Abstract
LINE1s (L1s) are a class of mammalian non-LTR (long terminal repeat) retroelements that make up nearly 20% of the human genome. Because of the difficulty of studying the mobilization of endogenous L1s, an exogenous cell culture retrotransposition assay has become integral to research in L1 biology. This assay has allowed for investigation of the mechanism and consequences of mobilization of this retroelement, both in cell lines and in whole animal models. In this paper, we outline the genesis of in vitro retrotransposition systems which led to the development of the L1 retrotransposition assay in the mid-1990s. We then provide a retrospective, describing the many uses and variations of this assay, ending with caveats and predictions for future developments. Finally, we provide detailed protocols on the application of the retrotransposition assay, including lists of constructs available in the L1 research community and cell lines in which this assay has been applied.
Keywords: LINE1 (L1), non-LTR retrotransposons, retrotransposition, cell culture assays, transgenic mice
Introduction
Class I mobile elements are also called retrotransposons, an apt name that carries within it a description of their unusual mechanism of action. The “retro” recalls “retrovirus”, and, like retroviruses, mobilization of retrotransposons involves reverse transcription of an RNA intermediate. Both retroelements and retroviruses are transcribed using the cell's own polymerases and associated factors. As a consequence, these elements are processed a similar manner to other nuclear genes, including the removal of sequences flanked by canonical intron splice donor and acceptor sites. The ability of retrotransposition intermediates to undergo splicing has proved serendipitous in the development of a reporter system that can track novel insertion events precipitated by an exogenously-provided retroelement.
Endogenous L1 retrotransposons are actively mobile in mammals, as evidenced by eighteen human diseases caused by recent insertions [1, 2]. In addition, many elements are polymorphic in human populations, though non-disease causing; [3-6] to list a few studies. However, because endogenous elements are typically inactive, new insertions from pre-existing active elements are difficult to detect in routine cell culture. An exogenous transfection system was developed to overcome many of the limitations of studying endogenous elements [7]. Here, a full-length L1 element is expressed from either its native or a CMV (cytomegalovirus) promoter. Reverse transcription and subsequent integration of the transcript into the genomic DNA is detected using a reporter gene that becomes functional only after transcription, splicing by the cellular machinery, and integration in a new location [Figure 1]. This assay has been used to generate and track the rate, mechanism and consequences of mobilization of L1 retrotransposons in cultured cells and whole animals. Variations on this method have been developed to track other retroelements, both autonomous and non-autonomous, in different cellular environments. In this paper, we will first discuss the history of the retrotransposition reporter assay, primarily as it pertains to L1 elements - its uses, successes, limitations and future directions. In the second section, we will describe protocols and constructs available for practical application of this assay in the reader's research lab.
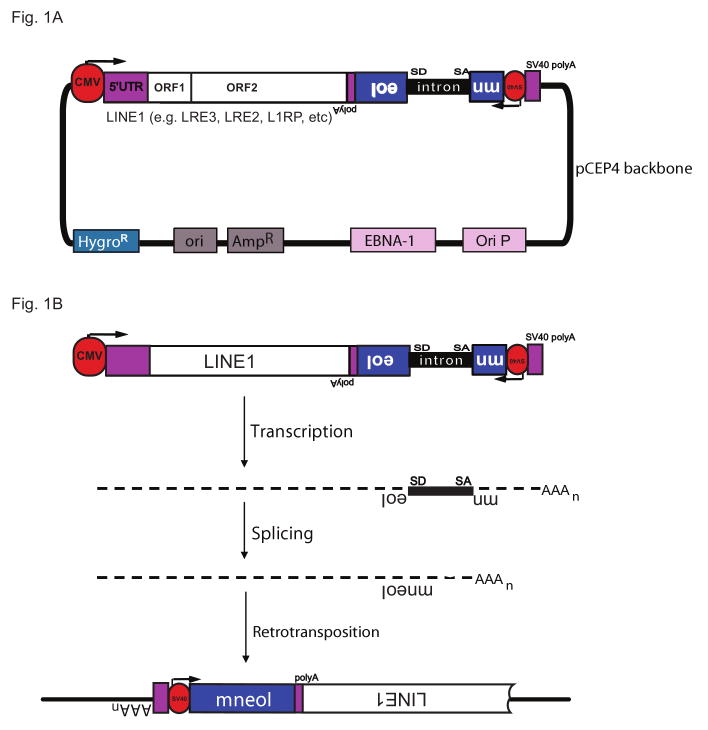
Figure 1.
Canonical LINE1 retrotransposition construct in pCEP4 vector backbone (e.g. JM101). The LINE1 insert may correspond to naturally-occurring active L1s such as LRE3 or L1RP or a variety of mutated or epitope-tagged derivatives. Purple boxes indicate LINE1-derived 5′ and 3′ UTR. mneoI = neomycin phosphotransferase. This selectable marker may be replaced by blasticidin, EGFP or no functional gene at all. Intron = γ-globin intron. SD is a splice donor and SA is splice acceptor sites present on the same orientation as the LINE1. This sequence may be replaced by group I self splicing intron. The antisense mneoI gene is driven by an SV40 promoter and terminates in a polyadenylation signal (polyA). The LINE1 can either be transcribed from a CMV promoter (as shown) or the native promoter located in the 5′ UTR. This transcript terminates in an SV40 polyadenylation signal (SV40 polyA), as indicated. The vector also encodes resistance to hygromycin (HygroR ); variants encode resistance to puromycin. The vector backbone encodes EBNA-1 (Epstein-Barr virus nuclear antigen 1) and OriP (Epstein-Barr virus origin of replication), which allow replication in primate cells. The vector can also be propagated in prokaryotic cells, as it contains an E. coli origin of replication (ori) and resistance to ampicillin (AmpR). Please refer to Table 1 for a comprehensive list of variants. B. A transcript is produced that comprises both the full-length 6 kb LINE1 and an mneoI gene in antisense, disrupted by an intron in the sense orientation. After reverse transcription, splicing and integration into the genome (retrotransposition), the mneoI gene can be expressed, resulting in resistance to G418. The LINE1 element may be 5′ truncated upon insertion. Figure adapted from Moran et al.[7].
History
Episomal systems for detecting retrotransposition were first developed twenty years ago in yeast and flies, starting with the confirmation in 1985 that an intron artificially inserted into a yeast Ty retroelement could be successfully spliced out upon retrotransposition of the element [8]. Heidmann et al. [9] took advantage of this discovery by developing a neo (neomycin phosphotransferase) gene cassette (neoRT) in which the gene was disrupted by an artificial intron containing polyadenylation signals so that, when unspliced, the neoRT transcript would prematurely terminate and not produce functional protein. When cells were selected on the drug G418, only those expressing a functional neo gene would survive. This strategy allowed for the selection of cells that had successfully undergone transcription, splicing, and reverse transcription and genomic integration of the vector sequence – in Heidmann's case, a murine leukemia virus. Critically, the retroviral transcript and the artificial intron were in the opposite orientation to the neo gene itself, so that removal of the intron was independent of transcription of the gene and entirely dependent on transcription of the retrovirus. This particular orientation of the intron and the selectable marker would be adapted in all future variations of this technique. Within a few years, Heidmann's neoRT cassette would be used to track mobilization of the Drosophila I retroelement [10] and the mammalian IAP (intracisternal A particle) element [11]. It would also be used to assay intron-less pseudogene formation in mice and human cells [12-14].
As the decade progressed, the original neoRT cassette [9] inspired other intron-containing reporters such as a Syrian hamster-derived intron in neo, a his3 auxotrophic marker and a lacZ colorimetric indicator [15-18]. The L1 retrotransposition cell culture assay developed by Moran et al.[7] used an mneoI cassette containing a γ-globin intron flanked by splice donor and acceptor sites [19]. Moran's constructs feature full-length 6 kb L1 elements known to be recently active in humans [20, 21] with the mneoI cassette inserted in the antisense orientation within the 3′ UTR [Figure 1]. The entire L1-mneoI fusion transcript is expressed from a CMV promoter and terminates in an SV40 polyadenylation signal. The vector backbone, pCEP4, encodes resistance to hygromycin. Typically, cells are transfected with the L1 construct, subjected to hygromycin and then to G418 selection. Colonies that survive these two serial selections are propagated. DNA is isolated from these cells and the insertion products are investigated by Southern blot or PCR and DNA sequencing analysis. Over the ensuing decade, further innovations to this protocol included the use of a self-splicing intron within the neo cassette [22], replacement of the mneoI indicator with a different drug selection, blasticidin S deaminase [23], a visual marker such as EGFP (enhanced green fluorescent protein) [24] or a naked intron with no marker gene at all [25]. These and other variations will be discussed in further detail below.
Uses of the L1 Retrotransposition Assay
The last several years have witnessed the L1 retrotransposition assay used in all aspects of the study of L1 biology, including mechanism, regulation and mutagenesis. Indeed, it seems as though the majority of publications on L1 retroelements feature some use of this assay. Here, we will highlight some of the successes of this assay, focusing on the technological innovations that accompanied each increase in biological understanding.
1. Process of retrotransposition and structure of products
One of the most useful variations on the vector used in the retrotransposition assay was the addition of a prokaryotic promoter and origin of replication to the mneoI cassette along with removal of a cryptic splice signal [26, 27]. This modification allowed for expression of the neo gene in prokaryotic cells, which caused those cells to be resistant to kanamycin. Therefore, new retrotransposition events containing the spliced, active neo gene could now be rescued in E. coli and the flanking sequence determined. This strategy of creating novel retrotransposition events in vitro became a fruitful area of research, with many studies tracing the structure of retrotransposition insertions, including the target sites for both endonuclease-containing and mutated elements, production of chimeric integrants and assays of the proportion of full-length insertions [26-31]. Hypothesis testing could also be conducted, either in cell culture or in animal models: for instance, to see whether the bias for AT sequences in the genomic DNA flanking preexisting L1 insertions applied equally to newly mobilized elements [25, 32].
Despite the many successes, these studies of insertion sites were inherently limited: only insertions that are longer than the selectable marker could be recovered. L1 retrotransposition often produces short 5′ truncated inserts, and these would be missed. In order to overcome this size bias, Babushok et al. [25] made use of a marker-less cassette in transgenic mice, where removal of the intron was detected purely by a PCR reaction, and insertion site flanking sequences were cloned using TAIL-PCR [33]. This construct allowed for the isolation of insertions as small as 400 bp.
Wei et al.[34] explored whether the neo cassette could be spliced and retrotransposed in trans if provided on a different plasmid from the L1 driver element. They found that retrotransposition was more efficient in cis, where both driver and retrotransposition reporter were provided on the same construct. Nevertheless, transfected L1s are able to drive the retrotransposition of sequences in trans as well, such as cellular genes co-transfected and then assayed by PCR for a loss of an intron sequence [12]. Recently, Garcia-Perez has investigated the formation of L1/snRNA fusion pseudogenes by pooling G418 positive colonies and amplifying products using primers specific for the U6 sequence [35].
In the traditional L1 retrotransposition assay, selection on the drug G418 results in the death of death of those cells that are not carrying de-novo retrotransposition events, with the sole survivors being cells that contain retrotransposition of the mneoI cassette. In order to more accurately trace the kinetics of retrotransposition against a background of retrotransposition negative cells, Ostertag et al. switched out the neo gene with an γ-globin intron-containing EGFP gene [24]. This variant allows for the easy measurement of retrotransposition frequency using a flow sorting of fluorescent cells.
Another development useful for investigating the process of retrotransposition was the creation of a vector which is a hybrid between a retrotransposon and a gutted adenovirus [36, 37]. Use of this gene delivery system demonstrated that retrotransposition could occur in infected primary cells in the absence of cell division. Single events per cell were the norm, with their frequency dependent on the copy number of the construct.
2. Regulation of Retrotransposition
The L1 retrotransposition assay has also proven to be a powerful tool to investigate the sequence requirements for both the repression and promotion of retrotransposition. L1 elements had been known to be cytosine methylated [38]; however, by methylating HpaII sites in the promoter of the L1 construct in vitro, Yu et al. conclusively demonstrated that DNA methylation suppresses retrotransposition [39]. Moreover, adding GAL4 DNA-binding sites upstream of the L1 element on the construct and co-transfection with the methyl-CpG binding protein MeCP2 confirmed that this suppression occurred using a canonical chromatin regulatory pathway. Another cellular strategy to regulate L1s makes use of the APOBEC3 family of cytidine deaminases: here, co-transfections were instrumental in determining that Apo3A and 3B suppressed L1 retrotransposition, although conflicting results were obtained for Apo3F for reasons that remain unclear [40-45]. Alu, a non-autonomous element, could be mobilized when co-transfected with the L1 construct; the retrotransposition of Alu, but not that of the L1 driver, was specifically blocked by Apo3G [41, 46]. Most recently, it was found that the AID (activation-induced deaminase) family of proteins, which are phylogenetically related to APOBEC, can also inhibit L1 retrotransposition; of note, this inhibition does not require deaminase function or DNA binding [47].
SiRNAs derived in vivo from double-stranded RNA antagonize the transcription of retrotransposons in a wide range of organisms, both plants and animals [48]. When it came to studying this phenomenon in mammalian cells, the cell culture assay proved once again to be central When L1 constructs were co-transfected with siRNAs to L1 sequence, retrotransposition was indeed reduced [49]. Subsequently, Yang et al. generated 5′ truncations of the L1 element that abrogated activity of the antisense promoter located ∼ +500 in the 5′UTR [50]. These mutations caused a modest increase in retrotransposition of the construct, which may be due to the loss of double-stranded RNA production.
L1 activity causes genomic instability, and the retrotransposition assay investigated the ways in which the cell responds to various insults and fights back. G418 positive colonies were increased in a line carrying a p53 mutation [51]; the authors of this study suggested that L1 activity may induce apoptosis in p53+ cells. L1 mobilization also precipitated DNA double-stranded breaks which attracted the histone variant γ-H2AX [52, 53], and led to recruitment of the DNA repair pathway [54, 55].
In addition to repression, the cell culture assay has been helpful in characterizing exogenous factors, both genic and environmental, that promote L1 retrotransposition. By cotransfecting Runx3 with an L1 construct carrying mutations at putative RUNX3 binding sites, Yang et al.[56] showed that this transcription factor is able to increase both expression and retrotransposition of the element. Moreover, mutating putative binding sites for the transcription factor YY1 resulted in changes in transcriptional initiation with little difference in retrotransposition [57]. Genotoxins such as γ-radiation and benzopyrene [58] and heavy metals (Hg, Ni, Cd) [59, 60] stimulated increased levels of retrotransposition in the cell culture assay.
3. Finding highly active naturally-occurring L1s
Potentially active L1 elements are occasionally found retrotransposed into protein-coding genes or as polymorphic elements in natural populations. One of the first and more popular ongoing uses of the retrotransposition assay has been to quantify the activity levels of newly isolated polymorphic L1s. In fact, Moran et al.'s [7] original study compared multiple full-length L1 elements; subsequent studies demonstrated that L1RP had a relatively high level of activity [61, 62]. Using flow cytometry to determine EGFP reporter activity, Brouha et al. isolated and measured the activity of another highly active element, LRE3 [63]. L1RP and LRE3, among other elements such as L1.3 [21, 62], remain popular L1 drivers for use in the retrotransposition assay.
The assay was later expanded to look at a large number of full-length L1s from the most evolutionarily recent family, Ta [4, 64, 65]. This survey allowed for an estimate to be made of how many L1s in the entire genome might be active [4]. In addition, taking this assay to naturally-occurring alleles of the same element demonstrated that different individual variants had different levels of activity – a “hot” L1 in one population might therefore not be as active in another [66, 67]. This use of the retrotransposition assay was not limited to testing human elements in human cell lines: mouse elements from different L1 subfamilies have been examined as well [68-70], in both mouse LTK- and human cell lines.
