Summary
Electron crystallography is arguably the only electron cryomicroscopy (cryoEM) technique able to deliver an atomic-resolution structure of membrane proteins embedded in the lipid-bilayer. In the electron crystallographic structures of the light driven ion pump, bacteriorhodopsin, and the water channel, aquaporin-0, sufficiently high resolution was obtained and both lipid and protein were visualized, modeled and described in detail. An extensive network of lipid-protein interactions mimicking native membranes is established and maintained in two-dimensional (2D) crystalline vesicles used for structural analysis by electron crystallography. Lipids are tightly integrated into the protein's architecture where they can affect the function, structure, quaternary assembly and the stability of the membrane protein.
Introduction
Lipids and membrane proteins co-evolved to form biological membranes. The proteins perform an array of functions such as drug and neurotransmitter transport, water and nutrient transport, signaling, cell-to-cell communication and adhesion among other things. Lipids intimately influence the structure and function of membrane proteins. Extensive lipid-protein interactions occur within the lipid bilayer as lipids fit into crevices and irregularities on the protein surface to maintain an electrochemical seal across the membrane. To fully understand the interdependence of membrane proteins with their surrounding lipids necessitates structural information in the context of a lipid bilayer. Electron crystallography is uniquely suited for this task, since the membrane protein of interest is crystallized in two-dimensions (2D) within a lipid bilayer. High-resolution electron crystallography is then used to determine the structure of the protein. Moreover, the function of the protein of interest can be assayed directly from the 2D crystals allowing the unique opportunity to directly couple both membrane protein structure and function from the very same membranes.
Electron crystallography became a bona fide method for determining the structures of membrane proteins following the pioneering work by Henderson and Unwin in the mid 1970s, who provided the first structural information of a membrane protein, bacteriorhodopsin (bR) where for the first time membrane traversing α-helices were visualized [1]. Henderson and Unwin's work was extended in later years, with gradual increases in resolution to 3Å [2,3••], and provided the first “atomic-level” structural information on specific lipid-protein interactions determined by electron crystallography. More recently, the 1.9Å resolution structure of the aquaporin AQP0 was determined by electron crystallography, providing a detailed look at non-specific lipid protein interactions [4••]. In this review we focus in particular on the high-resolution structures of bR [3••] and AQP0 [4••] to describe the different lipid-protein interactions that were identified and discuss examples of how these interactions influence membrane protein stability, structure, oligomeric assembly and function.
Lipid-protein interactions studied by electron crystallography
Only two examples exist to date where sufficiently high resolution could be obtained by electron crystallography to visualize and model the lipid-bilayer surrounding a membrane protein. Those are the structures of bR and AQP0, where lipids were modeled and fit directly into the experimental density maps [3••,4••]. In the case of bR, native lipids are functionally required and are woven into the structural framework of the protein, while in the case of AQP0, the lipids interact non-specifically with the protein and the lipids are not required for the function of the channel. Despite these differences, the interactions that occur between the membrane protein and the surrounding lipids exhibit startling similarities that will be discussed in this section.
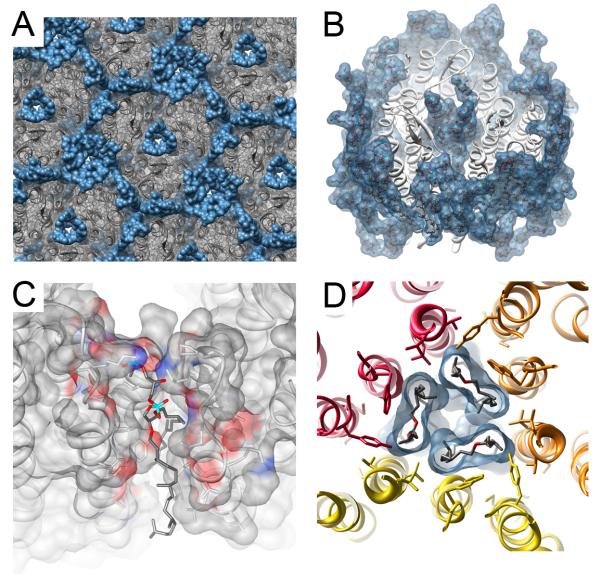
Bacteriorhodopsin is a homotrimeric light driven ion pump that assembles into hexagonal arrays in the purple membrane (PM) of the archaea halobacterium salinarum [5,6]. bR specifically requires native lipids for both assembling PM-like membranes in vitro [7] and for its function as a proton pump [8]. Early reconstitution experiments demonstrated that when bR was delipidated and reconstituted with the synthetic lipid dimyrostylphosphatidylcholine (DMPC), the protein did not form hexagonal arrays unless native PM lipids were added to the reconstitution [7]. When bR is reconstituted with native PM lipids, bR trimers form and are arranged in highly ordered hexagonal arrays with dimensions identical to the bR crystalline patches found in the PM (Figure 1A). The structure determined by electron crystallography shows that each bR monomer consists of 7 transmembrane α-helices with the N-terminus located on the extra-cellular side of the membrane and the C-terminus on the intra-cellular side. A total of 24 lipids were identified surrounding bR [3••]. 18 lipids contribute to a nearly complete lipid shell (annulus) surrounding the bR trimer (Figure 1B) while 6 lipids are found buried within the protein.
