined from three or more independent experiments. B, as for A, except that HeLa cells were used. C, 293T cells were transfected with expression constructs encoding V5 epitope tagged CMV-RRP41 or -RRP42 and either empty vector or ADAR1 p150. Extracts were harvested at 48 h after transfection and analysed by Western blot with antibody against V5 (RRP), CBP (ADAR1), or β-actin. D, HeLa cells were transfected with siRNA targeting ADAR1 (siADAR1) or firefly luciferase (siLucFF) as a control, and then analyzed for Renilla firefly luciferase expression using the CMV-RL reporter plasmid. Western analysis was with antibody against ADAR1 or β-actin.
In addition to measuring firefly or Renilla luciferase enzymatic activity as the reporter (Fig. 1A, B), we also analyzed the expression of additional cotransfected genes directly by Western blot analysis. Expression of two V5 epitope tagged RNA-binding proteins, RRP41 and RRP42, in 293T cells was extremely low in the absence of ADAR1, but was enhanced by co-expression of ADAR1 p150 (Fig. 1C). Quantification showed that the expression of RRP41 and RRP42 was enhanced ~50 to ~80 fold compared to the vector control. Taken together, these results establish that ADAR1 p150 co-expression enhances exogenous plasmid-based gene expression by a mechanism not only independent of the nature of the promoter used to drive gene transcription, but independent as well of the nature of the specific gene (firefly and Renilla luciferase; RRP41 and RRP42) and assay (enzymatic, Western) examined.
To further test the role of ADAR1 on plasmid-based gene expression, we used siRNA to transiently knock down endogenous ADAR1 protein. The siRNA targeted an exon 2 sequence, and hence knockdown both size forms of ADAR14, p110 and p150 (Fig. 1D). Knockdown of endogenous ADAR1protein reduced the expression of the CMV promoter driven Renilla luciferase reporter by ~70% compared to an siRNA against firefly luciferase used as a control. Thus, in HeLa cells, overexpression of ADAR1 p150 increased CMV-RL plasmid-based expression (Fig. 1B), and siRNA knockdown of ADAR1 reduced CMV-RL reporter expression (Fig. 1D).
The dsRNA binding domain of ADAR1 is sufficient for translational enhancement of exogenous gene expression
Full-length ADAR1 p150 protein includes a repeated Z-DNA binding domain, in addition to three copies of the dsRNA binding domain and the deaminase catalytic domain that are present in both p150 and p110.4,8 Asan approach to testing whether the gene expression enhancing activity of ADAR1 was a unique property of the inducible p150 protein, or whether the constitutively expressed p110 form of ADAR1 or even the Z-DNA or dsRNA binding domain regions also enhanced gene expression, various ADAR1 isoforms containing a C-terminal CBP-SBP epitope tag were constructed (Fig. 2A) and tested (Fig. 2B–C).
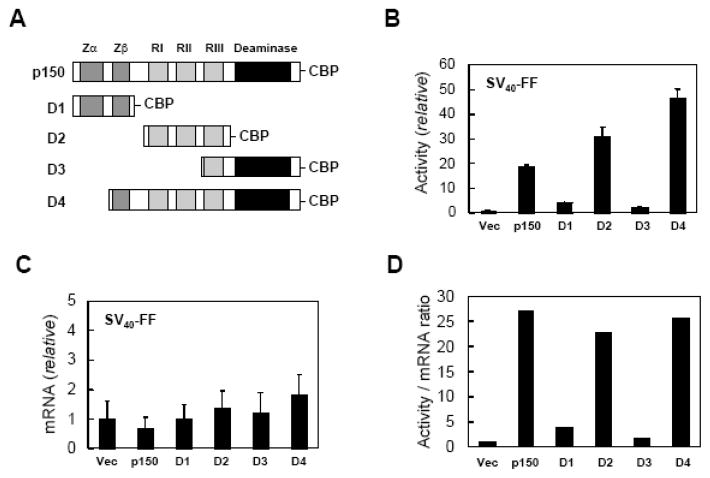
Figure 2. Reporter transcript levels are similar in the presence and absence of ADAR1 but reporter activity is upregulated.
A, Schematic representation of ADAR1 constructs possessing a C-terminal CBP epitope. Dark gray, the repeated Z-DNA binding motifs (Zα, Zβ); light gray, the three dsRNA binding motifs (RI, RII, RIII); black, the deaminase catalytic domain. B, 293T cells were transfected with the pGL3-control luciferase reporter (SV40-FF) and either empty vector or an ADAR1 domain construct (D1 to D4) as indicated. Extracts were prepared 24 h after transfection and analysed for luciferase activity. C, 293T cells were transfected with the pGL3-control luciferase reporter (SV40-FF) and either empty vector (Vec) or ADAR1 construct (p150, D1–D4) as indicated. RNA was isolated 24 h after transfection and quantified by real time PCR. Relative luciferase mRNA levels are shown for cotransfection with an ADAR1 construct compared to empty vector; the mean values and standard deviation were determined from three independent experiments. D, Luciferase activity per transcript amount at 24 h post transfection. Luciferase levels are from Figure 2B; transcript amounts are from Figure 2C.