3. Structure/Function Studies
The retrotransposition cell culture assay can test not only natural alleles of L1 elements, but is also an ideal system for examining artificially-generated mutations in the protein-coding regions of the element. These structure/function studies have been central in deciphering the mechanism of retrotransposition. Early uses of this assay showed the importance of expression of L1-encoded ORF1 and ORF2, particularly the endonuclease activity of ORF2p, in effective retrotransposition [7, 71]. The effects of mutations within the endonuclease domain have been investigated more closely recently, including characterizing the structure of altered proteins and cloning out the insertion breakpoints of endonuclease-defective retrotransposition [29, 72].
Knowledge of ORF1p's role as a nucleic acid chaperone has especially benefited from this method [73, 74]. By epitope-tagging mutated versions of the element, Kulpa and Moran [75] were able to show that ORF1 was important for RNP complex formation. Other studies showed that the initial AUG and other intergenic sequences present upstream of ORF2 were dispensable for function [76, 77].
Alterations of the L1 element do not have to be deleterious; indeed, some changes can increase retrotransposition competency. Boeke's lab has synthetically engineering a novel L1 element through codon optimization of mouse L1Spa [78]. This element, named ORFeus, is 24% divergent but two hundred-fold more active than its L1Spa element progenitor. An optimized human element has been developed as well which is somewhat more active than its progenitor element [54, 79].
4. Animal Models
Although most research using the retrotransposition assay has been carried out in cell culture (e.g. human HeLa), whole animal transgenics have become a burgeoning area of innovation. Whole animal retrotransposition assays can be used to both determine the stage and tissue-specificity of retrotransposition in vivo and as a mutagenesis tool for gene discovery. In 2002, Ostertag et al. created mice carrying an EGFP-tagged human L1 transgene [80]; these animals expressed green fluorescence upon mobilization of the retrotransposon. Integrated transgenes continued to drive low rates of retrotransposition in the germline of the descendants of the founder animals. A second-generation construct, using a highly active mouse RNA polymerase II large subunit promoter with CMV driving EGFP expression, was used to track retrotransposition visually through monitoring fluorescence in different mouse tissues [81]. These mice featured very little somatic L1 movement, with a strong signal only in the testis. Contrasting results were obtained a couple of years later, when retrotransposition was detected in rat neuronal precursor cells and in the brain of transgenic mice [82]. New data using human elements in mice and rats point to the likelihood that the bulk of L1 mobilization occurs in the early embryo rather than the germ lineage (Hiroki Kano, personal communication). In fact, the codon-optimized, CAG (chicken actin promoter)-driven ORFeus element is retrotranspositionally active in both germline and somatic tissues [32].
A recent development has been the possibility of using L1 as an insertional mutagen for gene discovery [32, 83]. Transposon-tagged genes can be easily cloned based on sequences flanking the element, giving this form of random mutagenesis a distinct advantage over methods based on chemicals such as ENU (ethylnitrosourea). An and Boeke have created an L1 ORFeus construct in which the promoter is separated from the transgene by a β-geo (β-galactosidase/neomycin fusion) cassette flanked by LoxP (locus of X-over P1) sites. Mating mice transgenic for this construct to those expressing the Cre (cyclization recombination) protein results in excision of this cassette and activation of the retroelement in the offspring. The LoxP transgene has been successfully combined in this manner with Pdx1 (pancreatic and duodenal homeobox 1)-Cre and Zp3 (zona pellucida glycoprotein 3)-Cre to drive retrotransposition specifically in the pancreas and female germ cells, respectively [84].
5. New Directions
The L1 retrotransposition assay is a flexible and powerful tool for the study of the mechanism and consequences of retrotransposition. In coming years, we expect greater variations on this technology – perhaps as a gene delivery vector or a means of mutagenesis in specific tissues or organisms. These developments may be aided by the creation of optimized elements. Once mobilized, mutagenic retrotransposition might be suppressed by RNAi – this could be accomplished by using a Cre-Lox strategy to activate an RNA polymerase III-driven L1 hairpin delivered in cis on the same construct as the L1 driver [85].
Thus far, most research has focused on mouse or human L1 elements; however, retrotransposition assays have also been developed for two mammalian LTR (long terminal repeat) class retroelements, IAP and ERV, using intron-containing neo reporter cassettes originally created for L1 mobilization [86-89]. Non-human or mouse L1 can be mobilized as well – rat elements have been successfully retrotransposed in HeLa cells, rat cells and whole animals [83, 90]. More excitingly, this assay has been expanded to test retrotransposition of LINEs in two species of fish [91, 92], and a variation using intron-containing URA3 or HIS3 genes as indicators was recently used to study mobilization of the Zorro3 element in yeasts [93, 94]. We look forward in the future to seeing similar studies of retrotransposition of less well-studied elements in diverse species of birds, bats and fish.
Another variant of the canonical retrotransposition assay features the use of L1 as a driver of trans-mobilization of non-autonomous retroelements such as Alu or SVA. Kajikawa and Okada [91] were the first to successfully co-transfect an L1 and a SINE element on different plasmids: in their case, an eel L1 promoted retrotransposition of an eel SINE carrying the mneoI reporter cassette. Because SINE elements are transcribed by RNA polymerase III which is not normally linked to the nuclear spliceosomal machinery, more recent studies of trans-mobilization use a modified form of the neo reporter cassette in which the γ-globin intron is replaced by a group I self-splicing intron [22]. This modified cassette, neoTet has been inserted into an Alu element on an episome in order to investigate the requirements for L1-driven retrotransposition of this element. Splicing and genomic integration of the Alu element occurred with an efficiency corresponding to the length of the poly(A) tail [95, 96]. Functional ORF2 was absolutely necessary for mobilization; however, ORF1p appeared only necessary for enhanced levels of retrotransposition [41, 97].