Figure 1. Specific lipid-protein interactions in the structure of bR.
(A) Hexagonal lattice in the two-dimensional (2D) crystals of bR [3••]. bR is colored in grey and purple membrane (PM) lipids in blue surface representation. (B) Each bR trimer (white ribbon) is surrounded by a nearly complete lipid bilayer. Three additional lipids were resolved at the 3-fold axis of the bR trimer. (C) Zoom view of specifically bound PM lipid integrated into the monomer-monomer interface of bR. Lipids are shown as a ball and stick and colored by heteroatom and bR is displayed with electrostatic surface representation (colored grey – neutral, blue – positive and red – negative). An acyl chain from this specific lipid inserts into a hydrophobic crevice and mediates interfacial α-helical packing. (D) The three PM lipids located at the 3-fold axis of bR are surrounded by a symmetrical array of tryptophans and hydrophobic residues. The three bR monomers in the trimer are colored red, orange and yellow.
The lipids that were identified buried in bR form extensive charge networks and appear to be critical for the function of the protein. Out of the 6 buried lipids, 3 are found cytoplasmically located between monomer-monomer interfaces, and 3 are found at the cavity at the 3-fold axis of the bR trimer (Figure 1 C and D). A deep hydrophobic crevice is formed in between two bR monomers on the cytoplasmic side of the protein. A single PM lipid (3 in total for the trimer) inserts one of its two acyl chains into this crevice to mediate interfacial α-helical packing of the protein (Figure 1C). A phosphoryl head group from this lipid in turn forms a salt bridge with the side chain amine from lysine 40 on bR. At the 3-fold axis of bR, three PM lipids were observed to plug the central cavity on the extracellular side. The head groups of these three central lipids are vertically lower by ∼5Å then the extracellular lipid leaflet surrounding bR. The displaced position of these head groups is dictated by a ring of tryptophans and hydrophobic residues originating from α-helices that are arranged symmetrically around the 3-fold axis of the protein (Figure 1D). Tryptophan residues are often used to dictate lipid head group positioning because the positively charged indole ring on this amino acid may simultaneously interact with the electronegative glycerol backbone and acyl lipid tail [9]. It has now been determined that the central lipids are likely to be the archaeal PM glycolipid - sulfated triglycosylarchaeol (STGA) [6]. When STGA is added to reconstituted lipid vesicles, the ion pumping kinetics of bR are greatly enhanced [8], supporting a specific functional role for these specifically bound lipids. By examining low-resolution electron diffraction data of bR 2D crystals, Mitsuoka and co-workers showed that the electrical charge of amino acid side chains and lipid head groups may be visualized [3••]. By using this technique it will be possible to map directly the proton pumping pathway in bR during the photocycle and determine the role lipids have during this process.
The structure of AQP0 determined by electron crystallography to 1.9Å resolution provides a unique example of how membrane proteins interact non-specifically with annular lipids [4••]. AQP0 forms a water channel in the eye lens, and like all other aquaporins, each monomer consists of 6-transmembrane α-helices arranged as in a barrel forming a central pore for water (reviewed in [10,11•]). Four AQP monomers assemble into tetramers although each monomer forms its own channel. It was suggested that tetramerization is required for the stability of the protein in the lipid bilayer and more recently it was shown that the oligomeric assembly is required for the binding of regulatory proteins to AQP0 [12•]. However, there is no data to suggest that the function of the channel is affected by the composition of the lipid bilayer. Unlike bR, which shows a strong structural and functional dependency on native PM lipids, AQPs do not require native lipids to support their native structures or functions in vitro. In fact, AQP permeability studies are routinely performed using a variety of different lipids including studies in oocyte membranes [13], and using reconstituted proteoliposomes [14] or 2D crystals composed of various lipids including E. coli total lipid extracts [15].