As shown in Figure 2B, when 293T cells were cotransfected with the luciferase reporter pGL3 and ADAR1-CBP constructs encoding p150, the p110-like form of ADAR1 (D4), or the dsRNA binding domain region (D2), luciferase activity was substantially increased. By contrast, neither the Z-DNA binding domain region (D1) nor the catalytic domain region (D3) of ADAR1, when tested as CBP-domain fusions, significantly enhanced luciferase activity (Fig. 2B). Firefly luciferase transcript levels present in the transfected 293T cells when quantified by real time PCR showed less than a 2-fold difference among vector, ADAR1 p150 and D1 to D4 transfected cells (Fig. 2C). The increased luciferase enzymatic activity per amount of luciferase transcript seen with p150, D2 and D4 suggests that the up-regulation of reporter expression mediated by ADAR1 is predominantly a translational effect (Fig. 2D). Furthermore, ADAR1 p150, the p110-like form (D4) and the dsRNA binding domain of ADAR1 alone (D2) all were capable of mediating the translational up-regulation. Similar results were obtained when the RRP RNA-binding proteins were utilized; p150, D2 and D4 enhanced their expression, but D1 and D3 did not (data not shown).
Decreased phosphorylation of eIF-2α in ADAR1-transfected cells
One mechanism of translational control involves the phosphorylation of protein synthesis initiation factor eIF-2 on the α subunit.26 When eIF-2α is phosphorylated on Ser 51, translation is inhibited. To test whether the phosphorylation status of eIF-2α is altered under conditions of up-regulation of gene expression by ADAR1, we compared the eIF-2α phosphorylation level in 293T cells transfected with the pcDNA6-RRP42 together with either empty vector or ADAR1 p150. As shown in Figure 3A, the level of phosphorylated eIF-2α was decreased and the expression of RRP42 increased by co-expression of ADAR1 p150 compared to vector. Similar results were obtained for RRP41. Increased RRP41 expression was observed when ADAR1 p150 was coexpressed, which correlated with a decrease in phosphorylated eIF-2α (data not shown).
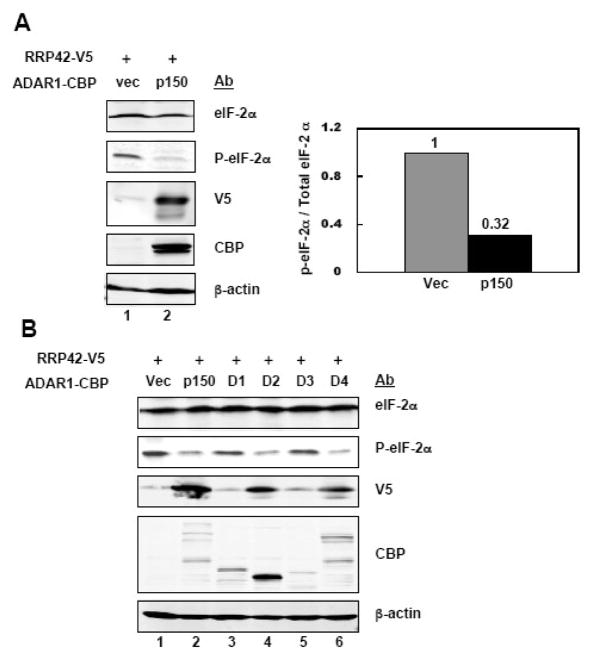
Figure 3. Phosphorylation of eIF-2α is decreased by ADAR1.
A, 293T cells were transfected with pcDNA6-RRP42-V5 and either empty pCTAP vector (Vec) or pCTAP ADAR1 p150 (p150). Extracts were prepared 48 h after transfection and analysed by Western blot. The right panel shows the normalized quantification of the level of phospho-eIF-2α relative to the level of total eIF-2α. B, 293T cells were transfected with pcDNA6-RRP42-V5 and empty vector (Vec), p150 or an ADAR1 domain construct (D1–D4) as indicated. Cells were harvested 48 h after transfection and analysed by Western blot with the indicated antibodies.
We next examined 293T cells cotransfected with pcDNA6-RRP42 and the ADAR1 domain constructs. We found that enhanced RRP42 expression, as measured by the V5 epitope tag, correlated with decreased phosphorylation of eIF-2α seen in cells transfected with the p150, D2 or D4 ADAR1 construct (Fig. 3B). By contrast, the D1 and D3 ADAR1 constructs neither enhanced RRP42 expression nor decreased eIF-2α phosphorylation in transfected cells (Fig. 3B). These results establish that p150 ADAR1, the p110-like ADAR1 (D4) and the dsRNA binding domain of ADAR1 (D2), all of which enhanced luciferase (Fig. 2) and RRP42 (Fig. 3) expression, also decreased eIF-2α phosphorylation in plasmid-transfected cells.
Activation of PKR is antagonized in ADAR1 transfected cells
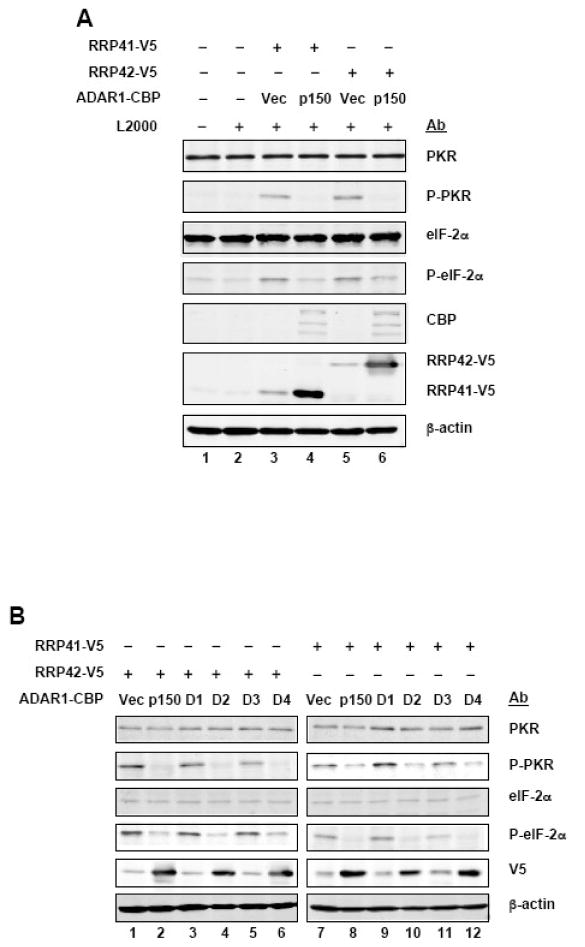
Among the protein kinases that catalyze eIF-2α phosphorylation is the RNA-activated protein kinase PKR. The level of PKR protein was similar in 293T cells transfected with pcDNA6-RRP41 or -RRP42, either with or without ADAR1 cotransfection, and cells not transfected (Fig. 4A). The phosphorylation of both PKR and eIF-2α were increased in vector-transfected cells and reduced phosphorylation of both PKR and eIF-2α were seen in ADAR1 p150 transfected cells (Fig. 4A). This decreased phosphorylation and activation of PKR correlated both with decreased phosphorylation of eIF-2α and with increased RRP41 and RRP42 expression. Furthermore, the p150, p110-like (D4) and dsRNA binding domain (D2) ADAR1 constructs led to decreased activation of PKR, decreased phosphorylation of eIF-2α, and increased RRP protein expression (Fig. 4B).
Figure 4. Activation of PKR kinase is decreased by ADAR1.
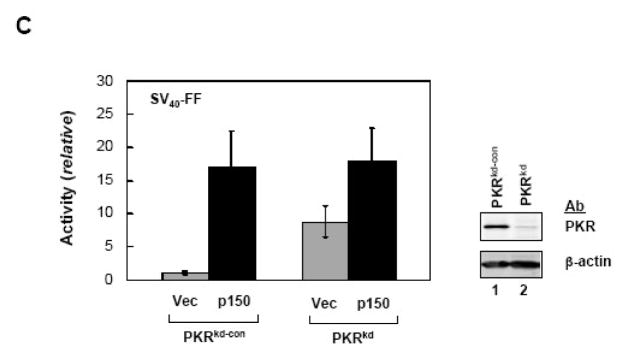
A. 293T cells were transfected with pcDNA6 constructs encoding RRP41-V5 or RRP42-V5, together with either empty pCTAP vector (Vec) or pCTAP ADAR1 p150 (p150). Extracts were prepared 48 h after transfection and analysed by Western blot as indicated for phospho-Thr446-PKR (P-PKR); PKR, phospho-Ser51-eIF-2α (P-eIF-2α); eIF-2α, ADAR1 (CBP); RRP41 and 42 (V5); and, β-actin. B, 293T cells were transfected with pcDNA6 constructs encoding RRP41-V5 or RRP42-V5 and empty pCTAP vector (Vec) or an ADAR1 pCTAP construct (p150, D1 to D4) as indicated. Extracts were prepared 48 h after transfection and analysed by Western blot as for A. C, HeLa cells stably knocked down for PKR (PKRkd) or PKR-sufficient control (PKRkd-con) cells were transfected with the SV40-FF luciferase reporter construct and either empty pCTAP vector (Vec) or pCTAP ADAR1 p150 (p150). Extracts were prepared at 24 h after transfection and analysed for luciferase activity and Western blot for PKR and β-actin. Transfections were repeated a minimum of three times and the mean and standard deviation are shown.
To further test the role of PKR protein as a determinant of gene expression efficiency, we utilized HeLa cells stably knocked down for PKR expression (PKRkd). These cells (Fig. 4C) have less than 5% of the PKR protein present in PKR-sufficient parental or knockdown control (PKRkd-con) cells.35 Luciferase reporter pGL3 activity was nearly 10-fold higher in PKRkd cells than in PKR-sufficient PKRkd-con cells when cotransfected with vector, while pCTAP ADAR1 p150 cotransfection gave comparably high levels of reporter activity in both PKRkd and PKRkd-con cells (Fig. 4C). A further enhancement of ~2-fold of reporter activity gene expression was observed in the PKRkd cells following ADAR1 p150 coexpression (Fig. 4D) that correlated with a modest reduction in eIF-2α phosphorylation (data not shown).
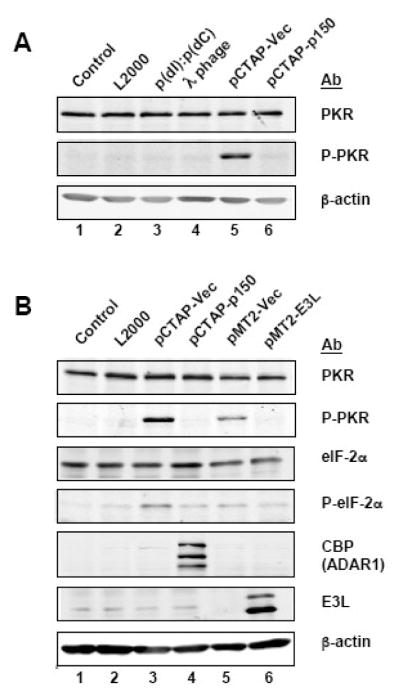
Tranfection with plasmid DNA but not synthetic DNA or phage λ DNA activates PKR
Since we observed that lipid-based transfection of pCTAP vector plasmid DNA activated PKR and eIF-2α phosphorylation robustly (Fig. 3, 4), we examined whether transfection with other DNAs would activate PKR. As shown in Figure 5A, neither synthetic dsDNA polydI:polydC nor λ-phage DNA when transfected in 293T cells activated PKR phosphorylation while the plasmid pCTAP vector DNA did (Fig. 5A). This result further suggests that either an immunostimulatory motif present in the plasmid, or possibly RNA expressed by the plasmid template, activated PKR. The empty plasmid vector pMT2 (Fig. 5B, lane 5), like the pCTAP plasmid vector (lane 3), also activated PKR phosphorylation. The vaccinia virus E3L protein is known to antagonize PKR, either by sequestering dsRNA or interacting with PKR directly.36,37 When the E3L protein was expressed (lane 6), the phosphorylation of PKR was decreased as was seen when ADAR1 p150 was overexpressed (Fig. 5B, lane 4).
Figure 5. Transfection of plasmid DNA but not synthetic or phage λ DNA activates PKR.
A. 293T cells were transfected with the synthetic poly(dI):poly(dC) dsDNA; bacteriophage lambda DNA (λ-phage); pCTAP vector; or, the ADAR1 construct (pCTAP-p150). Extracts were prepared 48 h after transfection and analysed by Western blot for phospho-Thr446-PKR (P-PKR); PKR; and, β-actin. B. 293T cells were not treated (control); treated with lipofectamine and no DNA (L2000); or transfected with pCTAP vector, ADAR1 construct (pCTAP-p150), pMT2 vector or vaccinia virus E3L construct (pMT2-E3L). Extracts were prepared 48 h later and analysed by Western blot for phospho-Thr446-PKR (P-PKR); PKR; phospho-Ser51-eIF-2α (P-eIF-2α); eIF-2α; ADAR1 (CBP); E3L; and, β-actin.
DISCUSSION
ADAR1 is firmly established as an RNA editing enzyme that catalyzes the deamination of adenosine to yield inosine, which can alter RNA structure and function since I is recognized as G instead of A.6,7 The results reported herein also implicate ADAR1 as a key positive effector of exogenous gene expression in transfected cells. We found that ADAR1 increased expression of four different plasmid-based reporters, independent of the promoter used to drive their expression in two different human cell lines. Furthermore, the enhanced expression was predominantly if not exclusively at the translational level: reporter protein expression was increased more than 20-fold under conditions of ADAR1 co-expression, whereas the level of reporter mRNA transcript changed less than 2-fold. The phosphorylation of PKR and eIF-2α was greatly reduced in ADAR1 transfected cells.
What is the mechanism of ADAR1 enhancement of exogenous gene expression in transfected cells? We found that the steady state level of protein was increased several fold while the steady state level of mRNA transcript largely remained unchanged, suggesting a translational control response modulated by ADAR1. Because phosphorylation of eIF-2α is a universally recognized mechanism of translational control,27,28 eIF-2α phosphorylation status was examined. Furthermore, because ADAR1 was originally discovered as a dsRNA unwinding protein,2,5 we further considered the possibility that the enhancing effect on gene expression was mediated at the level of dsRNA-dependent activation of PKR.22 Indeed, we found that plasmid transfection resulted in enhanced PKR Thr-446 phosphorylation and eIF-2α Ser-51 phosphorylation. ADAR1 p150 protein co-expression greatly decreased both PKR phosphorylation and eIF-2α phosphorylation, and these changes in phosphorylation status correlated with increased gene expression. In HeLa cells stably deficient in PKR kinase, reporter expression in the absence of ADAR1 co-expression was increased nearly 10-fold compared to PKR-sufficient control cells. The ~2-fold further increase in reporter expression seen in PKRkd cells in the presence of ADAR1 coexpression may reflect impairment of the residual PKR present in the cells.
Two lines of evidence indicate that only plasmid-based gene expression and not chromatin-based gene expression is enhanced by ADAR1. First, when we transfected ADAR1 p150 or p110 construct into 293 cells that stably overexpress NF-κB driven firefly luciferase, no reporter upregulation was seen (data not shown). Consistent with this notion that chromatin-based gene expression is unaffected by ADAR1, neither endogenous PKR, alpha subunit of eIF-2 or β-actin protein levels were detectably altered by ADAR1 p150 overexpression (Fig. 3 and 4). Previous studies have also shown a distinction between the effect of transfection on plasmid-based versus chromatin-based gene expression.38–40
We found that both size isoforms of ADAR1 protein, the IFN-inducible p150 protein and the constitutive p110-like protein,8 both enhanced plasmid-based gene expression consistent with the observations of others.38,41 We further found that enhanced gene expression correlated with reduced PKR activation and reduced eIF-2α phosphorylation in plasmid-transfected cells. Because the ADAR1 p150 and p110 proteins are both active deaminases8,13,17 and because both are dsRNA-binding proteins, at least two different mechanisms could account for these observations. By one mechanism, sequesteration of dsRNA structures that function as activators of PKR would abolish the PKR cascade. The other mechanism would entail RNA editing by adenosine deamination that would destabilize dsRNA structures and reduce the effective concentration of activator dsRNA. When subdomains of ADAR1 were tested in isolation of the whole protein, we found that neither the N-terminal Z-DNA binding domain nor the C-terminal catalytic domain alone were able to enhance gene expression, but that the central dsRNA-binding domain of ADAR1 was as effective as the whole p150 or p110 ADAR1 protein in enhancing gene expression. Furthermore, the vaccinia virus E3L protein which possesses one Z-DNA and one dsRNA binding domain also enhanced exogenous gene expression. These results indicate that dsRNA binding activity is sufficient to mediate the enhanced gene expression in transfected cells. But, they do not exclude a further contribution from A-to-I conversions by the active p150 and p110 deaminases that would destabilize dsRNA structures. What is clear is that dsRNA binding proteins including ADAR1 p150, ADAR1 p110 and E3L as shown herein, and E3L42 and PACT43 as shown independently, can all enhance exogenous gene expression.
That plasmid-based transfection leads to activation of PKR and eIF-2α phosphorylation is an interesting and known phenomenon.39,44,45 Our results presented herein establish that ADAR1 enhances exogenous gene expression by inhibiting the PKR and eIF-2α phosphorylation. PKR is well established as an RNA-dependent kinase that alters the translational pattern in cells by phosphorylation of eIF-2α.1,22 The dsRNA binding domains of PKR, however, do not bind DNA, and DNA is not an activator of PKR.1,22,46 Neither the synthetic dsDNA poly(dI):poly(dC) DNA nor phage-λ DNA when transfected into 293T cells led to PKR activation. PKR activation was not dependent upon plasmid DNA replication following transfection, as the effect was seen in both 293T and HeLa cells, with and without stably expressed of SV40 large T antigen respectively, as well as with both linearized (data not shown) and circular superhelical plasmid DNA forms. The extent of PKR activation following plasmid DNA transfection also varied with different plasmid backbones (data not shown).
What then is the source of the PKR activating moiety, presumably dsRNA produced in plasmid-transfected cells? Recent findings suggest two possibilities. RNA polymerase III has been identified as a cytosolic DNA sensor which generates 5-triphosphate containing dsRNA from transfected plasmid templates and poly (dA-dT) that is sufficient to trigger an innate immune response measured by IFN-β induction.47 Conceivably these polymerase III-generated dsRNAs, which activate RIG-I signaling, also activate PKR as we too recently described short synthetic 5′-triphosphate-containing ssRNA and dsRNA that activate both IFN-β induction and PKR phosphorylation.48 Short antisense strand RNAs also have been identified that are produced near the promoter regions of genes transcribed by RNA polymerase II in eukaryotic cells.49,50 Conceivably this occurs with transfected plasmid vectors as well, even without a cDNA insert in a manner in which there is sufficient overlap of some short transcripts made from the abundant plasmid templates to yield dsRNA activators of PKR that are otherwise sequestered or inactivated in the presence of overexpressed ADAR1. Recent evidence suggests that some bacterial-based plasmid DNAs display immunostimulatory effects when exposed to the cytosol,51,52 independently of CpG motifs recognized by endosomal TLR9.53 ADAR1 might function as a potent negative regulator of such a DNA-mediated signaling response. Consistent with this notion, ADAR1 p150 has recently been reported as a DNA sensor and suppressor of DNA-mediated activation of innate immunity in mouse embryonic fibroblast cells.54
Plasmid DNA transfection is a broadly utilized method for gene expression, with applications ranging from gene expression-function analyses in basic research to DNA vaccine vehicles for the prevention and treatment of disease.55,56 Our findings demonstrate that the regulation of PKR activation in transfected cells is an important factor that must be considered in gene expression studies. It is now of utmost importance to establish more precisely the mechanism by which ADAR1 functions to enhance exogenous plasmid-based gene expression through down-regulation of PKR kinase activity.
MATERIALS AND METHODS
Cell lines and maintenance
Human 293T and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 μg/ml of penicillin and 100 units/ml streptomycin (Invitrogen) and fetal bovine serum (Hyclone), either 10% (v/v) for 293T cells or 5% (v/v) for HeLa cells. HeLa cells stably knocked down for PKR expression by shRNA interference were as previously discussed;35,37 they were maintained in the above medium containing 1 μg/ml puromycin (Sigma). Cell growth was maintained at 37°C in a 5% CO2 atmosphere.
Plasmid constructs
The pCTAP vector (Stratagene) was used to construct pCTAP ADAR1 p150 with a C-terminal calmodulin binding peptide (CBP) epitope as follows. The cDNA encoding the 1226 amino acid human full-length ADAR1 p150 protein8,10 with M296L and UAG stop1227L mutations was subcloned as a HindIII fragment from pcDNA6 ADAR1 p150 (generously provided by A. Toth of this laboratory) at the HindIII site to generate pCTAP ADAR1 p150. Using pCTAP ADAR1 p150 as template, domain selective ADAR1 fragments D1–D3 were generated by PCR (Fig. 2A) and then inserted into the TOPO vector (Invitrogen). The integrity of the recombinant TOPO plamids was confirmed by sequence analysis; the corresponding ADAR1 fragments were then subcloned as either EcoRI (D1 and D3) or BamHI (D2) fragments into the pCTAP vector. The following primers were used for amplifying ADAR1 fragments: for D1 (Zα, Zβ), forward primer 5′-GCAGCGGAATTCACCACCATGAATCCGCGG-3′, reverse primer 5′-GCAGCGGAATTCGATTGCAGCTGGAGC-3′; for D2 (RI, RII, RIII), forward primer 5′-GGATCCACCATGGATGACTTGAATAGT-3′, reverse primer 5′-GGATCCGAAGGTGCTGCCAGTGAG-3′; for D3 (RIII, catalytic) forward primer 5′-GCAGCGGAATTCACCACCATGAGATACCTG-3′, reverse primer 5′-GCAGCGGAATTCCAATAGTGGGCAGAG-3′. To generate D4, which corresponds to the constitutive ADAR1 p110-like protein, the BamHI fragment was deleted from the pCTAP ADAR1 p150 construct.8
The pcDNA6/V5-His vector (Invitrogen) was used to generate the pcDNA6-RRP41 and pcDNA6-RRP42 constructs following PCR amplification of the open reading frame regions corresponding to human RRP41 (Genbank NM019037) and RRP42 (Genebank NM015004) using random-primed HeLa cell cDNA as a template. PCR fragments were initially cloned using the TOPO vector, DNA sequence analysis was performed, and then the RRP41 and RRP42 cDNA inserts were subcloned to the pcDNA6 vector. The primers used for PCR amplification of the RRP41 and RRP42 inserts were as follows: RRP41 forward 5′-GACACCATGGCGGGGCTGGAGCTC-3′, reverse 5′-ACCACAGTCCCCCAGCAAGATAGAGGC-3′; RRP42 forward 5′-GGCACCATGGCGTCCGTGACGCTG-3′, reverse 5′-AGAACATCCCAGGAATCCAACTTTCTG-3′.
The luciferase constructs were as follows: the SV40 firefly luciferase (SV40-FF) construct, pGL3 control, was from Promega; the NF-κB-dependent firefly luciferase construct was generously provided by I. Verma (Salk Institute, La Jolla, CA); the interferon regulatory factor-dependent firefly luciferase construct was generously provided by T. Fujita (Kyoto University, Japan); and the CMV Renilla luciferase (CMV-RL) construct, pRL, was from Promega. All plasmids were propagated in E. coli (XLI-Blue) grown in LB medium and purified using the MaxiPrep protocol procedure (Qiagen).
Transient transfection
293T or HeLa cells were transfected at 70 to 80% confluence using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Briefly, the plasmid DNA and Lipofectamine 2000 were diluted and mixed in Opti-MEM (Invitrogen), incubated for 20 min at room temperature, and then the DNA-Lipofectamine 2000 complexes were applied to the monolayer cultures in either 12-well (Western analyses) or 24-well (luciferase assay) plates. For transient knockdown of ADAR1, chemically synthesized siRNA with dTdT overhangs and the following validated sense-strand sequence as described by Jayan and Casey57 was obtained from Dharmacon: 5′-CGCAGAGUUCCUCACCUGUATT-3′. SiRNA against firefly luciferase was used as a negative control and was previously described.58 Sequential siRNA transfection was utilized to achieve maximal ADAR1 knockdown. Briefly, HeLa cells at ~50% confluency were transfected with 50nM of siRNA with Lipofectamine 2000 on day 1; the cells were transfected a second time with 50nM of siRNA on day 2, and then on day 3 were reseeded into 24-well plate for reporter plasmid transfection and western analysis on day 4.
Luciferase measurement
Cell-free extracts were prepared from 293T or HeLa cells at 24 h after transfection using passive lysis buffer (Promega). Luciferase activity was measured according to the manufacturer’s protocol (Promega) using a GEM Biomedical OPTOCOMP I luminometer. Protein concentrations of extracts were determined by the modified Bradford method (Bio-Rad). The luciferase activity was normalized to the protein concentration.
Western blot analysis
Extracts were prepared from 293T or HeLa cells at 48 h after transfection; the lysis buffer (50mM HEPES, pH 7.5, 300mM NaCl, 1mM DTT, 0.5% NP-40) contained 1% (vol/vol) protease inhibitor cocktail (Sigma). Proteins were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, blocked in Tris-buffer saline containing either 2% (wt/vol) bovine serum albumin (for detection of phosphoproteins) or 5% (wt/vol) nonfat milk (for detection of all other proteins) for 1 h at room temperature, and then incubated overnight at 4°C with the indicated primary antibody. Rabbit polyclonal antibodies were used to detect calmodulin binding peptide epitope (Millipore), human PKR (Santa Cruz Biotechnology), PKR phosphorylated at Thr 446 (Santa Cruz Biotechnology), eIF-2α (Cell signaling), eIF-2α phosphorylated at Ser51 (Cell signaling), and human ADAR1 (K88 number 2 as previously described8). Mouse monoclonal antibodies were used to detect β-actin (Sigma) and V5 peptide epitope-tagged proteins (Invitrogen). Rabbit polyclonal antibody against vaccinia virus E3L was as described.37 Western blot detection was done with IRDye 800-conjugated anti-rabbit immunoglobulin G or IRDye 680-conjugated anti-mouse immunoglobulin G secondary antibody according to the manufacturer’s protocols (Li-Cor). Immunoreactive bands were detected and quantified using a Li-Cor Odyssey infrared imaging system.
Reverse transcription and real time PCR analysis
Total RNA was purified from 293T or HeLa cells using an RNeasy kit (Qiagen). In order to eliminate residual DNA, on-column RNase-free DNase digestion was performed. Reverse transcription was carried out using random hexamer oligonucleotide primers and SuperScript II (Invitrogen) reverse transcriptase. Quantitative real-time PCR analyses were performed in triplicate, using iQ SybrGreen PCR Supermix (Bio-Rad) and a Bio-Rad Icycler MyIQ multicolor real time PCR instrument. Luciferase levels were normalized to glyceraldehydes-3- phosphate dehydrogenase (GAPDH) values. The primers used were: forward 5′-GGTGGTCTCCTCTGACTTCAACA-3′, reverse 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ for GAPDH and forward 5′-GCCTGAAGTCTCTGATTAAGT-3′, reverse 5′-ACACCTGCGTCGAAGATGT-3′ for firefly luciferase.
Acknowledgments
This work was supported in part by the research grants AI-12520 and AI-20611 from the National Institute of Allergy and Infectious Diseases, U.S. Public Health Service, and grant W911NF09D0001 from the U.S. Army Research Office. We thank the members of Samuel Laboratory for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 3.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 5.Rebagliati MR, Melton DA. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 6.Samuel CE. RNA editing minireview series. J Biol Chem. 2003;278:1389–1390. doi: 10.1074/jbc.R200032200. [DOI] [PubMed] [Google Scholar]
- 7.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 10.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Samuel CE. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Athanasiadis A, Placido D, Maas S, Brown BA, Lowenhaupt K, Rich A. The crystal structure of the Zbeta domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J Mol Biol. 2005;351:496–507. doi: 10.1016/j.jmb.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Emeson RB, Samuel CE. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274:18351–18358. doi: 10.1074/jbc.274.26.18351. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Samuel CE. Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274:5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- 17.Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 18.Gandy SZ, Linnstaedt SD, Muralidhar S, Cashman KA, Rosenthal LJ, Casey JL. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J Virol. 2007;81:13544–13551. doi: 10.1128/JVI.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattaneo R, Schmid A, Eschle D, Baczko K, ter M, V &, Billeter MA. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79:6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zahn RC, Schelp I, Utermohlen O, von LD. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J Virol. 2007;81:457–464. doi: 10.1128/JVI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr Top Microbiol Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 23.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 24.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular framework for the activation of RNA-dependent protein kinase. J Biol Chem. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 25.Samuel CE. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc Natl Acad Sci U S A. 1979;76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 27.Samuel CE. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 28.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 29.Chen JJ. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramer G, Cimadevilla JM, Hardesty B. Specificity of the protein kinase activity associated with the hemin-controlled repressor of rabbit reticulocyte. Proc Natl Acad Sci U S A. 1976;73:3078–3082. doi: 10.1073/pnas.73.9.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wek RC. eIF-2 kinases: regulators of general and gene-specific translation initiation. Trends Biochem Sci. 1994;19:491–496. doi: 10.1016/0968-0004(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 32.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 33.Scheper GC, Proud CG, van der Knaap MS. Defective translation initiation causes vanishing of cerebral white matter. Trends Mol Med. 2006;12:159–166. doi: 10.1016/j.molmed.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Samuel CE. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J Virol. 2007;81:8192–8200. doi: 10.1128/JVI.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romano PR, Zhang F, Tan SL, Garcia-Barrio MT, Katze MG, Dever TE, Hinnebusch AG. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol. 1998;18:7304–7316. doi: 10.1128/mcb.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P, Jacobs BL, Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J Virol. 2008;82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gommans WM, Maas S. Characterization of ADAR1-mediated modulation of gene expression. Biochem Biophys Res Commun. 2008;377:170–175. doi: 10.1016/j.bbrc.2008.09.109. [DOI] [PubMed] [Google Scholar]
- 39.Kaufman RJ, Murtha P. Translational control mediated by eucaryotic initiation factor-2 is restricted to specific mRNAs in transfected cells. Mol Cell Biol. 1987;7:1568–1571. doi: 10.1128/mcb.7.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laing KG, Elia A, Jeffrey I, Matys V, Tilleray VJ, Souberbielle B, Clemens MJ. In vivo effects of the Epstein-Barr virus small RNA EBER-1 on protein synthesis and cell growth regulation. Virology. 2002;297:253–269. doi: 10.1006/viro.2002.1354. [DOI] [PubMed] [Google Scholar]
- 41.Nie Y, Ding L, Kao PN, Braun R, Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies MV, Chang HW, Jacobs BL, Kaufman RJ. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Sen GC. PACT-mediated enhancement of reporter gene expression at the translational level. J Interferon Cytokine Res. 2003;23:689–697. doi: 10.1089/107999003772084806. [DOI] [PubMed] [Google Scholar]
- 44.Terenzi F, deVeer MJ, Ying H, Restifo NP, Williams BR, Silverman RH. The antiviral enzymes PKR and RNase L suppress gene expression from viral and non-viral based vectors. Nucleic Acids Res. 1999;27:4369–4375. doi: 10.1093/nar/27.22.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufman RJ, Davies MV, Pathak VK, Hershey JW. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol Cell Biol. 1989;9:946–958. doi: 10.1128/mcb.9.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAllister CS, Samuel CE. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2009;284:1644–1651. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 53.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ewert K, Slack NL, Ahmad A, Evans HM, Lin AJ, Samuel CE, Safinya CR. Cationic lipid-DNA complexes for gene therapy: understanding the relationship between complex structure and gene delivery pathways at the molecular level. Curr Med Chem. 2004;11:133–149. doi: 10.2174/0929867043456160. [DOI] [PubMed] [Google Scholar]
- 56.Weide B, Garbe C, Rammensee HG, Pascolo S. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol Lett. 2008;115:33–42. doi: 10.1016/j.imlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 57.Jayan GC, Casey JL. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J Virol. 2002;76:12399–12404. doi: 10.1128/JVI.76.23.12399-12404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Structure and gene silencing activities of monovalent and pentavalent cationic lipid vectors complexed with siRNA. Biochemistry. 2007;46:4785–4792. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]