Despite the many successes of the retrotransposition assay, caution must be taken with some interpretations. An episomal system can only approximate the mobilization of elements through mammalian evolution, for an endogenous element that becomes transcribed in one genomic context and jumps to another has a very different life history than a sequence delivered on an episome. Moreover, an element containing an artificial intron, such as in the retrotransposition constructs, may also be processed in a different manner than an endogenous intronless L1. Indeed, new insertions in tissue culture have a high rate of rearrangement and unusually long target site duplications [26]. These unusual features may point to a selection process that had occurred in existent elements over several generations; or, it may indicate differences between the mechanism of retrotransposition in cultured cells and whole organisms. Support for the latter hypothesis comes from studies in transgenic animals, where new insertions were more similar structurally to elements in the reference genome [25].
There are two more caveats to interpreting data from the retrotransposition assay. When providing an L1 element in trans to a non-autonomous element such as SVA, recombination between the two plasmids might result in a fusion construct and false positives in the assay (John Goodier, personal communication). Finally, when using a selectable marker, the retrotransposed sequence must be longer than the marker gene (e.g. neo, 1.4 kb) to be detected. Because retrotransposition commonly produces severely truncated elements, a significant proportion of insertions from the cell culture assay may be lost as false negatives.
Happily, some limitations of this assay as it has often been used in the past can be easily remedied. The L1 retrotransposition assay has often in the past been carried out in HeLa cells, which support a high level of retrotransposition: see [7] among others. However, HeLa are transformed cells known to carry many genetic and genomic abnormalities relative to normal somatic cells [98, 99], changes which might effect the regulation of retrotransposition in these cells. In order to obtain results closer to normal biology, we suggest that future research should focus on whole organisms or non-tumor cell lines where possible (see Table 2 for a list of cell lines that have been tested for retrotransposition competency). In addition, using native promoters to drive the L1 and matching the species of the element to the species source of the cell line or organism should be preferred to ensure that the retrotransposition assay mimics naturally-occurring retrotransposition as closely as possible.
Table 2.
Cell lines successfully used in the retrotransposition assay
| Cell Line (organism) | Description | Reference |
|---|---|---|
| 143B TK-(human) | Osteosarcoma | [56] |
| 293T (human) | Embryonic kidney with large T antigen | [36] |
| CC-2509 (human) | Primary fibroblasts | [36] |
| CC-2591 (human) | Primary hepatocytes | [36] |
| CHO-UV20 (hamster) | Defective in DNA repair (ERCC1-) | [54] |
| CRL-12203 (chicken) | Embryonic fibroblasts | [101] |
| DLD-1 (human) | P53 defective | [51] |
| Gli36 (human) | Glioma | [36] |
| H1, H7, H9, BG01,HSF-6, Cyth25 (human) | Human embryonic stem cells | [102] |
| H460 (human) | Lung adenocarcinoma | [103] |
| HCT116 (human) | Colon cancer | [31] |
| HEK293 (human) | Embryonic kidney | [44] |
| Hep3B (human) | Hepatocellular carcinoma | [36] |
| HT1080 (human) | Fibrosarcoma, both p53 wild-type and defective | [51] |
| IMR-90 (human) | Primary fibroblasts, only work when dividing | [104] |
| LTK- (mouse) | L cells lacking thymidine kinase | [7] |
| MCF-7 (human) | Breast adenocarcinoma | [51] |
| MCF-7-bcl2 (human) | Apoptosis suppressed | [101] |
| NIH3T3 (mouse) | Fibroblasts, transient assay | [78] |
| RCL (rat) | Chloroleukemia | [90] |
| SW480 (human) | p53 defective | [51] |
| T47D (human) | Carcinoma | [52] |
| V3, XR-1 CHO (hamster) | Defective in telomeres and non-homologous end joining or DNA repair | [29] |
| YZ-5 (human) | Competent for altered telomere maintenance | [53] |
Description of Methods
There are essentially two ways to do a retrotransposition assay in cell culture using a neo selection cassette: fast (transient) or slow (traditional). Another variation involves using FACS (fluorescent-activated cell sorting) for screening of EGFP expression.
1. Traditional
In the original retrotransposition assay [7], HeLa cells are seeded at 2 × 105 to 4 × 105 cells/well in 6-well plates and grown in DMEM-complete media to 70% confluence. Transfection is performed with 1 μg plasmid DNA and 4 μl lipofectamine reagent (BRL) in 1 ml of Opti-mem (BRL). After 5 hours at 37°C and 7% CO2, the cultures are supplemented with 1 ml of DMEM containing 20% FCS. This medium is again replaced after 16 hours. Two days later, cells can be trypsinized and seeded into 75 cm2 flasks and grown in 200 μg/ml hygromycin to select for the presence of the vector which encodes hygromycin-resistance. (Vectors encoding puromycin instead of hygromycin resistance are selected using 5 μg/ml puromycin.)
Cells are grown under hygromycin or puromycin selection for 10-14 days, after which they are trypsinized, counted with a hemocytometer and re-seeded at known densities in media supplemented with 300-400 μg/ml G418. This procedure selects for cells that have undergone retrotransposition and now express the neo selectable marker. 10-14 days later, cells are fixed and stained with 0.4% Giemsa. Retrotransposition efficiency is calculated based on the number of cells before and following G418 selection. Refer to Moran et al.[7] for more details on this protocol. Kill curves are suggested to optimize the dose and time of the antibiotic selections.
2. Transient Assay
In the transient variation, two sets of transfected cells are plated in parallel. One set serves as a transfection efficiency control, the denominator for retrotransposition efficiency. The second set is grown on G418 to select for cells in which the intron within the neo gene has been removed by splicing and the L1 construct has integrated into the genome by retrotransposition. More information on this assay can be found elsewhere [100]. A brief description of a protocol for HeLa cells follows below. This protocol must be optimized for different cell lines; the reader is encouraged to refer to publications using those cell lines for more details.
HeLa cells are plated at a density of 2×105 per well of a 6-well tissue culture dish. 3 μl of Fugene non-liposomal transfection reagent (Roche) is used for transfection along with 0.5-1.0 μg of plasmid and equal amounts of pGreen Lantern reporter plasmid as a measure of transfection efficiency. Cells are grown for three days and then trypsinized. Flow cytometry is used to measure the amount of green fluorescence. Wells that were not subject to trypsinization (test wells) are exposed to 400 μg/ml G418 for 12 days. G418 positive foci are fixed in 2% formaldehyde, 0.2% glutaraldehyde and 1 × PBS at 4°C for 30 minutes and then stained with 0.1% Bromophenol blue or 0.4% Giemsa for easy visualization. In parallel, single colonies can be propagated and removed for DNA isolation and PCR or Southern blot analysis.
This protocol can be scaled to 75 or 175 cm2 tissue culture flasks, using proportionately greater quantities of cells, Fugene and DNA. For instance, 2×106 HeLa cells, 24 μl Fugene and 8 μg DNA are suggested for use in 75 cm2 flasks.
3. EGFP selection
Transfections using constructs featuring an EGFP retrotransposition marker were not subjected to G418 selection and are instead examined for fluorescence. Following hygromycin or puromycin selection as described above (approximately day 8 post-transfection), cells are washed with 2 ml 1 × PBS and then incubated in 5 mM EDTA in PBS. The retrotransposition-defective JM111 construct [7] can be transfected in parallel to transfections with the L1-containig construct and used as a negative control to determine background levels of fluorescence. The cell suspension is transferred through cell strainer caps (Falcon) to polystyrene tubes and kept on ice. This solution is analyzed with a FACScan machine (Becton Dickinson) with a blue argon 488nm laser and fluorescein filters of 530.30 bandpass. Typically, between 10,000 and 20,000 events are analyzed per sample. Live-dead gating is performed based on the forward scatter versus side scatter profile. Analysis of living cells for fluorescence intensity can be conducted using the CellQuest software package. An arbitrary threshold of intensity such as <0.05% can be used as a value for background fluorescence, with the proportion of JM111 transfected cells fluorescing above this value set as the false positive rate. Refer to Ostertag et al.[24] for more details on this protocol.
4. Constructs
Traditionally, most constructs used for retrotransposition feature a highly active naturally-isolated L1 element, such as L1RP or LRE3 [7]. A neo gene cassette, mneoI, is disrupted by a γ-globin intron in the opposite orientation [19]. A later variation used an EGFP cassette disrupted by this same intron [24]. Vectors allowing propagation in bacterial cells can be used for easy recovery of novel insertion sites in E. coli [26, 27, 31]. A codon-optimized mouse element, ORFeus, is advantageous for gene-trapping and Cre-Lox conditional activation [32, 78, 84]. An alternative strategy for transfection utilizes a helper-dependent adenovirus backbone for retrotransposon delivery [36, 37]. Other variations include an ORF1 epitope tag [75], an RNAi cassette [85], and the use of a group I self-splicing intron in the retrotransposition marker [22]. There are also versions that include a blasticidin antibiotic selection marker [23] and even no selectable marker [25]. Refer to Figure 1A for a diagram of a canonical construct and to Table 1 for details and complete references.
Table 1.
Major constructs used for the L1 retrotransposition
| Construct | Features | Use | Reference | |
|---|---|---|---|---|
| pJM101 related others containing different L1 sequences | mneoI cassette with γ-globin intron, can contain L1 elements: L1.2, L1RP, L1.3, LRE2, LRE3 | High frequency retrotransposition | [7] | |
| pJM111 | pJM101 with two missense mutations in L1.2 ORF1 | <1% pJM101/L1.2 retrotransposition, used as negative control | [7] | |
| pAD5/L1-3 pAD6/L1-13 |
pJM101/L1.3 replaced with rat sequences L1-3 and L1-13 | Retrotransposition of rat L1 elements | [90] | |
| pL1.3-EGFP pL1RP-EGFP pL1.3-EGFP(puro) pL1RP-EGFP(puro) |
pJM101 with EGFP cassette disrupted by γ-globin intron, may contain puromycin resistance on the backbone | Visual selection for retrotransposition, may be puromycin-resistant | [24] | |
| pK7/L1.3mneoI400/ColE 1 pCEP4/L1.3mneoI400/ColE1 pJM140/L1.3/delta2K7 pBSSP140/L1.3K7 pSP140/L1.3K7 |
Neo cassette contains the EM7 promoter and Shine-Delgarno sequence, loss of cryptic splice site, for expression of neo (KanR) in bacterial cells, also a ColE1 prokaryotic ori | Recovery of sequence from insertion sites in bacterial cells | [26, 27] | |
| pDES89 | Encodes trimethoprim (Tmp) resistance, dhfr, and bacterial origin of replication ori | Recovery of sequence from insertion sites in bacterial cells | [31] | |
| pCEPsmL1 (ORFeus) | Codon optimized ORF1 and ORF2 of mouse element, GFP cassette retrotransposition marker | High frequency retrotransposition and mutagenesis | [78] | |
| ORFeusLSL | PCEPsml1 with L1 coding sequences disrupted by loxP-β-geo-stop-loxP cassette | Cre-lox activated retrotransposition | [84] | |
| SynL1_optORF1_neo SynL1_optORF1_optNeo |
L1.3 with cryptic polyadenylation sites, 5′ UTR removed. OptNeo contains a codon-optimized neo cassette. | Optimized for high frequency retrotransposition | [54, 79] | |
| pSMKL1.3mneoI | Helper-dependent adenovirus(HDAd) vector Containing the retrotransposition reporter CMV-L1.3mneoI and CMV-GFP as a reporter | Gene therapy vector that infects a range of cell types like an adenovirus but which integrates into the genome using retrotransposition | [37] | |
| pA/RT-pgk-L1RP-EGFP | pSTK6 HDAd vector containing the retrotransposition reporter pgk-L1RP-EGFP and SV40-β-gal | Second generation adenovirus-L1 delivery system | [36] | |
| pCEP-Hsp70-2-L1LRE3-mless | Driven by mouse Hsp70 promoter, contains γ-globin intron without full-length neo gene | No selectable marker; PCR required to isolate inserts | [25] | |
| pDK101 | pJM101/L1.3 modified to contain a C-terminus T7 gene 10 epitope tag on ORF1p | ORF1p tagged for biochemical studies | [75] | |
| pL1 -Silencer | L1RP-neo followed by an RNAi-cassette (H1 promoter, 19 bp sense and antisense sequences of a specific gene and an RNA polymerase III terminator) | Retrotransposition-based RNAi delivery system | [85] | |
| 99-PUR-RPS-pBlaster1 | L1RP with a blasticidin S deaminase retrotransposition marker and puromycin-resistant vector backbone | Features blasticidin selection which is faster than G418 selection | [23] | |
| pL1.3neoIII | T. thermophila 23S rRNA group I intron disrupting the neo selection cassette | Self-splicing intron detects retrotransposition that is not dependent on RNA polymerase II transcriptional processing | [22, 35] | |
5. Cell Lines
HeLa cells were the first human cell line used in the assay [7], and remain one of the most efficient. Other cell lines, both transformed and non-transformed, from a variety of organisms, have also been tested for retrotransposition competency. Note that chicken cells lack endogenous L1. These lines are listed in Table 2.
Conclusion
In this paper, we review the development of the L1 retrotransposition cell culture assay and describe its many successes in deciphering L1 biology over the last thirteen years. This assay has been central to the study of the mechanism and consequences of retrotransposition in mammalian cells and, increasingly, whole animal models. We also provide detailed protocols and a list of plasmid constructs in order to aid the reader in applying this technique to their own studies of retrotransposition. We look forward to further improvements and applications of this powerful research tool.
Acknowledgments
We thank John Goodier for impassioned discussions and critiques. Research in the Kazazian lab is funded by the National Institutes of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Babushok DV, Kazazian HH., Jr Hum Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 2.Belancio VP, Hedges DJ, Deininger P. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 3.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, Jorde LB, Batzer M. A Am J Hum Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. Hum Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 8.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 9.Heidmann T, Heidmann O, Nicolas JF. Proc Natl Acad Sci U S A. 1988;85:2219–2223. doi: 10.1073/pnas.85.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen S, Heidmann T. Embo J. 1991;10:1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidmann O, Heidmann T. Cell. 1991;64:159–170. doi: 10.1016/0092-8674(91)90217-m. [DOI] [PubMed] [Google Scholar]
- 12.Esnault C, Maestre J, Heidmann T. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 13.Maestre J, Tchenio T, Dhellin O, Heidmann T. Embo J. 1995;14:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tchenio T, Segal-Bendirdjian E, Heidmann T. Embo J. 1993;12:1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curcio MJ, Garfinkel DJ. Proc Natl Acad Sci U S A. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derr LK, Strathern JN, Garfinkel DJ. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz DA, Dahm MW, Bai L, Carnie S, Norris JS. Gene. 1993;127:233–236. doi: 10.1016/0378-1119(93)90725-i. [DOI] [PubMed] [Google Scholar]
- 18.Tchenio T, Heidmann T. J Virol. 1992;66:1571–1578. doi: 10.1128/jvi.66.3.1571-1578.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman JD, Goodchild NL, Mager DL. Biotechniques. 1994;17:46, 48–49, 52. [PubMed] [Google Scholar]
- 20.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 21.Dombroski BA, Scott AF, Kazazian HH., Jr Proc Natl Acad Sci U S A. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esnault C, Casella JF, Heidmann T. Nucleic Acids Res. 2002;30:e49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert N, Lutz S, Morrish TA, Moran JV. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert N, Lutz-Prigge S, Moran JV. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 28.Farley AH, Luning Prak ET, Kazazian HH., Jr Nucleic Acids Res. 2004;32:502–510. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 30.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 31.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 32.An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Proc Natl Acad Sci U S A. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YG, Whittier RF. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 34.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Jr, Kasahara N. Proc Natl Acad Sci U S A. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soifer H, Higo C, Kazazian HH, Jr, Moran JV, Mitani K, Kasahara N. Hum Gene Ther. 2001;12:1417–1428. doi: 10.1089/104303401750298571. [DOI] [PubMed] [Google Scholar]
- 38.Hata K, Sakaki Y. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 39.Yu F, Zingler N, Schumann G, Stratling WH. Nucleic Acids Res. 2001;29:4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 44.Stenglein MD, Harris RS. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 45.Turelli P, Vianin S, Trono D. J Biol Chem. 2004;279:43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 46.Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macduff DA, Demorest ZL, Harris RS. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girard A, Hannon GJ. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soifer HS, Kasahara N. Curr Gene Ther. 2004;4:373–384. doi: 10.2174/1566523043346084. [DOI] [PubMed] [Google Scholar]
- 50.Yang N, Kazazian HH., Jr Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 51.Haoudi A, Semmes OJ, Mason JM, Cannon RE. J Biomed Biotechnol. 2004;2004:185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gasior SL, Wakeman TP, Xu B, Deininger PL. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gasior SL, Roy-Engel AM, Deininger PL. DNA Repair (Amst) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Athanikar JN, Badge RM, Moran JV. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farkash EA, Kao GD, Horman SR, Prak ET. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El-Sawy M, Kale SP, Dugan C, Nguyen TQ, Belancio V, Bruch H, Roy-Engel AM, Deininger PL. J Mol Biol. 2005;354:246–257. doi: 10.1016/j.jmb.2005.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kale SP, Moore L, Deininger PL, Roy-Engel AM. Int J Environ Res Public Health. 2005;2:14–23. doi: 10.3390/ijerph2005010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 62.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 63.Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH., Jr Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boissinot S, Chevret P, Furano AV. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 65.Skowronski J, Singer MF. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):457–464. doi: 10.1101/sqb.1986.051.01.055. [DOI] [PubMed] [Google Scholar]
- 66.Lutz SM, Vincent BJ, Kazazian HH, Jr, Batzer MA, Moran JV. Am J Hum Genet. 2003;73:1431–1437. doi: 10.1086/379744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seleme MC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH., Jr Proc Natl Acad Sci U S A. 2006;103:6611–6616. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Nat Genet. 1998;20:288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- 69.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. Embo J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 72.Repanas K, Zingler N, Layer LE, Schumann GG, Perrakis A, Weichenrieder O. Nucleic Acids Res. 2007;35:4914–4926. doi: 10.1093/nar/gkm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. J Mol Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 74.Martin SL, Bushman D, Wang F, Li PW, Walker A, Cummiskey J, Branciforte D, Williams MC. Nucleic Acids Res. 2008;36:5845–5854. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kulpa DA, Moran JV. Hum Mol Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 76.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li PW, Li J, Timmerman SL, Krushel LA, Martin SL. Nucleic Acids Res. 2006;34:853–864. doi: 10.1093/nar/gkj490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han JS, Boeke JD. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 79.Gasior SL, Palmisano M, Deininger PL. Anal Biochem. 2006;349:41–48. doi: 10.1016/j.ab.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 81.Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Proc Natl Acad Sci U S A. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 83.Ostertag EM, Madison BB, Kano H. Genome Biol. 2007;8 1:S16. doi: 10.1186/gb-2007-8-s1-s16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.An W, Han JS, Schrum CM, Maitra A, Koentgen F, Boeke JD. Genesis. 2008;46:373–383. doi: 10.1002/dvg.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang N, Zhang L, Kazazian HH., Jr Nucleic Acids Res. 2005;33:e57. doi: 10.1093/nar/gni056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Nat Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- 87.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horie K, Saito ES, Keng VW, Ikeda R, Ishihara H, Takeda J. Genetics. 2007;176:815–827. doi: 10.1534/genetics.107.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. Genome Res. 2008;18:597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, Lower R, Buschmann C, Horn AV, Traub P, Schumann GG. Nucleic Acids Res. 2008;36:648–665. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kajikawa M, Okada N. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 92.Sugano T, Kajikawa M, Okada N. Gene. 2006;365:74–82. doi: 10.1016/j.gene.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 93.Goodwin TJ, Busby JN, Poulter RT. BMC Genomics. 2007;8:263. doi: 10.1186/1471-2164-8-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dong C, Poulter RT, Han JS. Genetics. 2009;181:301–311. doi: 10.1534/genetics.108.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dewannieux M, Esnault C, Heidmann T. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 96.Dewannieux M, Heidmann T. Genomics. 2005;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 97.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsu TC, Moorhead PS. Ann N Y Acad Sci. 1956;63:1083–1094. doi: 10.1111/j.1749-6632.1956.tb32124.x. [DOI] [PubMed] [Google Scholar]
- 99.Macville M, Schrock E, Padilla-Nash H, Keck C, Ghadimi BM, Zimonjic D, Popescu N, Ried T. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]
- 100.Wei W, Morrish TA, Alisch RS, Moran JV. Anal Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 101.Wallace NA, Belancio VP, Deininger PL. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 103.DeBerardinis RJ. PhD. University of Pennsylvania; Philadelphia: 1998. Cell and Molecular Biology; p. 204. [Google Scholar]
- 104.Shi X, Seluanov A, Gorbunova V. Mol Cell Biol. 2007;27:1264–1270. doi: 10.1128/MCB.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]