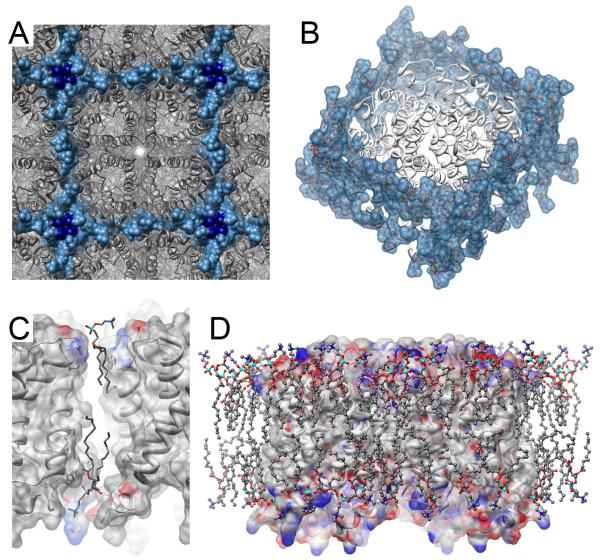
The AQP0 2D crystals were grown with the synthetic lipid DMPC [4••]. AQP0 tetramers were arranged in the 2D crystals as a square lattice having the same unit cell dimensions as AQP0 square arrays found in the eye lens (Figure 2A). A monolithic lipid bilayer of 28 DMPC lipids was observed surrounding the AQP0 tetramer. The lipids formed a continuous annular shell around the protein and mediated crystal contacts through extensive protein-lipid-protein interaction networks (Figure 2B). These contacts include both electrostatic and hydrophobic packing interactions as lipids are sandwiched between neighboring AQP0 tetramers (Figure 2C). Because of the aforementioned reasons, the observed lipid-protein interactions in the AQP0 2D crystals are considered inherently non-specific. In addition to the 28 annular lipids that make direct contact with AQP0, 8 additional DMPC lipids per tetramer were resolved at the four-fold crystallographic axis located between adjacent tetramers (dark blue, Figure 2A). These additional lipids make no direct contacts with AQP0 and therefore represent bulk lipids.
Figure 2. Non-specific lipid-protein interactions in the AQP0 structure.
(A) Square lattice in the 2D crystals of AQP0 [4••]. AQP0 is colored in grey and synthetic DMPC lipids in blue surface representation. (B) Each AQP0 tetramer is surrounded by a lipid annulus composed of 28 DMPC lipids. 8 additional bulk lipids are found at the tetramer interface, colored dark blue in (A). (C) Zoom view of protein-lipid-protein interactions that mediate crystal packing. AQP0 displayed with electrostatic surface representation (colored grey – neutral, blue – positive and red – negative) and lipids shown as ball and stick and colored by heteroatom. (D) Extensive lipid-protein interactions in the AQP0 2D crystals involve van der Waals interactions with the AQP0 hydrophobic belt and a network of charge complementation between lipid and protein.
Notably, even though the interactions between AQP0 and the synthetic lipid DMPC are non-specific, the types of hydrophobic packing and electrostatic contacts the lipids make with the protein are similar to the contacts made by specifically bound lipids such as in the case of bR. Typically an extensive network of charge complementation is established and maintained between lipids and protein. The electrostatic environment presented by residues on the surface of AQP0 determines the vertical positioning of the annular lipids in the bilayer (in fact bulk lipids which are not affected by protein electrostatics are vertically displaced from the annular lipids bilayer by as much as ∼5Å). The apolar lipid acyl chains are molded against the protein transmembrane hydrophobic belt, while the lipid glycerol backbone and phosphatidylcholine (PC) polar head groups interact with the hydrophilic domains of AQP0 and are supported by electrostatic interactions (Figure 2D). It appears that the lipids accommodate the protein to maintain a hydrophobic and electrical seal across the membrane. A significant conformational heterogeneity of lipid morphologies is observed in the context of the AQP0 2D crystals. Only a few lipid acyl chains are found in an extended conformation while most lipids adopt contorted structures induced by shallow hydrophobic grooves displayed by the AQP0 hydrophobic belt. In these cases, the lipid acyl chains bend and buckle to fit into irregularities on the protein. Similarly, most PC head groups are found with highly bent structures, again induced by positively charged amino acid sidechains on AQP0 (Figure 2C and D).
Lipid stabilization of membrane protein structures and assemblies
Electron crystallography may provide unique insight on how lipid interactions contribute to both the local and global stability of membrane proteins. The annular lipid environment may influence the dynamics of membrane proteins. The crystallographic B-factor (or temperature factor) represents the statistical disorder (dynamics) of the protein structure. Crystallographic B-factors observed in both X-ray and electron crystallographic structures are typically higher (more dynamic) for the lipids then for the protein amino acid sidechains with which lipids interact [16,17••].
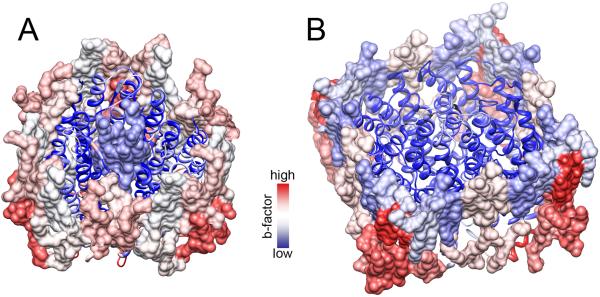
Generally, the more tightly bound the lipid to the protein, the lower the lipid crystallographic B-factor. In the electron crystallographic structure of bR, the three specific lipids incorporated into the trimer at the 3-fold axis have B-factor values as low as the B-factor values for the protein. Other lipids in the bR 2D crystals have significantly higher B-factor values then the protein especially at the lipid head group positions (Figure 3A). Similarly, in the electron crystallographic structure of AQP0 the annular lipids also have higher B-factors then the rest of the protein, however the bulk lipids that do not directly interact with the protein have the highest B-factor values (Figure 3B). These observations suggest that the protein stabilizes the lipids through extensive interactions. A recent study conducted an in-depth statistical analysis of the crystallographic B-factors for both AQP0 and bR by directly comparing the X-ray and electron crystallography studies [17••]. In general, overall B-factors were found to be lower in the electron crystallographic structures then those obtained by X-ray crystallography. Importantly, AQP0 residues that in 2D crystals interacted with lipids were found to interact with detergent molecules in 3D crystals used for X-ray analysis. However, these residues had significantly lower B-factor values when they were coupled to lipids rather then to detergents. This was attributed to lipid stabilizing interactions in the 2D crystals, suggesting that even non-specific lipid-protein interactions have a significant stabilizing effect on membrane proteins [17••].
Figure 3. Lipid stability in 2D crystals of bR and AQP0.
Ribbon representation of (A) bR and (B) AQP0 with associated lipids (surface representation) and colored by average crystallographic B-factor values (blue – low and red – high). The crystallographic B-factor is a term that reflects the statistical disorder (or dynamics) of a structural model. (A) Lipids are generally more dynamic then the protein [17••] while the dynamics of the specifically bound PM lipids at the 3-fold axis of bR are restricted by the surrounding protein. (B) Bulk lipids show the highest disorder in the AQP0 2D crystals because they make no direct contact with the protein.
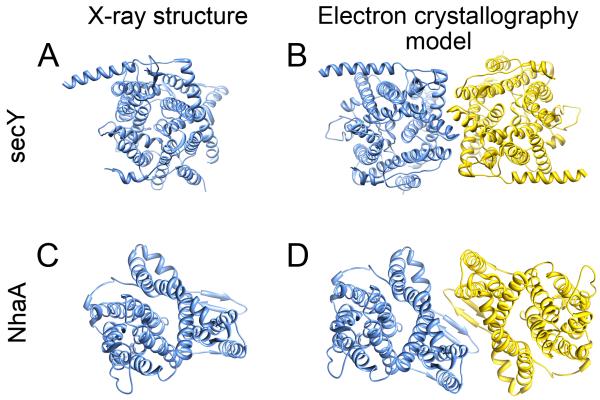
Lipid-protein interactions may also play an integral role in stabilizing quaternary architecture of membrane proteins. In fact the oligomeric assembly of many membrane proteins are disrupted upon delipidation by detergent extraction that is required for purification prior to crystallization. Bacteriorhodopsin does not assemble into trimeric hexagonal arrays unless native PM lipids are present [7]. Often however, the native oligomeric state of membrane proteins can be reconstituted in 2D lipid-protein crystals grown for electron crystallography. There are examples in the literature where membrane proteins were crystallize in 3D in an oligomeric state that differs from the oligomeric state found when the protein was incorporated into membranes and crystallized in 2D. For example, the protein translocation complex secY and the proton coupled sodium transporter NhaA, are both monomeric in 3D crystals but form dimers in membranes [18,19•] (Figure 4). Electron diffraction data from 2D crystals (even at low resolution) can therefore provide important structural insights into how membrane protein assemblies are organized within the membrane.
Figure 4. Lipid-protein interactions can influence the quaternary assembly of membrane proteins.
Two examples are presented, secY [24] (top) and NhaA [25] (bottom). (A) and (C) In both cases the membrane proteins crystallized as monomers in 3D crystals used for X-ray crystallography. (B) and (D) 2D crystals used for electron crystallography contained dimeric secY and NhaA, respectively [18,19•]. In the case of secY, there is evidence that protein translocation is mediated by the secY multimers although each monomer forms a channel [26]. The atomic resolution X-ray structures of secY and NhaA were combined with density maps obtained by electron crystallography to model the pseudo-atomic structures of these transporters in their membrane-embedded dimeric forms.
Functional analysis in 2D protein-lipid crystals
The function of a channel or a transporter can be probed directly by electrophysiology or fluorescence studies using 2D crystals if these crystals are vesicular. For example, AQP1 purified from red blood cell membranes was reconstituted into vesicles and channel permeability determined directly from resulting 2D crystals [20]. During the reconstitution experiments, vesicles were loaded with carboxyfluorescein, and then transferred from iso-osmolar buffer to hyper-osmolar buffer while fluorescence quenching was monitored by stopped-flow techniques. Initial permeability studies used high lipid-to-protein ratios yielding small unilamellar vesicles, while later experiments used low lipid-protein ratios resulting in highly ordered 2D crystals of AQP1. In both cases, the AQP1 channels remained fully functional, and vesicles shrank completely in less than 20 ms even in the tightly-packed, highly ordered 2D crystals. The calculated coefficient of osmotic water permeability was similar in magnitude to the water permeability of intact red blood cells.
Because 2D crystals closely mimic biological membranes they are physically robust and can accommodate modest structural changes in the protein as channels open and close in response to changing pH, binding substrates, activators or inhibitors, while maintaining crystallinity. This allows the biological function of the protein to be directly coupled with structural information obtained by electron diffraction from the very same membrane preparations. Some examples include the time-resolved (millisecond time-scale) studies following the light induced activation pathway of bR [21] and the substrate-induced activation of the acetylcholine receptor [22].
Conclusions
Lipids can influence the structure and the function of membrane proteins [23], probably because of the nature of the extensive lipid-protein interactions that are established and maintained in the context of a biological membrane. In electron crystallography the protein of interest is crystallized in 2D in the context of a lipid bilayer. The composition of this lipid bilayer can vary from purely synthetic to entirely native lipid compositions such as PM lipids. The interdependency of membrane protein structure and function on lipid compositions can therefore be probed directly by electron crystallography, and given sufficient resolution the structure of protein and of the surrounding lipid bilayer can be determined.
An emerging strength in electron crystallography is the ability to directly visualize the electrical charge of protein residues [3••]. In this way the pathways for proton and ion transport could be monitored directly and help decipher how for example transporters use electrochemical gradients to couple the transport of ions to a variety of substrates. Moreover, the intricacies of electrostatic interactions that occur between lipids and membrane proteins may be directly probed to understand how lipid electrostatics stabilize the structure and promote the function of membrane proteins.
Acknowledgements
We thank the Murdock Charitable Trust and the Washington Research Foundation for generous support of our electron cryomicroscopy facility. Research in the Gonen laboratory is supported by the National Institutes of Health GM079233 and the American Diabetes Association Award # 1-09-CD-05. T.G. is a Howard Hughes Medical Institute Early Career Scientist. SLR is supported by the National Institutes of Health F32 EY18768.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that none have a financial interest related to this work.
References
- 1.Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 2.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 3••.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y. The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. J Mol Biol. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. This paper describes the 3Å resolution electron crystallographic structure of bacteriorhodopsin. Specifically bound lipids are resolved and lipid-protein interactions discussed. Charges on protein amino acid residues are also visualized by electron microscopy. [DOI] [PubMed] [Google Scholar]
- 4••.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison S, Walz T. Lipid-protein interactions in double-layered two-dimensional crystals of aquaporin-0. Nature. 2005;438:633–638. doi: 10.1038/nature04321. This paper describes the 1.9Å resolution electron crystallographic structure of aquaporin-0. Non specific lipid-protein interactions are described. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramaniam S, Hirai T, Henderson R. From structure to mechanism: electron crystallographic studies of bacteriorhodopsin. Philos Transact A Math Phys Eng Sci. 2002;360:859–874. doi: 10.1098/rsta.2001.0971. [DOI] [PubMed] [Google Scholar]
- 6.Cartailler JP, Luecke H. X-ray crystallographic analysis of lipid-protein interactions in the bacteriorhodopsin purple membrane. Annu Rev Biophys Biomol Struct. 2003;32:285–310. doi: 10.1146/annurev.biophys.32.110601.142516. [DOI] [PubMed] [Google Scholar]
- 7.Sternberg B, L'Hostis C, Whiteway CA, Watts A. The essential role of specific Halobacterium halobium polar lipids in 2D-array formation of bacteriorhodopsin. Biochim Biophys Acta. 1992;1108:21–30. doi: 10.1016/0005-2736(92)90110-8. [DOI] [PubMed] [Google Scholar]
- 8.Hojeberg B, Lind C, Khorana HG. Reconstitution of bacteriorhodopsin vesicles with Halobacterium halobium lipids. Effects of variations in lipid composition. J Biol Chem. 1982;257:1690–1694. [PubMed] [Google Scholar]
- 9.Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 10.Andrews S, Reichow SL, Gonen T. Electron crystallography of aquaporins. IUBMB Life. 2008;60:430–436. doi: 10.1002/iub.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Gonen T, Walz T. The structure of aquaporins. Q Rev Biophys. 2006;39:361–396. doi: 10.1017/S0033583506004458. This is A thorough review on aquaporin structures determined both by electron and X-ray crystallography. [DOI] [PubMed] [Google Scholar]
- 12•.Reichow SL, Gonen T. Noncanonical binding of calmodulin to aquaporin-0: implications for channel regulation. Structure. 2008;16:1389–1398. doi: 10.1016/j.str.2008.06.011. Oligomerization in aquaporins is important for binding of regulatory proteins such as calmodulin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 14.Zeidel ML, Ambudkar SV, Smith BL, Agre P. Reconstitution of functional water channels in liposomes containing purified red cell CHIP28 protein. Biochemistry. 1992;31:7436–7440. doi: 10.1021/bi00148a002. [DOI] [PubMed] [Google Scholar]
- 15.Walz T, Smith BL, Zeidel ML, Engel A, Agre P. Biologically active two-dimensional crystals of aquaporin CHIP. J Biol Chem. 1994a;269:1583–1586. [PubMed] [Google Scholar]
- 16.Hunte C. Specific protein-lipid interactions in membrane proteins. Biochem Soc Trans. 2005;33:938–942. doi: 10.1042/BST20050938. [DOI] [PubMed] [Google Scholar]
- 17••.Hite RK, Gonen T, Harrison SC, Walz T. Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Arch. 2007 doi: 10.1007/s00424-007-0353-9. The temperature B-factors of bacteriorhodopsin and aquaporin-0 determined by both electron and X-ray crystallography are systematically compared and contrasted. Special attention is given to amino acid residues that are interacting with lipid or detergent molecules. An overall stabilizing effect of lipids on protein is observed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemons WM, Jr., Menetret JF, Akey CW, Rapoport TA. Structural insight into the protein translocation channel. Curr Opin Struct Biol. 2004;14:390–396. doi: 10.1016/j.sbi.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 19•.Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kuhlbrandt W. Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol. 2009;386:351–365. doi: 10.1016/j.jmb.2008.12.042. The ion channel NhaA crystallizes as a monomer in 3D crystals but dimers are physiologically important and indeed 2D crystals contain dimers. This paper suggests that crystallographic protein-protein contacts may inhibit the NhaA transport mechanism. [DOI] [PubMed] [Google Scholar]
- 20.Walz T, Smith BL, Zeidel ML, Engel A, Agre P. Biologically active two-dimensional crystals of aquaporin CHIP. J Biol Chem. 1994;269:1583–1586. [PubMed] [Google Scholar]
- 21.Subramaniam S, Lindahl M, Bullough P, Faruqi AR, Tittor J, Oesterhelt D, Brown L, Lanyi J, Henderson R. Protein conformational changes in the bacteriorhodopsin photocycle. J Mol Biol. 1999;287:145–161. doi: 10.1006/jmbi.1999.2589. [DOI] [PubMed] [Google Scholar]
- 22.Berriman J, Unwin N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy. 1994;56:241–252. doi: 10.1016/0304-3991(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 23.Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 25.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 26.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]