Abstract
Background
Selenoproteins contain the twenty-first amino acid, selenocysteine, and are involved in cellular defenses against oxidative damage, important metabolic and developmental pathways, and responses to environmental challenges. Elucidating the mechanisms regulating selenoprotein expression at the transcriptional level is key to understanding how these mechanisms are called into play to respond to the changing environment.
Methods
This review summarizes published studies on transcriptional regulation of selenoprotein genes, focused primarily on genes whose encoded protein functions are at least partially understood. This is followed by in silico analysis of predicted regulatory elements in selenoprotein genes, including those in the aforementioned category as well as the genes whose functions are not known.
Results
Our findings reveal regulatory pathways common to many selenoprotein genes, including several involved in stress-responses. In addition, tissue-specific regulatory factors are implicated in regulating many selenoprotein genes.
Conclusions
These studies provide new insights into how selenoprotein genes respond to environmental and other challenges, and the roles these proteins play in allowing cells to adapt to these changes.
General Significance
Elucidating the regulatory mechanisms affecting selenoprotein expression is essential for understanding their roles in human diseases, and for developing diagnostic and potential therapeutic approaches to address dysregulation of members of this gene family.
Keywords: selenoprotein, selenium, transcription, oxidative stress
Introduction
Twenty-five selenoprotein genes encoding the amino acid, selenocysteine, have been identified in the human genome. Many are involved in oxidative stress protection or in maintaining cellular redox balance. Their promoter activities are of particular interest as many are differentially expressed in specific tissues and developmental stages, and in response to various environmental stimuli. In addition, reduced activity of selenoproteins can result in a compensatory increase of non-selenium dependent antioxidants to counteract the damaging effect of oxidative stress. These observations suggest that selenoproteins participate in multiple molecular pathways and that their expression may be tightly associated with complex regulatory networks and signaling. Understanding regulatory networks at the transcriptional level will provide insights into cellular roles of selenoproteins and their connections with other protective antioxidant pathways. The goal of this review is to provide an overview of the current state of knowledge of transcriptional regulation and putative regulatory pathways for selenoprotein genes. The review is divided into two major components. First, we provide an overview of published studies on transcriptional regulation of selenoprotein expression, based on classical experimental approaches and focusing primarily on the human and rodent genes. The second component is based on our in silico analysis of the genomic sequences in the promoter regions of the 25 human selenoprotein genes. The availability of this information lays the groundwork for future experimental investigation to validate predictions made from bioinformatics approaches.
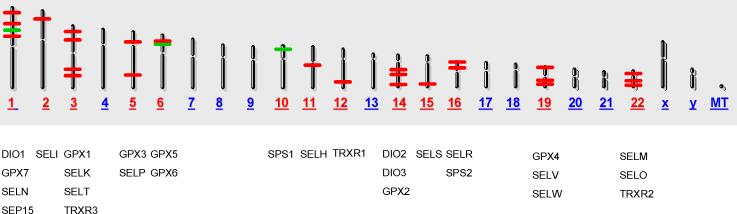
Table 1 lists the protein names, gene names and synonyms, chromosome location, gene IDs and RefSec numbers of the twenty-five known human selenoprotein genes. The same information is given in Table S1 for cysteine orthologs of human selenoproteins, and for known factors involved in selenoprotein biosynthesis. The chromosome locations of the human selenoprotein genes and cysteine orthologs are also depicted in Fig. 1. Of note, close proximity is seen for some closely related selenoprotein genes and orthologs, including two glutathione peroxidases, GPX5 and GPX6, driven by divergent promoters on chromosome 6, two iodothyronine deiodinases, DIO2 and DIO3, on chromosome 14, and the related selenoproteins, SELV and SELW, on chromosome 19. This proximity is consistent with the related genes arising by gene duplication and divergence. Table 2 provides an overview of gene regulatory elements identified and investigated experimentally for four glutathione peroxidases, the three iodothyronine deiodinases, and five additional selenoproteins. Experimental evidence for most of these reported regulatory elements is provided below.
Table 1.
Gene identifiers and chromosome location of human selenoprotein genes.
| Gene name | Gene ID | Chromosome location | Gene Symbol | Synonyms | RefSec |
|---|---|---|---|---|---|
| glutathione peroxidase 1 | 2876 | 3p21.3 | GPX1 | GSHPX1, MGC14399, MGC88245 | NM_000581 |
| glutathione peroxidase 2 (gastrointestinal) | 2877 | 14q24.1 | GPX2 | GI-GPx, GSHPX-GI | NM_002083 |
| glutathione peroxidase 3 (plasma) | 2878 | 5q23 | GPX3 | GPx-P, GSHPx-3, GSHPx-P | NM_002084 |
| glutathione peroxidase 4 (phospholipid hydroperoxidase) | 2879 | 19p13.3 | GPX4 | MCSP, PHGPx, snGPx, snPHGPx | NM_002085 |
| glutathione peroxidase 6 (olfactory) | 257702 | 6p22.1 | GPX6 | CL683, FLJ14777, NPGPx | NM_182701 |
| iodothyroninedeiodinase type I | 1733 | 1p33-p32 | DIO1 | 5DI, DIO1, MGC130050, MGC130051, TXDI1 | NM_000792 |
| iodothyroninedeiodinase type II | 1734 | 14q24.2-q24.3 | DIO2 | 5DII, D2, SelY, TXDI2 | NM_013989 |
| iodothyroninedeiodinase type III | 1735 | 14q32 | DIO3 | 5DIII, D3, DIOIII, TXDI3 | NM_001362 |
| selenophosphate synthetase 2 | 22928 | 16p11.1 | SEPHS2 | SPS2 | NM_012248 |
| 15 kDa selenoprotein | 9403 | 1p31 | SEP15 | Sep15 | NM_004261 |
| selenoprotein H | 280636 | 11q12.1 | SELH | C11orf31 | NM_170746 |
| selenoprotein I | 85465 | 2p23.3 | SELI | KIAA1724 | NM_033505 |
| selenoprotein K | 58515 | 3p21.31 | SELK | HSPC030, HSPC297, MGC17057 | NM_021237 |
| selenoprotein M | 140606 | 22q12.2 | SELM | MGC40146, SEPM | NM_080430 |
| selenoprotein N | 57190 | 1p36.13 | SEPN1 | RSS, SELN, MDRS1, RSMD1, FLJ24021 | NM_020451 |
| selenoprotein O | 83642 | 22q13.33 | SELO | SELO | NM_031454 |
| selenoprotein P | 6414 | 5q31 | SEPP1 | SELP, SeP | ENST00000361970 |
| selenoprotein R | 51734 | 16p13.3 | SEPX1 | SELR, SELX, SEPX, MsrB | NM_016332 |
| selenoprotein S | 55829 | 15q26.3 | SELS | AD-015, MGC2553, SBBI8, VIMP | NM_203472 |
| selenoprotein T | 51714 | 3q25.1 | SELT | SeIT | NM_016275 |
| selenoprotein V | 348303 | 19q13.2 | SELV | SeIV | NM_182704 |
| selenoprotein W | 6415 | 19q13.3 | SEPW | SEPW1 | NM_003009 |
| thioredoxin reductase 1 | 7296 | 12q23-q24.1 | TRXR1 | MGC9145, TR1, TRXR1, TXNR, TXNRD1 | NM_182742 |
| thioredoxin reductase 2 | 10587 | 22q11.21 | TRXR2 | SELZ, TR-BETA, TRXR2,TXRND2 (formerly TR3) | NM_006440 ENST00000361682 |
| thioredoxin reductase 3 | 114112 | 3q21.3 | TRXR3 | TRXR3, TXNRD3, TGR, (formerly TR2) | XM_001130163 |
Figure 1. Human selenoprotein family gene map.
Red bars depict the positions of selenoprotein encoding genes and green bars depict cysteine ortholog encoding genes. Red numbers indicate selenoprotein or cysteine ortholog genes present, blue numbers indicate their absence.
Table 2.
Experimentally validated gene regulation of selenoproteins by transcription factors.
| Protein name | TFs involved in regulation of the selenoprotein gene expression | PubMed ID |
|---|---|---|
| glutathione peroxidase 1 | p53, p63 | 12893824, 15059885, 9792801 |
| glutathione peroxidase 2 (gastrointestinal) | Nrf2, beta-catenin/TCF, p63, Nkx3.1 | 18787804, 18559366, 17937616, 17210444, 16794261, 16446369, 16061659, 15923610, 17081103 |
| glutathione peroxidase 3 (plasma) | Nkx3.1, Sp1, HIF-1 | 16061659, 15096516 |
| glutathione peroxidase 4 (phospholipid hydroperoxidase) | C/EBPepsilon, NF-kappaB, EGR, SREB, Sp1/Sp3, NF-Y, Smad | 15225122, 12427732, 17688422, 17081103, 16223606, 12888488 |
| iodothyronine deiodinase, type I | HNF4alpha, KLF9, GATA4 , SP1, RXR | 18426912, 9492050, 18641053 |
| iodothyronine deiodinase, type II | TTF-1, Nkx-2.5, GATA,NF-kappaB, CREB, RXR, EGF | 11145743, 16728495, 12775767, 10803591, 15291742, 10614643, 18641053, 17991726 |
| iodothyronine deiodinase, type III | HIF-1alpha, Shh through Gli2, Smad2, Smad3, Smad4 | 18259611, 17720805, 16037131 |
| selenoprotein P | HNF4 alpha, HNF3beta, BRN-2, FoxO1a | 18163890, 17986386, 9687017 |
| selenoprotein S | NF-kappaB | 16574427 |
| selenoprotein W | MTF-1, Sp1 | 16221973, 15337603 |
| methionine sulfoxide reductase B1 | Sp1 | 17519015 |
| thioredoxin reductase 1 | Nrf2, Oct-1, Sp1, and Sp3 | 18445702, 18215477, 17942419, 16219762, 15521073, 11375392, 14980055 |
Glutathione peroxidases
Analysis of mammalian selenoproteomes identified five selenium-containing glutathione peroxidases (GPXs): cytosolic or classical GPX (GPX1), gastrointestinal GPX (GPX2), plasma or extracellular GPX (GPX3), phospholipid hydroperoxide GPX (GPX4), and in humans, an olfactory system GPX (GPX6). Two cysteine containing GPXs, GPX5 and GPX7, have also been identified. GPXs reduce hydroperoxides to the corresponding alcohols, using glutathione (GSH) as cofactor. Cytosolic GPX clearly functions as an antioxidant, as convincingly demonstrated by the sensitivity of knockout mice to hydroperoxide challenge. Gastrointestinal GPX, found primarily in the epithelial lining of the gastrointestinal tract, was originally suggested to act as a barrier against intestinal hydroperoxide absorption. Plasma GPX is expressed at highest levels in kidney, and is directed to extracellular compartments and tissues in contact with body fluids, e.g., kidney, ciliary body, and maternal/fetal interfaces. It is thought to be an efficient extracellular antioxidant. Phospholipid hydroperoxide GPX is highly abundant in testes, and is indispensable for sperm maturation and embryogenesis. Thus, each of the GPXs appear to have distinct roles, particularly in cellular defense mechanisms [1].
Cytosolic glutathione peroxidase
(GPX1, cGPX) was, for more than a decade the only known mammalian selenoprotein, resulting in many of the properties of selenium being attributed to this enzyme. Despite this distinction, transcriptional regulation of this isoform is not well understood. The promoter was reported to contain an oxygen responsive element and some cell types upregulate GPX1 in response to hyperoxia. An Oxygen Responsive Element Binding Protein (OREBP) was purified and characterized. In addition to binding the GPX1 oxygen responsive element, OREBP was itself regulated by oxygen tension [2]. GPX1 is also upregulated by estrogens, but a typical estrogen-responsive element has not been detected. Estrogen responsiveness is thought to be the result of estradiol-mediated activation of nuclear factor kappa B (NFkB), and its consequent activation of GPX1, but the latter has not been experimentally validated [3]. Several groups have identified GPX1 as a direct target of p53 under different stress conditions such us hypoxia, DNA damage induced by ionizing radiation or adriamycin, or inhibition of topoisomerase II by etoposide [4-6]. The p53 binding site was mapped to the promoter [4, 5] and expression of endogenous GPX1 was significantly induced at both mRNA and enzyme activity levels by etoposide in U2-OS human osteosarcoma cells but not in p53-negative Saos-2 osteosarcoma cells. Furthermore, upon etoposide activation of p53, transactivation of the GPX1 promoter was blocked by a dominant negative p53 mutant [2]. The p53 enrichment at the promoter of the GPX1 was observed following DNA induced damage and hypoxia, although there was little effect of hypoxia treatment on the expression of GPX1. These results are consistent with previous publications showing that p53 preferentially interacts with the p300 coactivator during DNA damage but not during hypoxic stress [7, 8]. GPX1 mRNA levels were induced approximately four fold after 24 h of adriamycin treatment comparing the p53+/+ and p53-/- HCT116 colonic epithelial cells [6]. Thus p53 induction of GPX1 may partially protect cells from oxidative damage.
Gastrointestinal glutathione peroxidase
(GPX2, GI-GPX) while proposed as a barrier against intestinal hydroperoxide absorption has also been implicated in the control of inflammation and malignant growth. A putative antioxidant/electrophile response element (ARE/EpRE) that is highly conserved between mouse, rat, and human was identified in the promoter of GPX2 and demonstrated to be a target of the nuclear factor E2-related factor 2/Kelch-like ECH associating protein 1 (Nrf2/Keap1) system. The transcription factor, Nrf2 controls the expression of a number of protective genes in response to oxidative stress. In CaCo-2 human epithelial colorectal adenocarcinoma cells, GPX2 reporter gene constructs were induced by t-butyl hydroquinone, sulforaphane, and curcumin, antioxidants known to activate AREs via electrophilic thiol modification of Keap1, the cytosolic inhibitor of Nrf2 [9]. Overexpression of Nrf2 also activated the reporter constructs, and these effects were reversed by mutation of the ARE in the promoter and by overexpressed Keap1. Binding of Nrf2 to the ARE sequence in the authentic GPX2 gene was confirmed by chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSA). Thyme extract and quercetin, a plant derived flavenoid, also significantly activated the GPX2 promoter, effects that depended on a functional ARE [10], whereas resveratrol did not activate the isolated EpRE but enhanced the GI-GPX promoter activity. Thus, the presumed natural antioxidants sulforaphane and curcumin as well as dietary polyphenols predicted to be promising chemopreventive agents may exert their anti-inflammatory and anticarcinogenic effects not only by previously reported induction of phase 2 enzymes, but also by the up-regulation of the selenoprotein GPX2.
GPX2 is a direct target of the Est family transcription factor, PU.1, in neutrophil-like cells. Studies on PU.1 knockout mice have shown that it is required for the development of macrophages, B cells, neutrophils, and dendritic cells [11-13] but also regulates the expression of several components of the NADPH oxidase complex [3], used by these cells to fight invading pathogens. Two-fold induction in GPX2 expression was observed in the 503PU cell line re-expressing PU.1 compared with the parent IL-3-dependent 503 myeloid cell line, which was derived from the neonatal liver of PU.1 knockout mice. Northern blot analysis, EMSA, site directed mutagenesis and transfection studies confirmed that the two PU.1 binding sites located in the promoter and in the 3'UTR of the GPX2 locus are responsible for this upregulation, leading to speculation that a synergistic effect of the two sites in PU.1 regulation would result in higher GPX2 expression levels and protect neutrophils from reactive oxygen species [3]. These findings add GPX2 to the list of phase II enzymes, which are part of the adaptive response.
A study of pathology of smoking-related pulmonary emphysema in patients revealed that Nrf2 protein and GPX2 expression levels were significantly decreased in emphysema patients. The decrease in GPX2 was inversely correlated with airway obstruction and distension indexes, and increased macrophage expression of the lipid peroxidation product 4-hydroxy-2-nonenal [15]. In w.t. mice, cigarette smoke significantly induced expression of GPX2 in lungs at both the mRNA and protein level. Disruption of Nrf2 shortened the onset and increased the severity of emphysema in response to chronic cigarette smoke, and the lungs of these mice exhibited higher oxidative stress, as evident from increased levels of lipid peroxidation and oxidative DNA damage [16]. Nrf2 small interfering RNA (siRNA) decreased the expression of GPX2 in lung epithelial cells, whereas activation of Nrf2 by specific knockdown of Keap1 upregulated the expression of GPX2. This study showed GPX2 to be the major oxidative stress-inducible cellular GPX isoform in the lungs, and that it's basal as well as inducible expression is dependent on Nrf2.
The GPX2 promoter contains five putative beta-catenin/TCF binding sites. Accordingly, the promoter was active in two cell lines with a constitutively active Wnt pathway [4]. Promoter studies in cell lines with either constitutively active or silent Wnt signaling identified one site as sufficient for activation, and mutation of the site reduced GPX2 promoter activity by 50%. Overexpression of wild-type adenomatous polyposis coli (APC) tumor suppressor protein in cells harboring a mutated APC gene decreased basal GPX2 promoter activity. GPX2 is transiently up-regulated during development of gastrointestinal adenocarcinomas. Given that GPX2 is highly expressed in the proliferative area of the intestinal crypt-to-villus axis this finding suggests a function of GPX2 in the maintenance of normal renewal of the intestinal epithelium. Whether up-regulation of GPX2 during carcinogenesis supports tumor growth or can rather be considered as a counteracting effect remains to be investigated [4].
Some of the pleiotropic actions of cholesterol-lowering statins have been attributed to their antioxidant activity. A recent study reported that mRNA levels of GPX2 were induced upon activation of Nrf2 by simvastatin in Wistar rat liver [5]. Simvastatin triggered nuclear translocation of Nrf2 in rat liver and in primary rat hepatocytes in a mevalonate-dependent and cholesterol-independent way. In liver nuclear extracts from simvastatin-treated rats, the DNA-binding activity of Nrf2 was significantly increased and the mRNA of GPX2 was induced. Thus, activation of Nrf2/Keap1 signaling pathway by simvastatin might provide effective protection of the cell from the deleterious effects of oxidative stress.
GPX2 was found to be up-regulated by p63, a member of the p53 tumor suppressor family. A unique responsive element was found in the GPX2 promoter that is bound and activated by p63 but not by p53. Overexpression of GPX2 alleviates oxidative stress-induced apoptosis in MCF-7 breast carcinoma cells, and this protective function of GPX2 is p53 dependent. GPX2 deficiency increased susceptibility to oxidative stress-induced apoptosis. Given that the deltaN isoform of p63 is frequently overexpressed in tumor cells, these observations provide insight into the mechanism by which some isoforms of p63 serve as a pro-survival factor by up-regulating GPX2 to reduce the p53-dependent oxidative stress-induced apoptotic response [19].
Three putative retinoic acid response elements were identified in the GPX2 gene. In the absence of retinoic acid treatment, MCF-7 cells had very low levels of GPX2 mRNA and low GPX activity, whereas HT29 cells had high levels of both. Retinoic acid treatment increased GPX2 mRNA levels and GPX activity in MCF-7 cells. Neither GPX2 mRNA level nor GPX activity was increased in HT29 cells [6].
Mice mutated in Nkx3.1, a homeobox gene known to be required for prostatic epithelial differentiation and suppression of prostate cancer, were shown to display deregulated expression of several antioxidant and pro-oxidant enzymes, including GPX2 and GPX3, peroxiredoxin 6, and sulfhydryl oxidase Q6. Formation of prostatic intraepithelial neoplasia in these mutant mice is associated with increased oxidative damage of DNA, providing a molecular link between a gene involved in prostate carcinogenesis and oxidative damage of the prostatic epithelium [21].
Plasma glutathione peroxidase
(GPX3, plGPX, eGPX) deficiency has been associated with cardiovascular disease and stroke, but the regulation of GPX-3 expression is largely uncharacterized. A functional consensus site for the redox regulated transcription factor activator protein 1 (AP-1) was identified in the 5' promoter region of GPX3 [22]. Subsequently, a novel transcription start site (TSS) located downstream of the previously published site was identified and shown to exhibit a >25-fold increase in transcriptional activity. Analysis of the novel GPX-3 promoter identified functional stimulating protein 1 (Sp1) and hypoxia-inducible factor-1-binding sites, as well as the redox-sensitive ARE/EpRE and putative metal response element (MRE) [7]. Hypoxia-inducible factors (HIFs) respond to changes in available oxygen in the cellular environment; specifically, to decreases in oxygen, or hypoxia. Hypoxia was identified as a strong transcriptional regulator of GPX-3 expression.
Phospholipid hydroperoxide glutathione peroxidase
(GPX4, PHGPX) is characterized as an important enzyme for protecting cells from oxidative stress-induced apoptosis and suppressing cytokine-induced NFkB activation. PHGPX overexpression suppresses production of leukotrienes and prostanoids, and silences 5-lipoxygenase. The high expression level of PHGPX in testicular tissue suggested a more specific function during sperm maturation [8]. PHGPX is encoded for by a joint sperm nucleus/PHGPX gene (sn/PHGPX) and can be expressed as cytosolic, mitochondrial or nuclear isoforms. DNase protection assays in the putative promoter region indicated the presence of five distinct protein-binding regions, and EMSA and supershift experiments revealed binding of Sp1, nuclear factor Y (NF-Y) and members of the Smad family. Site-directed mutagenesis of the consensus binding sequences abolished in vitro transcription factor binding. Expression of reporter genes was most impaired when Sp1/Sp3 and NF-Y binding site-deficient constructs were tested. ChIP assays suggested the in vivo relevance of these transcription factors, which may contribute to differential regulation of expression of the mitochondrial and cytosolic PHGPX isoforms.
The snGPX form differs from the other PHGPXs due to the presence of an arginine-rich N-terminus conferring nuclear localization. This N-terminus is encoded by an alternative exon located in the first intron of the PHGPX gene. The expression of snGPX is controversial, having been attributed either to alternative pre-mRNA splicing or the presence of a distinct promoter region. Preliminary sequence analysis of the region located upstream of the alternative exon revealed potential DNA-binding sites, one of which is specific to the cAMP-response element modulator (CREM) transcription factors. Nuclear protein extract from highly purified rat spermatid cells and recombinant CREM-tau protein can specifically bind to this element in EMSAs. In transient transfection experiments, expression of CREM-tau can induce the activation of an snGPX-reporter gene in NIH-3T3 cell line. These results were confirmed by ChIP experiments, indicating that snGPX expression is mediated by the transcription factor CREM-tau, which acts as a cis-acting element localized in the first intron of the PHGPX gene [25]. In other studies, the putative snGPX promoter was reported to lack major promoter activity, but to suppress activity of the PHGPX promoter. Negative regulatory elements were identified in the first intron of the sn/PHGPX gene, and DNase protection assays revealed the existence of several protein-binding sites. In vivo binding of EGR1 and SREBP1 was shown by ChIP assay, providing evidence for the existence of intronic negative cis-regulatory elements in the sn/PHGPX gene [26].
PHGPX expression was upregulated after TNF alpha exposure in polymorphonuclear leukocytes and neutrophil-like cells that differentiated from the human promyelocytic leukemia cell line, HL60. No increase was seen in macrophage-like differentiated HL60 cells and other cell lines including HeLa human cervical carcinoma cells and HEK293 human embryonic kidney cells, suggesting that this regulation is cell type specific. Up-regulation of PHGPX was inhibited by treatment with the antioxidants, pyrrolidine dithiocarbamate, and N-acetyl-L-cysteine, and by inhibitors of NFkB and Src kinases [27]. Subsequent studies by the same group investigated the transcription factors involved in TNF alpha-induced up-regulation of PHGPX. Promoter activity was up-regulated by TNF alpha stimulation in cells transfected with a PHGPX promoter-luciferase reporter, and this was effectively abrogated by a mutation in the CCAAT/enhancer-binding protein (C/EBP)-binding sequence in this region. ChIP assays demonstrated that C/EBP epsilon bound to the promoter in HL60 cells. Increased binding of nuclear protein to the C/EBP-binding sequence was observed by EMSA in cells stimulated with TNF alpha, and it was inhibited by pre-treatment with an anti-C/EBP-epsilon antibody. Up-regulation of PHGPX mRNA was also detected in HEK-293 cells overexpressing C/EBP-epsilon as a result of TNF alpha stimulation. These results indicate that C/EBP-epsilon is a critical transcription factor in TNF alpha-induced up-regulation of PHGPX expression [9].
Thioredoxin reductases
The mammalian thioredoxin reductases (TRXRs) are selenoenzymes that catalyze the reduction of the active site disulfide of thioredoxin using NADPH. Thioredoxin regulates the redox status of the cells via participation in many different types of reactions, including synthesis of deoxyribonucleotides, redox control of transcription factors, reduction of peroxides, regulation of apoptosis, and extracellular immunoregulatory cytokine and chemokine activities. Perturbations of TRXR activity are implicated in a number of cell proliferative disorders including carcinogenesis, and in immunological diseases.
Thioredoxin reductase 1
(TRXR1) is a nearly ubiquitous, predominantly cytosolic enzyme. In mouse embryos, expression is highest in neuronal tissues, and lowest in heart, whereas in adult mice, expression is highest in liver and kidney [29]. Regulation of TRXR1 expression occurs via numerous pathways targeting transcriptional control, and via AU-rich element-mediated post-transcriptional RNA turnover. The core TRXR1 promoter lacks TATA or CCAAT boxes. It contains a POU motif binding the Oct-1 transcription factor and two sites binding Sp1 and Sp3, which were identified with EMSAs using crude nuclear extracts [30]. Analysis of the TRXR1 promoter region showed that mutations at Oct-1 and Sp1/Sp3 motifs decreased TRXR1 gene expression by ~50%, suggesting that other factors may play a role in this regulation [10]. TRXR1 has a single ARE motif located 9bp upstream of the TSS, and is regulated by Nrf2 [31]. TRXR1 expression is induced by cadmium, and deletion or site-directed mutation of the ARE abolished the response to cadmium. In contrast, overexpression of a dominant negative mutant of Nrf2 suppressed cadmium-induced activation of TRXR1 promoter through the ARE. ChIP assays confirmed binding of Nrf2 to the ARE in cadmium-treated cells. Thus, cadmium-induced TRXR1 gene expression is mediated by activation of Nrf2 and it's binding to the TRXR1 promoter ARE [32].
4-Hydroxynonenal (4-HNE), a major end product of lipid peroxidation, can induce oxidative stress, but has also been found to exert cytoprotective effects at low concentrations. This was shown to occur primarily through induction of TRXR1 via Nrf2. Pretreatment with 4-HNE at sublethal doses significantly protected PC12 cells against subsequent oxidative cell death induced by H2O2 and 6-hydroxydopamine. TRXR1 activity and mRNA levels were significantly elevated by 4-HNE, whereas the glutathione system did not change. TRXR1 siRNA treatment resulted in less resistance to oxidative stress, and the adaptive response was completely abolished. Nrf2 siRNA produced lower constitutive levels of TR1 and less resistance to oxidative stress, and the 4-HNE-induced TRXR1 expression and subsequent adaptive response were again abolished in such cells. These findings, taken together, suggest that stimulation with 4-HNE at sublethal concentrations induces cytoprotective effects, primarily through induction of TRXR1 via Nrf2 [33].
Regulation of TRXR1 gene expression appears to be particularly complex and involves the expression of 7 different transcript forms of mRNA. Regulation of gene expression is observed at the transcriptional level via usage of alternative promoters/TSS as well as at post-transcriptional levels including alternative splicing [11-14] and regulation of mRNA turnover TRXR1 mRNA levels are known to be post-transcriptionally modulated via a cluster of AU-rich motifs located in the 3' UTR of the TRXR1 mRNA. RNA instability elements are generally found in transiently expressed proto-oncogene, nuclear transcription factor, or cytokine mRNAs. Human TRXR1 was found to contain 7 AU-rich motifs, and deletion of 3 or 6 of these resulted in progressive stabilization of the mRNA [15].
Thioredoxin reductase 2
(TRXR2) is a mitochondrial enzyme, with highest expression in murine embryonic heart, followed by liver. In adult mice, expression is highest in spleen, followed by kidney, and intermediate levels in liver, heart, and testis. Little has been reported on regulation of TRXR2; however, evidence has been presented for TRXR2 having a role in regulation of HIF-1. HIF-1, a key regulator for adaptation to hypoxia consists of two subunits, constitutively expressed HIF-1 beta, and HIF-1 alpha, which is regulated both by hypoxia and under normoxia by stimuli including nitric oxide (NO). Overexpression of TRXR2 attenuated NO-evoked HIF-1 alpha accumulation and transactivation of HIF-1 in HEK293 cells. In contrast, TRXR1 enhanced HIF-1 alpha protein and activity upon NO treatment [35]. Evidence has also been presented for estrogen regulation of TRXR2 in perinatal rat brain. Treatment with estradiol on postnatal day 2 resulted in increased TRXR2 in female hypothalamus on postnatal day 5 [36].
Thioredoxin glutathione reductase
(TRXR3, TGR) is highly expressed in testes, being as abundant as beta-actin mRNA. While originally suggested to be testis-specific, TGR is expressed at levels comparable to or greater than TRXR2 in lung, kidney, heart and brain. Little has been reported on the regulation of TGR expression [29].
Methionine sulfoxide reductases
(Msr, SELR, SELX) consist of two distinct families of stereospecific enzymes that catalyze the reduction of oxidized methionine residues. MsrA is specific for the S epimer of methionine sulfoxide, while MsrB is specific for the R form. Of the MsrB proteins, MsrB1 is a selenoprotein in vertebrates, whereas MsrA is a selenoprotein in the unicellular green algae, C. reinhardtii. The MsrB1 gene promoter contains three Sp1 binding sites that are sufficient for maximal promoter activity in transient transfection experiments. A pivotal role for Sp1 in constitutive expression of the MsrB1 gene was demonstrated through transient expression of mutant MsrB1 promoter-reporter gene constructs and ChIP experiments. High levels of MsrB1 mRNA, protein and promoter activity were detected in low metastatic MCF-7 human breast cancer cells, whereas very low levels of MsrB1 mRNA and promoter activity were detected in highly metastatic MDA-MB231 cells. MDA-MB231 cells can be induced to express MsrB1 by treatment with 5-Aza-2'-deoxycytidine, a demethylating agent. Thus, MsrB1 promoter activity appears to be controlled by epigenetic modifications such as methylation [37].
Iodothyronine deiodinases
Three iodothyronine deiodinases (DIOs), reviewed in [16] are involved in the activation and inactivation of thyroid hormone. Types 1 and 2 iodothyronine deiodinases catalyze conversion of the prohormone, thyroxine or T4, to the active hormone, 3,5,3'-triiodothyronine or T3. Type 3 iodothyronine deiodinase comprises the major inactivating pathway that terminates the action of T3 and prevents activation of the prohormone T4.
Type 1 iodothyronine deiodinase
(DIO1), a selenoenzyme catalyzing the bioactivation of thyroid hormone, is highly expressed in the liver. Promoter analysis of the mouse DIO1 gene demonstrated that hepatocyte nuclear factor 4 alpha (HNF4 alpha) plays a key role in the transcriptional activation of the mouse DIO1 gene. Deletion and substitution mutation analyses demonstrated that a proximal HNF4 alpha response element is crucial for this regulation, and DIO1 mRNA and enzyme activity levels are markedly reduced in the livers of HNF4 alpha-null mice. HNF4 alpha, also known as NR2A1, is required for development of the liver and for controlling the expression of many genes specifically expressed in the liver and associated with a number of critical metabolic pathways [38] Human and rodent DIO1 mRNA levels are also stimulated by thyroid hormone, and two thyroid hormone response elements (TREs) were identified and functionally characterized in the human DIO1 gene promoter [39]. Retinoic acid increases the concentration of DIO1 in human thyroid carcinoma cell lines [17]. This can be accounted for by the TREs in the human DIO1 gene that also respond to retinoic acid [18-20]. An Sp1 binding site immediately 5' of the upstream TRE increases basal expression in the presence of unliganded thyroid hormone receptor, thus decreasing T3 responsiveness [20]. Mouse DIO1 is also stimulated by thyroid hormone, but a direct role for thyroid hormone receptor action has not been reported. Thyroid hormone-inducible Krüppel-like factor 9 (KLF9) stimulates the mouse DIO1 promoter very efficiently through two CACCC sequences that are located on either side of the HNF4 alpha-response element. Furthermore, KLF9 functions together with HNF4 alpha and GATA4 to synergistically activate the mouse DIO1 promoter, suggesting that DIO1 is regulated by thyroid hormone in the mouse through an indirect mechanism requiring prior KLF9 induction. Physical interactions between GATA4 and HNF4 alpha and between GATA4 and KLF9 are reported to be required for this synergistic response. These results suggest that HNF4 alpha regulates thyroid hormone homeostasis through transcriptional regulation of the mouse DIO1 gene with GATA4 and KLF9 [38].
Studies in the FRTL5 rat thyroid cell line have shown a 3-fold increase in DIO1 mRNA induced by TSH, which is replicated by dibutryl cAMP or forskolin. The effects of these agonists were additive to that of T3, the combination resulting in a 5-fold stimulation relative to control [21]. This could not be explained by an alteration in DIO1 mRNA disappearance rate, and the effect was blocked by cycloheximide, indicating that persistent protein synthesis is required for the effect. The mechanism for the stimulation of rat DIO1 transcription by cAMP has not been elucidated.
IL-1, IL-6, TNF alpha, and other cytokines have been postulated as potential mediators of the alterations in thyroid function that occur during severe illness [22-24]. TNF alpha, IL-1 beta, and interferon gamma decrease DIO1 activity and mRNA in FRTL5 cells, although TGF beta has no effect [25]. The effects of TNF alpha have been examined in hepatocytes and HepG2 cells with contradictory results. TNF alpha decreased the T3-stimulated DIO1 mRNA in HepG2 cells.
Type 2 iodothyronine deiodinase
(DIO2) mRNA is abundantly expressed in human thyroid but present at very low levels in adult rat thyroid. The human DIO2 sequence was activated 10-fold by transiently expressed thyroid transcription factor 1 (TTF-1) in COS-7 monkey kidney fibroblast cells, whereas the response of the rat DIO2 gene to TTF-1 was only 3-fold despite 73% identity of the two promoters. Four putative TTF-1 binding sites in human DIO2 were compared by EMSA using in vitro expressed TTF-1. Only two sites, both of which are absent in rat DIO2, had significant affinity. Functional analyses showed that both sites are required for the full response to TTF-1 [41]. DIO2 mRNA is also expressed at high levels in human heart but is barely detectable in the corresponding rodent tissue. The human DIO2 promoter is very sensitive to the cardiac transcription factors, Nkx-2.5 and GATA-4. Nkx-2.5 activates a human DIO2-reporter construct, whereas the rat DIO2 promoter is much less responsive to Nkx-2.5 induction. Two Nkx-2.5 sites were identified in the human promoter by EMSA. GATA-4 alone was a poor inducer of the human DIO2 promoter. However in synergy with Nkx-2.5, it activated DIO2 reporter gene expression in the human, but not the rat promoter. Functional analysis showed that both Nkx2.5 sites are required for the complete Nkx-2.5 response and for the Nkx-2.5/GATA-4 synergistic effect. Finally, a mutant Nkx-2.5 protein (N188K), which causes, in heterozygosity, congenital heart diseases, did not transactivate the DIO2 promoter and interfered with its activity in cardiomyocytes, possibly by titrating endogenous Nkx-2.5 protein away from the promoter [42].
The human DIO2 promoter is NFkB responsive, with a 600bp region exhibiting 15-fold induction. EMSAs identified two strong NFkB binding sites with very similar binding affinity, but site-directed mutagenesis and promoter assays indicated that only one site was activated in the presence of the p65 subunit of NFkB. Other cytokine mediators such as STAT3 or STAT5 did not induce transcription of the DIO2 gene [43]. Human DIO2 gene transcription is potently increased by cAMP in some tissues via a conserved cAMP response element (CRE) located in the promoter region [44]. In addition, several TATA box/TSS units are present in the promoter, suggesting the presence of different transcripts that might be characterized by different biological properties. Transient transfection studies showed that cAMP induces transcription from the most 5' TSS, located about 80 nucleotides from the CRE. Site-directed mutagenesis and deletion analysis showed that a single CRE/TATA box/TSS unit is needed to confer responsiveness to cAMP, and ChIP studies showed binding of transcription factor CRE binding protein (CREB) to the CRE [26]. Epidermal growth factor (EGF) was also shown to modulate transcription of DIO2 via the CRE, and does not involve the AP-1 site. EGF stimulation culminates with the assembly at the DIO2 CRE of a composite complex, consisting of c-Jun, c-Fos, and CREB [46].
Thyroid status controls DIO2 activity both at the pre- and posttranslational levels [27-29]. Deiodination of T4 increases in the cortex of hypothyroid rats, and hypothyroidism elevates DIO2 mRNA in the brain [30-34]. Treatment of hypothyroid rats shows that T3 decreases DIO2 mRNA, whereas T4 primarily decreases DIO2 activity, indicating that, in vivo, T3 and T4 can exert their suppressive effects on DIO2 activity by pre- and posttranslational mechanisms, respectively [34]. T3-induced DIO2 mRNA suppression is transcriptional, because 100 nM T3 does not affect the short DIO2 mRNA half-life, and this is a direct T3 effect. Although the presence of a negative TRE in the DIO2 5' UTR can be inferred, it has not yet been identified. Dexamethasone and TRH modestly increase DIO2 mRNA in GH4C1 cells. In marked contrast to T3, rT3 reduces DIO2 activity but does not affect DIO2 mRNA levels, indicating that its regulation of DIO2 is completely posttranslational [35].
An increase in pineal gland DIO2 activity induced by an endogenous beta-adrenergic mechanism correlates precisely with similar changes in DIO2 mRNA [36, 37]. DIO2 mRNA and activity are also increased several fold by hypothyroidism in somatosensory regions of the brain of postnatal rats, providing protection against the deleterious effects of insufficient T3 availability during brain development [38]. An effect of stress and traumatic brain injuries to increase DIO2 activity in the CNS has also been reported [39, 40].
Type 3 iodothyronine deiodinase
(DIO3) catalyzes inactivation of the active thyroid hormone, T3, by the sequential removal of iodine groups. DIO3 transcription is stimulated by TGF beta, acting via a Smad-dependent pathway. Combinations of Smad2 or Smad3 with Smad4 stimulate human DIO3 gene transcription only in cells that express endogenous DIO3 activity, indicating that Smads are necessary but not sufficient for DIO3 induction. Maximum stimulation of DIO3 by TGF beta also requires MAPK and is synergistic with phorbol ester and several mitogens known to signal through transmembrane receptor tyrosine kinases [47]. Because TGF beta3 is a target of hypoxia, hypoxia was investigated as a regulator of DIO3 expression. DIO3 activity and mRNA are increased by hypoxia and by hypoxia mimetics that increase HIF-1. Using ChIP, HIF-1 alpha was shown to interact specifically with the DIO3 promoter, indicating that DIO3 may be a direct transcriptional target of HIF-1. Endogenous DIO3 activity decreased T3-dependent oxygen consumption in both neuronal and hepatocyte cell lines, suggesting that hypoxia-induced DIO3 may reduce metabolic rate in hypoxic tissues. In a rat model of cardiac failure due to right ventricular hypertrophy, HIF-1 alpha and DIO3 proteins were induced specifically in the hypertrophic myocardium of the right ventricle, creating an anatomically specific reduction in local T3 content and action. These results suggest a mechanism of metabolic regulation during hypoxic-ischemic injury in which HIF-1 reduces local thyroid hormone signaling through induction of DIO3 [48]. It was recently shown that DIO3 responds to the Shh/Gli2 cascade [41]. DIO3 mRNA expression, protein levels and DIO3 enzymatic activity were significantly increased in HaCaT cells by the Shh transcriptional effector Gli2 and this induction was completely reversed by treatments with forskolin or cyclopamine, which both inhibit Shh signaling [42, 43].
In situ hybridization studies on DIO3 gene expression within the CNS revealed dramatic increases in DIO3 mRNA after a short term T3 treatment [44]. Further studies will be needed to determine whether this reflects T3-induced increases in gene transcription, mRNA stabilization, or a combination of these factors. In X. laevis, this effect is direct, i.e., not blocked by cycloheximide. DIO3 promoter analysis conducted on the human and rat DIO3 promoters shows a positive regulation by T3, although the magnitude of this regulation is modest compared with the effect of thyroid status on DIO3 activity [16]. DIO3 activity is not increased in the placenta of the hyperthyroid rat, unlike the situation in brain, indicating that this gene is differentially responsive to T3 in different tissues [45, 46].
In cultured astroglial cells, all-trans-retinoic acid (5 μM) causes a marked increase of up to 200-fold in DIO3 activity, producing an additive effect with thyroid hormones [47]. Furthermore, it was shown that the regulation of DIO3 expression by retinoids involves both RAR and RXR pathways and is cell type-specific [48].
Oxidative stress up-regulated DIO3 activity and DIO3 mRNA accumulation in primary cultures of rat astrocytes. Stimulation of DIO3 activity by H2O2 was synergistic with T4, phorbol ester, and also cAMP. N-Acetyl cysteine and selenium repletion, which respectively increase intracellular glutathione and glutathione peroxidase, inhibited DIO3 regulation by H2O2, whereas L-buthionine sulfoximine, which decreases intracellular glutathione, mimicked H2O2 effects. The ERK pathway was required in DIO3 regulation by oxidative stress and the p38 MAPK pathway was implicated in H2O2-induced DIO3. It was suggested that the expected decrease in T3 might modulate the cellular injury of oxidative stress in some pathological brain conditions [49].
Selenoprotein P
(SEPP1, SELP) is unique in that it contains 8 to 10 selenocysteines in mammals, 16 to 18 in fish and amphibians, and 28 in sea urchin. Selenoprotein P is expressed at highest levels in liver, but SELP mRNA is present in most tissues, with significant levels in testis, brain, gut, and hematopoietic cells. The murine SELP gene promoter contains four hepatic nuclear factor 3 beta (HNF3 beta) binding motifs, in accord with prominent expression in liver. Two BRN-2 motifs and multiple GATA-1 motifs are consistent with expression in brain and hematopoietic cells, respectively. SRY motifs present in the promoter region might explain detection of SELP in Leydig cells [50]. Human SELP was identified as a target of HNF4 alpha via expression profiling and RNA interference screening in HEK-293 cells [51].
The human SELP promoter responds to overexpression of FoxO1a, a member of the forkhead class O (FOXO) family of transcription factors. Two FOXO-responsive elements were identified and characterized by generation of point mutation and deletion constructs [52]. FOXO family members, including FOXO1, FOXO3a, and FOXO4, are implicated in the regulation of a variety of cellular processes, including cell cycle, apoptosis, DNA repair, stress resistance, and metabolism. The activities of FOXO proteins are regulated by oxidative stress, which induces their phosphorylation, translocation to the nucleus, and acetylation-deacetylation [53].
Selenoprotein S
(SEPS1, SELS) plays an important role in the production of inflammatory cytokines and its expression is activated by endoplasmic reticulum (ER) stress. Two NFkB sites were identified in the human SELS promoter. SELS gene expression, protein levels and promoter activity were all increased 2-3-fold by TNF alpha and IL-1 beta in HepG2 cells. A putative ER stress response element in the SELS proximate promoter is fully functional and responsive to ER stress. However, concurrent treatment of HepG2 cells with IL-1 beta and ER stress produced no additive effect on SELS gene expression. Thus, SELS appears to be a new target gene of NFkB. Together with previous findings that SELS may regulate cytokine production in macrophage cells, a regulatory loop between cytokines and SELS has been proposed to play a key role in control of the inflammatory response [54].
Selenoprotein W
(SEPW, SELW) is highly expressed in skeletal muscle, heart and brain. Although its function is unknown, a role for SELW as an antioxidant was proposed due to its ability to bind glutathione [54] and render cells resistant to hydrogen peroxide [55]. A role for SELW in cellular anti-cadmium defense was also suggested [56]. Transcriptional regulation of rat SELW was investigated by in vitro binding assays using nuclear extracts from rat C6 glial cell and oligonucleotides containing Sp1, TFII-1, MRE and AP-1 putative regulatory elements found in the rat SELW promoter. The Sp1 transcription factor was shown to bind to the Sp1 consensus sequence in the rat SELW promoter as well as to an MRE. Although competition analysis showed specific binding at a TFII-1 site, super-shift analysis using anti-TFII-1 antibody did not yield any super-shifted band. Therefore, the rat SELW gene may be a target for Sp1, whose binding to various regulatory sequences of the SELW promoter may activate or repress its transcription. Putative MRE, GRE, AP-1 and LF-A1 sites were also tested but no evidence was obtained for specific binding as indicated by lack of competition with unlabeled probes [57]. The same group showed that expression of a reporter gene fused to a rat SELW promoter fragment was induced two- to four-fold by copper and zinc but not cadmium in rat glial cells [58]. Further, this response was abolished by mutation of the MRE site, indicating that MTF1 is involved in the expression of rat SELW, even though initial studies failed to demonstrate MTF-1 binding to that sequence [57]. Subsequent studies identified three MREs in the mouse SELW promoter. Two of these, found in an inverted orientation and overlapping almost completely, are located in the core promoter and specifically bound to MTF-1, while the third located further upstream did not. In addition, microarray analysis demonstrates that SELW is significantly downregulated in livers from cadmium- and mock-treated Mtf1Mx-cre mice, strongly suggesting that MTF-1 is important for the basal expression of mouse SELW [56].
Initiation of transcription from TATA and TATA-less promoters
The expression of most protein-coding genes is controlled at the transcriptional level by mechanisms involving the regulation of initiation. Based on the presence or absence of the TATA sequence, gene promoters are divided into two major categories, TATA and TATA-less. Promoters in the first category contain a consensus TATA box, located approximately 30bp upstream of the TSS, and a strong initiator (Inr) element, which encompasses the transcriptional start site. Promoters in the second category lack the consensus TATA box, strong Inr element or both. In some TATA-less promoters, the TATA box is replaced by an AT-rich sequence and in others a functional promoter element at -30 appears to be lacking completely [59]. Computational analysis suggests that the prevalence of the TATA box has been overestimated in the past and that the majority of metazoan genes are TATA-less [60]. For example, approximately 50% of the transcribed genes in Drosophila melanogaster lack TATA sequences [61].
TATA motifs were identified in the mouse and human SELP promoters [50, 63, 64]. Deletion mapping and luciferase analysis showed that the TATA box and a putative Sp1 site are necessary for human SELP transcription [62]. The human DIO2 gene uses a single CRE/TATA box/TSS unit, verified by site-directed mutagenesis and promoter deletion analysis to confer cAMP responsiveness, as discussed above [26]. A TATA box identified in the 5' flanking sequence to the TSS of murine DIO3 gene, as well as CAAT and GC boxes, are critical for DIO3 transcription [63]. Previous studies on other selenoprotein gene promoters indicate that transcription of murine SELW [66, 67], rat GPX4 [68, 69], human TRXR1 [30], human Sep15 [64] and human MsrB1 [37] is driven by TATA-less promoters. Interestingly, human DIO2 has a TATA box while Fundulus heteroclitus DIO2 does not [65]. Similarly the mouse DIO3 has a TATA box [63] and human DIO3 does not. Thus, some selenoprotein genes may differentially fall into the TATA and/or TATA-less promoter category depending on species.
TATA-less promoters require Sp-binding sites for significant activity [72], and the degree of activation from Sp1 tends to be stronger in the context of TATA-less promoters than TATA-containing promoters [66]. This activation most likely involves Sp1 recruitment of TFIID to TATA-less promoters [74, 75]. Furthermore, in many of these promoters, Sp1 binding is intimately involved in the determination of the transcription start site or sites [76-79]. As discussed above, Sp1 is involved in the transcriptional regulation of GPX3, GPX4, DIO1, SELR, SELW, and TRXR1. Furthermore, we identified multiple Sp-binding sites in the proximal promoters for the rest of the selenoprotein genes, indicating that this factor may play a role in transcriptional activation of these genes. The Sp family of transcription factors is discussed further below.
Posttranscriptional regulation of selenoprotein gene expression
Alternative splicing of pre-mRNAs is a powerful and versatile regulatory mechanism contributing to proteomic diversity. It allows for switches in protein isoforms in the absence of permanent changes in the cell's genetic content, and without changes in transcriptional activity. Alternative splicing can affect almost all aspects of protein function, ranging from the determination of cellular and subcellular localization to quantitative control of gene expression and modulation of enzyme activity. Alternative splicing of untranslated (UTR) regions can also determine mRNA localization and stability, as well as efficiency of translation. Other, less frequent, complex events that give rise to alternative transcript variants include alternative transcription start sites and multiple polyadenylation sites [67-69].
Analysis of selenoprotein gene structure and mRNA variants using AceView [70] indicates alternative splice variants and alternative polyadenylation sites for the majority of the human selenoprotein genes. An experimental validation of alternative splice variants was reported for the following selenoprotein genes: DIO2 [71, 72], GPX4 [73, 74], SelN [75], SEPP1 [62], TRxR1 [11-14, 76-78], TRXR3 [77, 79]. Alternative transcripts resulting from alternative TSS/promoter usage have been identified for DIO2 [71], DIO3 [63], GPX4 [80] and TRxR1 [77]. Alternative transcripts resulting from alternative polyadenylation sites were proposed for DIO3 [16, 63]. Future studies will almost certainly reveal more variants and provide insights into their biological functions.
In summary regulation of selenoprotein gene expression at the posttranscriptional level contributes to selenoprotein diversity, leading to expression of proteins with different subcellular localization and tissue or developmentally specific expression patterns. An example is the GPX4 gene encoding mitochondrial PHGPx and cytosolic PHGPx isoforms via differential TSS usage or testis-specific nuclear protein snGPx via alternative splicing [73, 80].
In silico analysis of selenoprotein gene promoters
Genomic regions spanning 2000bp upstream and 1000bp downstream of the transcription start site (TSS) of the selenoprotein genes listed in Table 1 were extracted using Genomatix software “Gene to Promoter” (www.genomatix.de). Those sequences were analyzed for single promoter elements and complex modules using “MatInspector” and “Frame Worker” (www.genomatix.de). Analyses of the sequences spanning -250bp to +50bp relative to the TSS were carried out using “MatInspector” to examine for the presence of TATA boxes and core promoter elements. To examine the promoter regions for the presence of CpG islands, we used CpG finder and TSSW by SoftBerry (www.softberry.com/berry.phtml). It should be noted that in silico identified TF binding sites and TF modules do not prove the gene to be a target and require further experimental validation. Transcription factor modules are usually linked to at least one known biological function and make this gene a good candidate for further experimental verification. Detailed analysis of the predicted regulatory elements and their location relative to TSS of the individual transcript variants would be required. Predicted co-regulation patterns for selenoprotein gene expression identified in silico and discussed below can be used to guide studies on transcriptional regulation of selenoproteins and their spatial and temporal tissue specific expression. Identification of transcription regulatory networks will provides the links to specific biological pathways and help to direct the studies and uncover the biological functions of newly identified selenoproteins.
Single transcription factor sites and complex modules in selenoprotein genes
Clustering analysis of differentially regulated genes can reveal common regulatory patterns by specific transcription factors, thereby enabling transcription factor signatures to be established and potential common regulatory elements to be identified. We searched for common potential promoter elements within the human selenoprotein family members, and found 21 single transcription factor elements to be common to all family members (Table S2). Potential complex transcription factor modules consisting of two or more adjacent transcription factor binding sites were found for subsets of selenoprotein genes (Table S3) and some correlate with tissue expression profiles for those genes, suggesting co-regulation. Selenoprotein genes that exhibit widespread expression, e.g. TRXR1 and SELP also exhibit the highest numbers of potential regulatory elements, whereas those with tissue-restricted expression, e.g. GPX2, SELV, SELT and TRXR3 exhibit the lowest numbers of potential regulatory elements.
NFkB is one of the major stress responsive transcription factors. NFkB is found in almost all animal cell types and is involved in cellular responses to cytokines, free radicals, UV irradiation, oxidized LDL, and bacterial or viral antigens. NFkB plays a key role in regulating the immune response to infection. Consistent with this role, incorrect regulation of NFkB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. Given the known functions of selenoproteins in stress responsiveness, NFkB might be predicted to be involved in regulation of multiple selenoproteins, but to date, experimental evidence for regulation by NFkB has only been reported for DIO2, GPX4, and SELS. Our analysis revealed the presence of putative NFkB response elements in all 25 human selenoprotein genes (Fig. 2).
Figure 2. Selenoprotein gene organization and putative transcription factor binding sites.
Regions spanning 2kb upstream and 1kb downstream of putative transcription start sites (red arrows) were analyzed for putative MREs (red pentagonals), NFkB sites (blue ovals) and CpG islands (yellow boxes). Open boxes designate exons, solid black lines indicate introns, and dashed lines indicate putative untranscribed regions for these genes. Green arrows indicate putative translation start sites. Vertical lines indicate 500bp intervals.
MTF-1 is involved in the transcriptional response to heavy metal exposure, cellular responses to oxidative stress and hypoxia. MREs, the short, cis-acting sequences to which MTF-1 binds, have previously been identified in the promoter regions of GPX3, SELP and SELW, but their function has only been experimentally verified for SELW. Known links between metals and stress responsiveness might predict regulation of multiple selenoprotein genes by MTF-1 via MREs. Our analysis revealed the presence of putative MREs upstream of the TSS in 18 selenoprotein genes, and when the region spanning 1kb downstream of the TSS was included, only one selenoprotein gene, SELK, lacked a putative MRE (Fig. 2). Further analysis of the expression of several selenoprotein genes in MTF-1 overexpressing and MTF-1 knockout mouse embryo fibroblast cells reveal differential expression, indicating that this factor may play a role in their regulation (our unpublished data).
The Sp family of transcription factors have four members in humans: Sp1, Sp2, Sp3, and Sp4 [81]. Sp1 was identified first and is the most well characterized [82]. Sp1 and Sp3 are ubiquitous transcription factors, implicated in the control of a wide variety of genes [82, 83]. An activator Sp1 and repressor Sp3 are thought to compete for similar binding sites, and the relative rate of transcription is affected by the outcome of this competition [84, 85]. Sp2 is least similar to the other Sp family members, and very little is known about its function. Lastly, Sp4 has tissue-specific expression restricted to the brain and nervous system [84]. Our in silico analysis indicates that all selenoprotein genes except DIO2 have putative Sp transcription factor binding sites. Further, we found in silico that in 16 of the selenoprotein genes (GPX1, GPX2, DIO1, DIO3, TRXR1, TRXR2, SELI, SELN, SELO, SELR, SELS, SELT, SEP15, SELV, and SELW) Sp sites were present in the core promoter (identified as 40bp up- and downstream of the TSS). Thus, the Sp family of transcription factors is likely to play significant roles in regulating selenoprotein transcription, and possibly in tissue specific expression, as many selenoproteins are expressed in brain.
Sequence analysis of the putative transcription initiation regions of human selenoprotein genes demonstrated the presence of various potential regulatory sequences that may contribute to the initiation of transcription from TATA-less promoters (Table S4). We identified TATA boxes in the DIO2 and SELP genes, consistent with previously published data. We identified 2 putative TBP sites in the SPS2 gene at position -82/-66 and position -5/+11 relative to the TSS, but have not experimentally validated functionality. Putative AP1 transcription factor binding sites were found in SELM, SELN, SELV and the putative AP1R binding site in DIO2 core promoters.
Regulation of expression by methylation
A CpG island is defined as a sequence with a G+C content of greater than 60% and ratio of CpG to GpC of at least 0.6 [83]. CpG islands are frequently located within 5' regulatory regions of genes, with about 70 to 80% of these dinucleotides being methylated [86, 87]. In the case of CpG island-containing promoters, the lack of methylation is usually associated with the chromatin pattern of actively transcribed genes [88-90]. In contrast, genes without CpG islands are dependent on the methylation of single sites within their promoter regions, which prevents binding of specific factors. Such methylation dependency was described for transcription factor AP-2 [91], whereas for Sp1 the data are contradictory [92, 93]. Sixteen of the human selenoprotein promoters contain putative CpG islands (Fig. 2) suggesting a possible role of methylation in tissue-specific expression of these genes. Most of these span the TSS.
SUMMARY
Considerable efforts have gone into studying the transcriptional regulation of a number of selenoprotein genes, particularly those encoding proteins whose biological functions are at least partially understood. However, particularly for most of the recently identified selenoproteins of unknown function, expression profiling provides the only experimentally validated information available to date. Bioinformatics mining has identified many important leads to pursue experimentally, including investigation of the roles of stress responsive transcription factors such as NFkB, MTF-1 and Nrf2, as well as factors with roles in tissue specific or developmentally regulated expression. The importance of performing such analysis is indicated by the fact that published data for several transcription factors, such us NFkB, MTF-1, HIF-1, and the SP family (described above and summarized in Table 2), coincides with predictions based on bioinformatic approaches. Undoubtedly, future studies along these lines will provide important new insights into how selenoprotein genes respond, and the roles they play, in allowing cells to adapt to environmental and other challenges.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (DK47320, DK52963, NS40302 to MJB) and the Hawaii Community Foundation (No. 20071388 to ZRS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Brigelius-Flohe R. Biol Chem. 2006;387:1329–35. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- [2].Tan M, Li S, Swaroop M, Guan K, Oberley LW, Sun Y. J Biol Chem. 1999;274:12061–6. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- [3].Throm SL, Klemsz MJ. J Leukoc Biol. 2003;74:111–7. doi: 10.1189/jlb.0203061. [DOI] [PubMed] [Google Scholar]
- [4].Kipp A, Banning A, Brigelius-Flohe R. Biol Chem. 2007;388:1027–33. doi: 10.1515/BC.2007.137. [DOI] [PubMed] [Google Scholar]
- [5].Habeos IG, Ziros PG, Chartoumpekis D, Psyrogiannis A, Kyriazopoulou V, Papavassiliou AG. J Mol Med. 2008;86:1279–85. doi: 10.1007/s00109-008-0393-4. [DOI] [PubMed] [Google Scholar]
- [6].Chu FF, Esworthy RS, Lee L, Wilczynski S. J Nutr. 1999;129:1846–54. doi: 10.1093/jn/129.10.1846. [DOI] [PubMed] [Google Scholar]
- [7].Bierl C, Voetsch B, Jin RC, Handy DE, Loscalzo J. J Biol Chem. 2004;279:26839–45. doi: 10.1074/jbc.M401907200. [DOI] [PubMed] [Google Scholar]
- [8].Ufer C, Borchert A, Kuhn H. Nucleic Acids Res. 2003;31:4293–303. doi: 10.1093/nar/gkg650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hattori H, Imai H, Kirai N, Furuhama K, Sato O, Konishi K, Nakagawa Y. Biochem J. 2007;408:277–86. doi: 10.1042/BJ20070245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rundlof AK, Carlsten M, Arner ES. J Biol Chem. 2001;276:30542–51. doi: 10.1074/jbc.M101452200. [DOI] [PubMed] [Google Scholar]
- [11].Dammeyer P, Damdimopoulos AE, Nordman T, Jimenez A, Miranda-Vizuete A, Arner ES. J Biol Chem. 2008;283:2814–21. doi: 10.1074/jbc.M708939200. [DOI] [PubMed] [Google Scholar]
- [12].Rundlof AK, Fernandes AP, Selenius M, Babic M, Shariatgorji M, Nilsonne G, Ilag LL, Dobra K, Bjornstedt M. Differentiation. 2007;75:123–32. doi: 10.1111/j.1432-0436.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- [13].Rundlof AK, Janard M, Miranda-Vizuete A, Arner ES. Free Radic Biol Med. 2004;36:641–56. doi: 10.1016/j.freeradbiomed.2003.12.004. [DOI] [PubMed] [Google Scholar]
- [14].Damdimopoulos AE, Miranda-Vizuete A, Treuter E, Gustafsson JA, Spyrou G. J Biol Chem. 2004;279:38721–9. doi: 10.1074/jbc.M402753200. [DOI] [PubMed] [Google Scholar]
- [15].Gasdaska JR, Harney JW, Gasdaska PY, Powis G, Berry MJ. J Biol Chem. 1999;274:25379–85. doi: 10.1074/jbc.274.36.25379. [DOI] [PubMed] [Google Scholar]
- [16].Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Endocr Rev. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- [17].Schreck R, Schnieders F, Schmutzler C, Kohrle J. J Clin Endocrinol Metab. 1994;79:791–8. doi: 10.1210/jcem.79.3.8077363. [DOI] [PubMed] [Google Scholar]
- [18].Jakobs TC, Schmutzler C, Meissner J, Kohrle J. Eur J Biochem. 1997;247:288–97. doi: 10.1111/j.1432-1033.1997.00288.x. [DOI] [PubMed] [Google Scholar]
- [19].Schmutzler C, Kohrle J. Thyroid. 2000;10:393–406. doi: 10.1089/thy.2000.10.393. [DOI] [PubMed] [Google Scholar]
- [20].Zhang CY, Kim S, Harney JW, Larsen PR. Endocrinology. 1998;139:1156–63. doi: 10.1210/endo.139.3.5849. [DOI] [PubMed] [Google Scholar]
- [21].Toyoda N, Nishikawa M, Mori Y, Gondou A, Ogawa Y, Yonemoto T, Yoshimura M, Masaki H, Inada M. Endocrinology. 1992;131:389–94. doi: 10.1210/endo.131.1.1319323. [DOI] [PubMed] [Google Scholar]
- [22].Boelen A, Platvoet-Ter Schiphorst MC, Wiersinga WM. J Clin Endocrinol Metab. 1993;77:1695–9. doi: 10.1210/jcem.77.6.8263160. [DOI] [PubMed] [Google Scholar]
- [23].Chopra IJ, Sakane S, Teco GN. J Clin Endocrinol Metab. 1991;72:1113–6. doi: 10.1210/jcem-72-5-1113. [DOI] [PubMed] [Google Scholar]
- [24].van der Poll T, Romijn JA, Wiersinga WM, Sauerwein HP. J Clin Endocrinol Metab. 1990;71:1567–72. doi: 10.1210/jcem-71-6-1567. [DOI] [PubMed] [Google Scholar]
- [25].Pekary AE, Berg L, Santini F, Chopra I, Hershman JM. Mol Cell Endocrinol. 101(1994):R31–5. doi: 10.1016/0303-7207(94)90256-9. [DOI] [PubMed] [Google Scholar]
- [26].Canettieri G, Franchi A, Sibilla R, Guzman E, Centanni M. J Mol Endocrinol. 2004;33:51–8. doi: 10.1677/jme.0.0330051. [DOI] [PubMed] [Google Scholar]
- [27].Silva JE, Larsen PR. J Clin Invest. 1982;70:1110–23. doi: 10.1172/JCI110699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].St Germain DL. J Clin Invest. 1985;76:890–3. doi: 10.1172/JCI112049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].St Germain DL. Endocrinology. 1988;122:1860–8. doi: 10.1210/endo-122-5-1860. [DOI] [PubMed] [Google Scholar]
- [30].Croteau W, Davey JC, Galton VA, St Germain DL. J Clin Invest. 1996;98:405–17. doi: 10.1172/JCI118806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gereben B, Bartha T, Tu HM, Harney JW, Rudas P, Larsen PR. J Biol Chem. 1999;274:13768–76. doi: 10.1074/jbc.274.20.13768. [DOI] [PubMed] [Google Scholar]
- [32].Tu HM, Kim W, Salvatore D, Bartha T, Legradi G, Larsen PR, Lechan RM. Endocrinology. 1997;138:3359–68. doi: 10.1210/endo.138.8.5318. [DOI] [PubMed] [Google Scholar]
- [33].Burmeister LA, Pachucki J, St Germain DL. Endocrinology. 1997;138:5231–7. doi: 10.1210/endo.138.12.5602. [DOI] [PubMed] [Google Scholar]
- [34].Leonard JL, Kaplan MM, Visser TJ, Silva JE, Larsen PR. Science. 1981;214:571–3. doi: 10.1126/science.7291997. [DOI] [PubMed] [Google Scholar]
- [35].Kim SW, