Summary
Neuronal Calcium Sensor-1 (NCS-1) is a high-affinity, low-capacity Ca2+-binding protein expressed in many cell types. We previously showed that NCS-1 interacts with inositol 1,4,5-trisphosphate receptor (InsP3R) and modulates Ca2+-signaling by enhancing InsP3-dependent InsP3R channel activity and intracellular Ca2+ transients. Recently we reported that the chemotherapeutic agent, paclitaxel (taxol) triggers μ-calpain dependent proteolysis of NCS-1, leading to reduced Ca2+-signaling within the cell. Degradation of NCS-1 may be critical in the induction of peripheral neuropathy associated with taxol treatment for breast and ovarian cancer. To begin to design strategies to protect NCS-1, we treated NCS-1 with μ-calpain in vitro and identified the cleavage site by N-terminal sequencing and MALDI-mass spectroscopy. μ-calpain cleavage of NCS-1 occurs within an N-terminal pseudoEF-hand domain, which by sequence analysis appears to be unable to bind Ca2+. Our results suggest a role for this pseudoEF-hand in stabilizing the three functional EF-hands within NCS-1. Using isothermal titration calorimetry (ITC) we found that loss of the pseudoEF-hand markedly decreased NCS-1’s affinity for Ca2+. Physiologically, this significant decrease in Ca2+ affinity may render NCS-1 incabable of responding to changes in Ca2+ levels in vivo. The reduced ability of μ-calpain treated NCS-1 to bind Ca2+ may explain the altered Ca2+ signaling in the presence of taxol and suggests a strategy for therapeutic intervention of peripheral neuropathy in cancer patients undergoing taxol treatment.
Introduction
Neuronal Calcium Sensor-1 (NCS-1) is a calcium (Ca2+) binding protein important in intracellular signaling. NCS-1 is composed of four ‘helix-loop-helix’ EF-hand motifs, an ancestral structural EF hand domain (pseudoEF-hand) which has lost the ability to bind Ca2+ and three functional EF hands (EF hand 1, 2 and 3) which bind Ca2+ with varying affinities [1, 2]. Although structurally nearly identical to EF-hands 1,2, and 3, the pseudoEF-hand is not a functional Ca2+-binding site, and is unable to bind Ca2+ due to a lack of acidic amino acids (Asp or Glu) at the +X and −Z positions in the loop, which are required for Ca2+ coordination [1, 2]. Interaction of NCS-1 with downstream proteins is regulated by Ca2+ binding and N-terminal myristolation. Although both Ca2+ binding and myristolation induce conformational changes, myristolation is not required for the interaction between NCS-1 and the inositol 1,4,5-trisphosphate receptor (InsP3R) [3]. The binding of NCS-1 to the InsP3R enhances Ca2+ signaling [3].
Recently, we found that the chemotherapeutic drug paclitaxel (taxol) binds to NCS-1 and addition of taxol to cells further enhances the NCS-1 amplification of InsP3R dependent Ca2+ signaling [4]. We have shown that taxol in nanomolar concentrations induced oscillatory changes in cytosolic Ca2+ in an InsP3R-dependent manner, and increased binding of NCS-1 to the InsP3R, whereas knockdown of NCS-1 abrogated taxol-induced Ca2+ oscillations. In addition, taxol at a concentration between 80 and 800 ng/ml (937 nM) is sufficient to induce Ca2+ oscillations, and taxol binds to NCS-1 with an EC50 of 728 ± 44 ng/ml (557 ± 34 nM), all within the range observed in taxol treated patients (steady-state plasma concentrations in patients treated with taxol are between 85 and 850 ng/ml) [4]. The effects of taxol on Ca2+ signaling are potentially important because it is a drug used to treat a variety of tumor types including ovarian, breast, lung, head, and neck cancers (reviewed by [5]). Although it is clear that taxol exerts its chemotherapeutic effect through its action on microtubule assembly [6], taxol also induces an irreversible peripheral neuropathy in over 30% of treated individuals and the mechanism of this side effect is unclear [7]. Disturbed homeostasis of Ca2+ has been proposed as the cause of the taxol-induced peripheral neuropathy [8, 9]. In mouse models, pretreatment with μ-calpain antagonists abrogated the taxol-induced peripheral neuropathy [10]
In isolated cells, the immediate response to addition of taxol is the appearance of InsP3-mediated Ca2+ oscillations [4]. Exposure of cells to taxol for several hours, which more closely approximates the situation when taxol is used as a chemotherapeutic agent, abolishes InsP3-mediated Ca2+ signaling [11]. The sequence of events appears to be an immediate enhancement of Ca2+ release from intracellular stores which activates μ-calpain. In particular, μ-calpain activity was significantly higher in taxol- (800 ng/ml, 6 hours) than vehicle-treated cells, using concentrations of taxol within the therapeutic range [11]. The degradation of NCS-1 follows the activation of μ-calpain and the subsequent loss of NCS-1 leads to the attenuation of InsP3-mediated Ca2+ signaling [11]. The loss of NCS-1 is believed to result in a negative-feedback loop, leading to the cessation of Ca2+ oscillations and impaired phosphoinositide-mediated Ca2+ signaling [11] Taxol administration to mice also leads to decreased NCS-1 levels [11]. NCS-1 levels can be maintained in cells when inhibitors of μ-calpain are included [11].
In this study we show that specific proteolysis of NCS-1 by μ-calpain can occur in vitro. The location of the cleavage site alters the pseudoEF-hand site and the ability to bind Ca2+ is diminished. From structural considerations this cleavage could induce changes in exposed hydrophobic surface areas that could alter the specificity of protein-protein interactions. In this case, specific cleavage of NCS-1 by μ-calpain appears to create an altered NCS-1 protein with deficient Ca2+ binding properties. The altered Ca2+ binding will attenuate InsP3R-dependent Ca2+ signaling.
Experimental Methods
Overexpression of NCS-1
NCS-1 was produced by overexpressing rat NCS-1 cDNA sub-cloned into pET21-a+ bacterial expression vector (provided by Andreas Jeromin, Baylor College). NCS-1 purification protocol was modified from that described by Zozulya et al. [12]. The NCS-1 vector was transformed into Statagene BL21(DE3) Codon Plus RIL competent E. coli cells. Cells were grown at 37°C in 2L baffled flasks with 1L LB broth and ampicillin (100 μg/mL) and chloramphenicol (35 μg/mL). At an OD595nm of 0.5 to 0.7, overexpression was induced with 0.5mM isopropyl-D-thiogalactoside (IPTG) and cells were shifted to 18°C for ~16 hours. Cells were harvested by centrifugation (3,000 rpm, 30 min, 4°C) and resuspended in 10 mL of 50 mM HEPES, pH 7.5, 100 mM KCl, 1 mM dithiothreitol (DTT), 1 mM MgCl2, and 1 mM CaCl2.
Protein Purification by Hydrophobic Interaction Chromatography
Bacteria expressing recombinant NCS-1 were lysed with lysozyme (Sigma, 2 mg/mL) coupled with 3 freeze-thaw cycles using ethanol-dry ice. Cell lysate was homogenized by tip sonication (Branson Sonicator) for 2 minutes on ice using a 50% duty cycle. Homogenized lysate was clarified by centrifugation at 40,000 × g (20,000 rpm, 1 hr, 4°C) and the resulting supernatant further sonicated for 2 minutes at 50% duty cycle to reduce sample viscosity. Hydrophobic interaction chromatography (HIC) was used to purify NCS-1 as described [13]. Large scale purification of NCS-1 protein was performed using a GE Healthcare HiTrap Phenyl HP high substitution 5mL column. The lysates were loaded on a column equilibrated in 50mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 1 mM MgCl2, and 10 mM CaCl2. Following sample application the column was washed with 10 column volumes of 50 mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 1 mM MgCl2, and 10 mM CaCl2. Recombinant NCS-1 protein was eluted using 50mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 1 mM MgCl2, and 50 mM EDTA in 25 × 1 mL fractions using eppendorf tubes on ice. The protein was eluted with 50 mM EDTA to improve yield. Protein purification was monitored using SDS-PAGE. 1 mL elution fractions were collected (fractions 5 to 7 of 25 mLs) and pooled. The concentration of NCS-1 containing fractions (typically fractions 5 to 7 of 25 mLs) was determined using the Bradford Assay (Bio-Rad). Elution fractions were pooled if above A595nm = 0.200. Yields of 10–15 mg of pure NCS-1 are obtained using the above procedure.
Preparation of Ca2+-Free Protein
Purified recombinant NCS-1 was stripped of Ca2+ using a method modified from Fisher et al., [13]. Briefly, protein was buffer exchanged using a Bio-Rad Econo-Pac 10DG column equilibrated in 50 mM HEPES, 100 mM KCl, at pH 7.5. NCS-1 was then dialyzed against 10 mM EDTA at pH 2.0 in a Pierce Slide-A-Lyzer 7k dialysis cassette for 1 hr, followed by dialysis against Millipore deionized water for 1hr, and then 10 mM TRIS pH 7.4 for 1 hr. Last, the protein was dialyzed against 50mM HEPES, pH 7.5, containing 100 mM KCl, 1 mM DTT. Dialysis was done using all plastic containers. The protein was concentrated (up to 2.14 mg/mL or 100 μM) using a Millipore Ultracel − 10K Amicon Ultra-15 centrifugal filter device.
Digestion of NCS-1 by μ-calpain
We used μ-calpain was purchased from SIGMA (Calpain-1, Active From Human Plasma SIGMA C-6108) and was > 98% by SDS-PAGE according to the manufacturer. NCS-1 in vitro proteolysis trials were also conducted with Calpain-1 from Human Erythrocytes (Calbiochem Cat# 208713) at > 95% purity by SDS-PAGE with similar results (data not shown). The digestion of NCS-1 produced discrete cutting of the protein using 1:5 and 1:10 mass ratio of μ-calpain to NCS-1. The reaction volume was 14.9 μL. The digestion buffer was 10 mM HEPES, 10 mM DTT, 5 mM Ca2+, and 3.5 mM EDTA at pH 7.2 [4]. Reactions contained 10 μL (0.7 mg/mL) NCS-1, 7 μg total, or buffer for controls. The 1:5 reaction contained 1.4 μL of μ-calpain at a concentration of 1 mg/mL. The 1:10 reaction contained 1.4 μL of μ-calpain at a concentration of 0.5 mg/mL. Control reactions included undigested NCS-1 and μ-calpain without NCS-1 incubated with the digestion reactions. Immediately upon addition of μ-calpain the reaction was incubated at 37°C for 10 minutes. The reaction was quenched on ice after the addition of 14.9 μL SDS-PAGE buffer.
SDS-PAGE Gel Electrophoresis, N-terminal Sequencing of NCS-1 Digestion Products
SDS-PAGE of undigested and digested NCS-1 was performed on 15% polyacrylamide. The proteins were visualized using Invitrogen SimplyBlue SafeStain and scanned into digital image format for analysis. 1:5 and 1:10 NCS-1 μ-calpain cleavage reaction products were prepared. Approximately 7μg of digested protein was loaded in each lane. Protein was transferred to PVDF membrane, Immobilon-PSQ, for N-terminal sequencing using a wet western blot transfer apparatus and 15% SDS-PAGE at 250 mA for 1 hr. The PVDF membrane was rinsed in ddH2O and stained with coomassie brilliant blue. The coomassie blue stained bands of interest at 17.5 kDa and 13 kDa were cut out from the membrane and sent for N-terminal sequencing by Edman degradation at the Tufts Core Facility (Tufts Medical School, Boston, MA). The Tufts Core Facility uses an ABI 494 protein sequencer.
MALDI Mass Spectrometry
The NCS-1 digestion samples 1:5 and 1:10 were prepared in 7 μg quantities as described. In order to quench the reaction, 0.1 M HCl final concentration was added to the reaction tubes and samples were flash frozen at 80°C. The samples were dialyzed into 50 mM HCl using a SIGMA micro-Bio-Dialyzer. HCl, a volatile buffer, was chosen in order to maximize the MALDI-TOF signal from the protein sample. The samples were sent to Tufts Core Facility for MALDI-TOF Mass Spectrometry.
Ca2+ Titration by Isothermal Titration Calorimetry
Ca2+ titrations by ITC were done as previously described [14] [15]. Briefly, the Ca2+ titration experiment was run on a Microcal VP-ITC instrument. The protein sample was exchanged into ITC buffer, 50 mM TRIS, 100 mM KCl, and 0.5 mM DTT at pH 7.2 using a Bio-Rad Econo-Pac 10DG column. This exact buffer was used to dissolve Ca2+ to a final concentration used in experiments (2.5 and 5 mM) for ΔN(1–36)NCS-1 and NCS-1 respectively. The NCS-1 concentration in the sample cell was 100 μM. The experiment was run at 20°C with a total of 60 injections of 1.5 μL injection volume (3 second duration) and 240 second intervals. As a control, Ca2+ ligand solution was injected into buffer solution without protein under the same experimental conditions and resulted in a flat isotherm with negligible heat evolution. Data was processed using ORIGIN 7.0 (Microcal) and binding isotherms were fit using either a 2-sets of sites model (NCS-1) or a 3-site sequential binding model (ΔN(1–36)NCS-1). The dissociation constant (Kd), stoichiometry (n), and the enthalpy (ΔH) of binding were determined and the entropy (ΔS) was calculated.
Visualization of ΔN(1–36) NCS-1
Structural coordinates for NCS-1 were obtained from the pdb database (www.pdb.org) and visualized using the program PYMOL (www.pymol.org) [1]. ΔN(1–36)NCS-1 was compared with full length NCS-1 for loss of hydrophobic surface burial and hydrogen bonding.
Results
N-terminal sequencing coupled with MALDI mass spectroscopy show μ-calpain specifically cleaves NCS-1
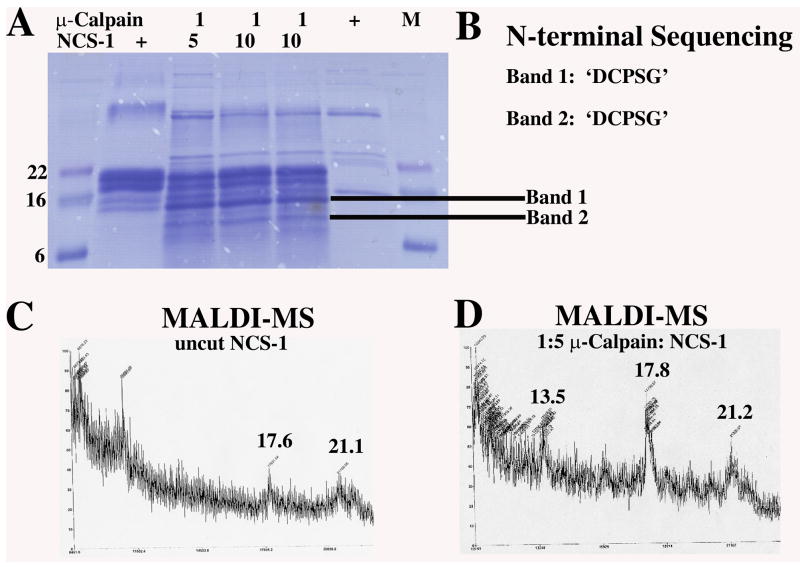
To determine if μ-calpain cleavage of NCS-1 is specific we conducted in vitroμ-calpain NCS-1 proteolysis reactions. Experiments were done using μ-calpain of >98% purity (SIGMA C-6108 from human plasma). Although trace contamination with m-calpain (calpain-2) in the μ-calpain (calpain-1) enzyme stock used is possible, m-calpain requires significantly more Ca2+ for activation than the micromolar amounts used in this study. NCS-1 in vitro proteolysis trials were also conducted with calpain-1 from Human Erythrocytes (Calbiochem Cat# 208713) at > 95% purity by SDS-PAGE with similar results (data not shown). Products were resolved by SDS-PAGE, transferred to a PVDF membrane and specific bands cut out for N-terminal sequencing. Identical NCS-1 reaction products were analyzed by MALDI mass spectroscopy (Figure 1). Uncut NCS-1 showed MALDI-MS peaks with the approximate masses of 17.6 kDa and 21 kDa. μ-calpain cleaved NCS-1 from the 1:5 reaction yielded peaks at ~13.5 kDa, 17.8 kDa, and 21 kDa. The 21 kDa peak is believed to be full length NCS-1. Full length NCS-1 digested at the site determined by N-terminal sequencing would result in a protein fragment of 17,545 Da, in agreement with our MALDI-MS results. The 13.5 kDa fragment may correspond to band 2 in the 1:10 reaction and may result from μ-calpain cleavage of the C-terminally truncated 17.6 kDa NCS-1, identified by MALDI-MS of untreated NCS-1.
Fig. 1. Cleavage of NCS-1 by μ-calpain in vitro.
A. Cleavage reactions of NCS-1 by μ-calpain resolved by SDS-PAGE. Digestion reactions were performed using 1:5 or 1:10 mass ratios of μ-calpain to NCS-1. Bands cut out for N-terminal sequencing are indicated. B. Results from 5 cycles of N-terminal sequencing of NCS-1 μ-calpain digestion products for Band 1 and Band 2. C and D. MALDI mass spectroscopy from samples of purified recombinant full length NCS-1 (panel C) and μ-calpain treated NCS-1 digestion products (1:5 mass ratio) (panel D). The ~21 kDa peak corresponds to uncut full length NCS-1 and the 17.8 kDa and 13.5 kDa peaks correspond to μ-calpain cleaved NCS-1. The 17.6 kDa peak in untreated NCS-1 corresponds to a C-terminally degraded NCS-1 product obtained during purification.
Two bands were cut from the SDS-PAGE gel and sent for N-terminal sequencing. From the 1:5 digestion a ~15 kDa band was chosen (Band 1) and from the 1:10 digestion a ~13 kDa band was cut (Band 2). Five cycles of N-terminal sequencing yielded frequencies for amino acids for each position at the cleavage site. Both bands yielded the same N-terminal sequencing results (Figure 1), identifying the N-terminus of μ-calpain cleaved NCS-1 as ‘DCPSG’. The five amino acid sequence was located within the N-terminus of NCS-1. Since both bands were found to contain the same N-terminal sequence it seems likely that cleavage at Lys36 of NCS-1 is specific. This seems to agree with the reported specificity of μ-calpain which prefers Leu, Val or Ile at the P2 position and Lys, Tyr, Arg, or Met at the P1 position [16, 17]. It is possible that additional faster time scale cleavage events occur in vitro undetectable using the methods described here, although under prolonged incubation (~1 hr) at 37°C we did not detect further degradation products. In vivo we were unable to detect intermediate NCS-1 proteolysis products and cleavage of NCS-1 by μ-calpain appears to destabilize the protein leading to complete degradation through the ubiquitin proteolytic pathway [11] [16].
Based on our N-terminal sequencing results, MALDI mass spectroscopy and the three-dimensional structure of human NCS-1, μ-calpain cleaves NCS-1 within the pseudoEF-hand motif after residue K36.
ΔN(1–36)NCS-1 displays a different elution profile than full length NCS-1 by Hydrophobic Interaction Chromatography
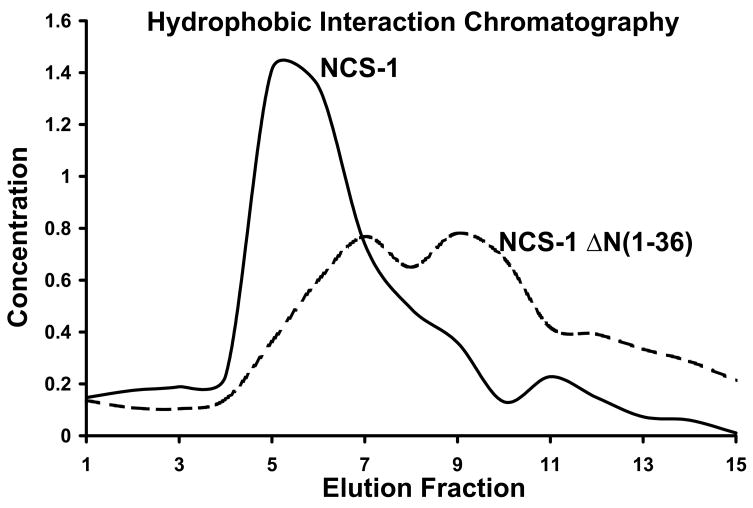
To test the effect of μ-calpain cleavage on NCS-1 structure and Ca2+ binding properties, we subcloned the μ-calpain NCS-1 cleavage product, ΔN(1–36)NCS-1, for recombinant expression and purification. Full length NCS-1 and ΔN(1–36)NCS-1 were subjected to Hydrophobic Interaction Chromatography (HIC) to determine conformational changes and differential exposure of hydrophobic surfaces. Proteins have varying levels of exposed hydrophobic patches on their surfaces depending on their three-dimensional conformation and HIC uses these patches by determining the differential absorption of proteins to resin substituted with hydrophobic groups such as phenyl [18]. The production of recombinant NCS-1 was optimized by modifying the purification procedure of Jeromin et al. [15] and utilizing a high capacity phenyl HIC column for purification, yielding approximately 5 mL of 100μM NCS-1 per 3 Liter E. coli growth. The Ca2+ bound form of NCS-1 binds tightly to phenyl substituted resin and elutes upon stripping of Ca2+ using the metal chelator EDTA [15], suggesting a Ca2+ dependent conformational change which results in burial of hydrophobic surface area. Full length NCS-1 elutes with a sharp Gaussian profile in the 1st and 2nd column volume upon addition of EDTA to the HIC column, whereas μ-calpain cleaved ΔN(1–36)NCS-1 elutes later with a much broader profile indicative of both a change in the exposed hydrophobic surface area and multiple conformational states (Figure 2).
Fig. 2. HIC chromatograms of NCS-1 and ΔNCS-1(1–36) show changes in exposed hydrophobic surface area upon μ-calpain cleavage.
NCS-1 (solid trace) and ΔNCS-1(1–36) (dashed trace) were purified by HIC and resulting elution fractions were quantified using Bradford protein assay. NCS-1 elutes as a sharp peak in the presence of EDTA whereas ΔNCS-1(1–36) displays a much broader elution profile indicative of multiple conformational states or changes in exposed hydrophobic surface area.
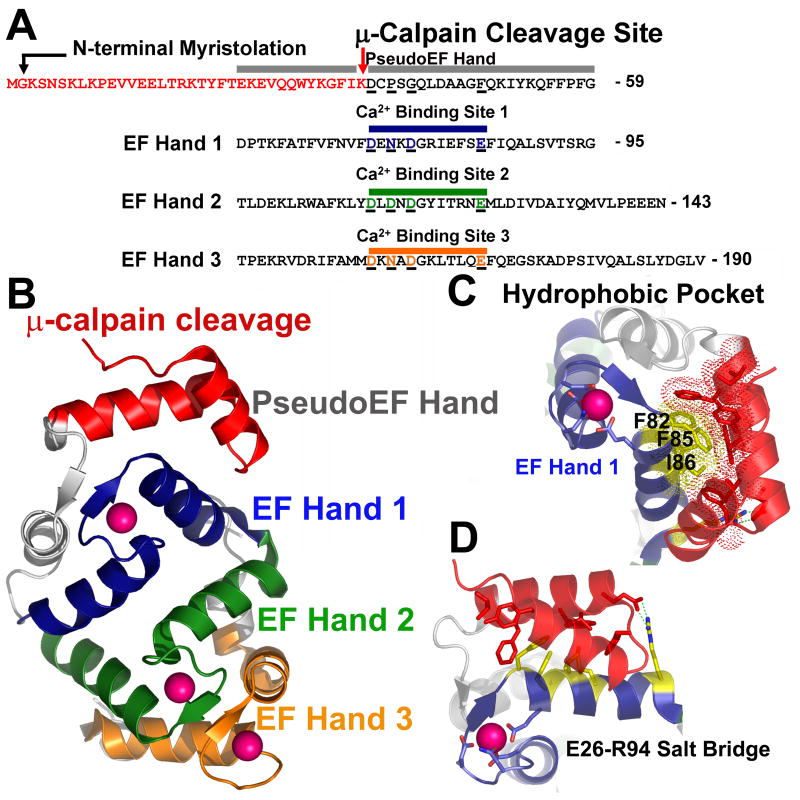
Examination of the crystal structure of Ca2+ bound human NCS-1 shows μ-calpain cleavage occurs within the loop of the helix-loop-helix pseudoEF-hand domain. Although this pseudoEF-hand domain is not expected to bind Ca2+, it contributes directly to the stability of the EF hand 1 where it forms one half of a four helix bundle and serves to bury significant hydrophobic surface area (Figure 4). Destabilization of EF hand 1 domain through cleavage of the pseudoEF-hand can be further expected to cause conformational changes in EF hand 2 and EF hand 3 which pack tightly against EF hand 1, and thus result in altered affinity of NCS-1 for Ca2+.
Fig. 4. μ-calpain cleavage of NCS-1 directly impacts Ca2+ binding through destabilization of EF-hand 1, exposure of hydrophobic surface, and possible disruption of the overall NCS-1 fold.
A. Schematic of the sequence of NCS-1. Colored red are residues removed by μ-calpain cleavage. Residues which encompass the pseudoEF hand, EF hand 1, EF hand 2, and EF hand 3, along with corresponding Ca2+ binding sites are aligned and color coded. The non-functional pseudoEF hand is colored grey. The Ca2+ binding loop of EF hand 1 is colored blue, EF hand 2 green and EF hand 3 orange. Acidic residues coordinating Ca2+ in each of these EF hand domains are underlined and colored accordingly. Corresponding residues in the pseudo-EF hand which do not bind Ca2+ are also aligned and underlined for comparison. B. Overall structure of NCS-1. Residues removed by μ-calpain within the pseudoEF hand are colored red. The pseudoEF hand is labeled grey. EF hand 1 is colored blue, EF hand 2 is colored green, and EF hand 3 is colored orange. Bound Ca2+ ions are depicted as pink spheres. C. Hydrophobic pocket centered on F82, F85 and I86 in NCS-1 (colored yellow and shown as sticks with dots indicating van der Waals contacts. Residues forming van der Waals contacts with F82, F85 and I86 which would be removed by μ-calpain cleavage are shown as red sticks. D. Salt bridge formed between R94 of EF hand 1 and E26 of the pseudoEF-hand would be disrupted by μ-calpain cleavage possibly destabilizing the conformation of EF hand 1.
μ-calpain cleavage significantly lowers the affinity of NCS-1 for Ca2+
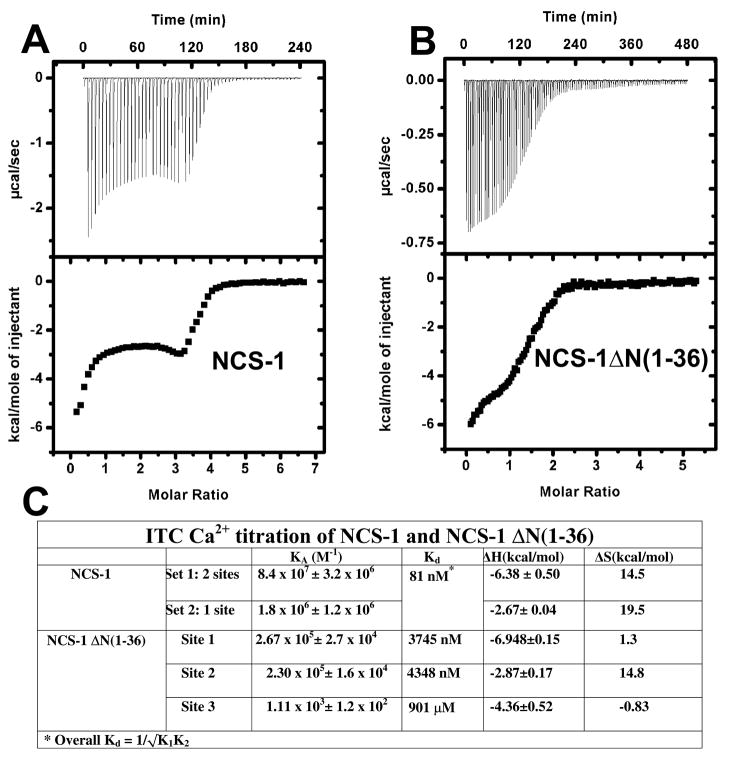
In order to test the hypothesis that μ-calpain cleavage alters the affinity of NCS-1 for Ca2+ we directly measured the Ca2+ binding properties of ΔN(1–36)NCS-1 and full length NCS-1 using isothermal titration calorimetry (ITC) (Figure 3). NCS-1 was stripped of Ca2+ using a modification of previously published protocols, [13, 15] and titrated with Ca2+ by ITC to measure the baseline affinity of full length NCS-1 for Ca2+. The resulting isotherm was best fit with a ‘two sets of sites model’ in agreement with previously published ITC studies on NCS-1 which showed that EF hand 1 binds Ca2+ independently of EF hand 2 and EF hand 3 [15]. An overall Kd of 81 nM was determined (Figure 3 Panel A) which compares well with previous results [15].
Fig. 3. Isothermal titration calorimetry of NCS-1 and ΔNCS-1(1–36) shows μ-calpain cleavage disrupts Ca2+ binding in NCS-1.
A. Full length NCS-1 was titrated with Ca2+ and the resulting isotherm fitted with a 2-site binding model. B. ΔNCS-1(1–36) was titrated with Ca2+ and the resulting isotherm fitted with a 3-sets of sites binding model using the program ORIGIN. C. ITC fitting results and derived thermodynamic parameters for NCS-1 and ΔNCS-1(1–36).
In order to determine the effect of μ-calpain cleavage on NCS-1 Ca2+ binding we purified ΔN(1–36) NCS-1 in parallel with full length NCS-1 and subjected it to the identical ITC protocol. ΔN(1–36) NCS-1 displays a significantly different binding isotherm than full length NCS-1, and could only be fit using a three site sequential binding model (Figure 3 Panel B). Site 1 and Site 2 bind Ca2+ with a Kd of ~ 4 μM, orders of magnitude weaker than full length NCS-1, whereas Site 3 binds Ca2+ with very low affinity (Kd~900 μM) (Figure 3 Panel C). It is not possible from ITC data to determine directly the location of each site within the structure of NCS-1 but given the location of the μ-calpain cleavage site, which is directly in contact with EF hand 1, it is likely that site 3 corresponds to EF hand 1. The fact that the ΔN(1–36) NCS-1 binding isotherm can be best fit with a sequential site binding model also strongly suggests that removal of the N-terminal 36 residues disrupts the global fold of NCS-1, resulting in multiple partially folded conformational states.
Discussion
Examination of the structure of NCS-1 suggests that μ-calpain cleavage would affect the global conformation and Ca2+ binding properties of NCS-1
In order to further determine the molecular basis of the reduced affinity for Ca2+ and apparent changes in exposed hydrophobic surface area for ΔN(1–36) NCS-1 we examined the structure of human NCS-1. The coordinates for the high resolution crystal structure of human NCS-1 were downloaded from the PDB database ([19], www.pdb.org, PDB ID: 1G8I). NCS-1 was visualized using the program PYMOL (www.pymol.org). ΔN(1–36) NCS-1 was visualized by removing the N-terminal 36 residues from the pdb file. The overall structure of NCS-1 contains four EF hand structural motifs in series which occur in pairs, pseudoEF-hand and EF hand 1 make up the first pair and EF hand 2 and EF hand 3 make up the second pair [1]. Each EF hand pair is organized around the Ca2+ binding sites comprising 2 symmetrical domains. Two molecules of Ca2+ are bound by EF hand 2 and EF hand 3, whereas only 1 molecule of Ca2+ is bound by EF hand 1 (Figure 4B). Examination of the Ca2+ binding loops of EF hands 1, 2 and 3 in comparison with the analogous loop in the pseudoEF-hand shows a lack of glutamate or aspartate residues in the pseudoEF-hand, which are necessary for Ca2+ binding (Figure 4 Panel A). μ-calpain cleavage occurs N-terminal to the sequence ‘DCPSG’ at the beginning of the loop in the pseudoEF-hand domain (Figure 4 Panel A). In full length NCS-1, the pseudoEF-hand domain stabilizes EF hand 1 through several tight hydrophobic interactions. In particular, phenylalanine 82 and 85 are buried in a tight hydrophobic pocket formed by the first helix of the helix-loop helix pseudoEF-hand domain. EF hand 1 is further stabilized by a salt bridge between glutamate 26 of the pseudoEF-hand domain and arginine 96 of the first helix of EF hand 1 (Figure 4 Panels C and D). μ-calpain cleavage of NCS-1 at K36 would remove the first helix of the pseudoEF-hand domain eliminating the salt bridge and exposing a large hydrophobic surface on EF hand 1, possibly explaining the increase in overall hydrophobicity observed for ΔN(1–36) NCS-1 during hydrophobic interaction chromatography. A change in exposed hydrophobic surface area in the pseudo-EF hand domain could destabilize EF hand 1, reducing its affinity for Ca2+. Loss of the E26-R96 salt bridge might destabilize the Ca2+ binding loop of EF hand 1 further affecting Ca2+ binding. In support of this full length NCS-1 seems to bind Ca2+ cooperatively while in ΔN(1–36) NCS-1 there are three independent Ca2+ binding sites.
Because EF hand 1 is involved in stabilizing the global fold of EF hands 2 and 3, the effect of μ-calpain cleavage of the pseudoEF-hand could propagate throughout NCS-1 protein possibly explaining both the observed reduction in overall affinity for Ca2+ in ΔN(1–36) NCS-1 and the change in binding mechanism from a two site mode of binding to a sequential binding mode.
Conclusion
In this study we identified the location of μ-calpain cleavage of NCS-1 and the functional consequences of this cleavage on Ca2+ binding. Remedies or preventative strategies for taxol induced peripheral neuropathy are needed. Agents that inhibit μ-calpain have limitations for this purpose because μ-calpains are expressed in most cell types. In addition, the development of inhibitors specific to only μ-calpain may be very difficult because calpain active sites are structurally very similar. However, an inhibitor based on the peptide sequence of NCS-1 which is cleaved by μ-calpain (and possibly m-calpain as well) might circumvent both of the above mentioned problems. The present results can be used to develop molecular peptidomimetics of the cleavage site [20]. This peptidomimetic drug based on the sequence of NCS-1 recognized by μ-calpain could compete with native NCS-1 as a μ-calpain substrate and possibly protect native NCS-1 from degradation. Pretreatment with a mimetic of the NCS-1 cleavage site should be able to specifically inhibit taxol-induced degradation of NCS-1 [21]. Overall, an in depth understanding of the molecular basis of μ-calpain dependent NCS-1 cleavage and the effect on Ca2+ binding would allow design of specific peptidomimetic μ-calpain inhibitors for use as a novel treatment strategy for taxol induced peripheral neuropathy.
Abbreviations used are
- NCS-1
neuronal calcium sensor 1
- ITC
isothermal titration calorimetry
- Ca2+
calcium
- MALDI-MS
matrix assisted laser desorption/ionization mass spectroscopy
Footnotes
We wish to thank Ewa Folta-Stogniew of the W.M. Keck facility at Yale University for help conducting the ITC experiments. Michael Berne and Jon P. DeGnore of Tufts University Medical School performed N-terminal sequencing and MALDI-MS. BEE is funded by P50 DK57328 and DK57751. ETP is supported by a NIH/National Cancer Institute T32 training grant (5T32CA009085).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bourne Y, et al. Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1) J Biol Chem. 2001;276(15):11949–55. doi: 10.1074/jbc.M009373200. [DOI] [PubMed] [Google Scholar]
- 2.Aravind P, et al. Regulatory and structural EF-hand motifs of neuronal calcium sensor-1: Mg 2+ modulates Ca 2+ binding, Ca 2+-induced conformational changes, and equilibrium unfolding transitions. J Mol Biol. 2008;376(4):1100–15. doi: 10.1016/j.jmb.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 3.Schlecker C, et al. Neuronal calcium sensor-1 enhancement of InsP3 receptor activity is inhibited by therapeutic levels of lithium. J Clin Invest. 2006;116(6):1668–74. doi: 10.1172/JCI22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehmerle W, et al. Paclitaxel induces calcium oscillations via an inositol 1,4,5-trisphosphate receptor and neuronal calcium sensor 1-dependent mechanism. Proc Natl Acad Sci U S A. 2006;103(48):18356–61. doi: 10.1073/pnas.0607240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mekhail TM, Markman M. Paclitaxel in cancer therapy. Expert Opin Pharmacother. 2002;3(6):755–66. doi: 10.1517/14656566.3.6.755. [DOI] [PubMed] [Google Scholar]
- 6.Jordan MA, Kamath K. How do microtubule-targeted drugs work? An overview. Curr Cancer Drug Targets. 2007;7(8):730–42. doi: 10.2174/156800907783220417. [DOI] [PubMed] [Google Scholar]
- 7.Scripture CD, Figg WD, Sparreboom A. Peripheral neuropathy induced by Paclitaxel: recent insights and future perspectives. Curr Neuropharmacol. 2006;4(2):165–72. doi: 10.2174/157015906776359568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siau C, Bennett GJ. Dysregulation of cellular calcium homeostasis in chemotherapy-evoked painful peripheral neuropathy. Anesth Analg. 2006;102(5):1485–90. doi: 10.1213/01.ane.0000204318.35194.ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao PC, et al. Involvement of endoplasmic reticulum in paclitaxel-induced apoptosis. J Cell Biochem. 2008;104(4):1509–23. doi: 10.1002/jcb.21730. [DOI] [PubMed] [Google Scholar]
- 10.Wang MS, et al. Calpain inhibition protects against Taxol-induced sensory neuropathy. Brain. 2004;127(Pt 3):671–9. doi: 10.1093/brain/awh078. [DOI] [PubMed] [Google Scholar]
- 11.Boehmerle W, et al. Chronic exposure to paclitaxel diminishes phosphoinositide signaling by calpain-mediated neuronal calcium sensor-1 degradation. Proc Natl Acad Sci U S A. 2007;104(26):11103–8. doi: 10.1073/pnas.0701546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zozulya S, Ladant D, Stryer L. Expression and characterization of calcium-myristoyl switch proteins. Methods Enzymol. 1995;250:383–93. doi: 10.1016/0076-6879(95)50086-3. [DOI] [PubMed] [Google Scholar]
- 13.Fisher JR, et al. Purification of myristoylated and nonmyristoylated neuronal calcium sensor-1 using single-step hydrophobic interaction chromatography. Protein Expr Purif. 2000;20(1):66–72. doi: 10.1006/prep.2000.1298. [DOI] [PubMed] [Google Scholar]
- 14.Celic A, et al. Domain mapping of the polycystin-2 C-terminal tail using de novo molecular modeling and biophysical analysis. J Biol Chem. 2008;283(42):28305–12. doi: 10.1074/jbc.M802743200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeromin A, et al. N-terminal myristoylation regulates calcium-induced conformational changes in neuronal calcium sensor-1. J Biol Chem. 2004;279(26):27158–67. doi: 10.1074/jbc.M312172200. [DOI] [PubMed] [Google Scholar]
- 16.Wang KK. Calpain and caspase: can you tell the difference? Trends Neurosci. 2000;23(1):20–6. doi: 10.1016/s0166-2236(99)01479-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang KK, Villalobo A, Roufogalis BD. Calmodulin-binding proteins as calpain substrates. Biochem J. 1989;262(3):693–706. doi: 10.1042/bj2620693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Farrell PA. Hydrophobic interaction chromatography. Methods Mol Biol. 2004;244:133–8. doi: 10.1385/1-59259-655-x:133. [DOI] [PubMed] [Google Scholar]
- 19.Sussman JL, et al. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 6 Pt 1):1078–84. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 20.Fear G, Komarnytsky S, Raskin I. Protease inhibitors and their peptidomimetic derivatives as potential drugs. Pharmacol Ther. 2007;113(2):354–68. doi: 10.1016/j.pharmthera.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donkor IO, Korukonda R. Synthesis and calpain inhibitory activity of peptidomimetic compounds with constrained amino acids at the P2 position. Bioorg Med Chem Lett. 2008;18(17):4806–8. doi: 10.1016/j.bmcl.2008.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]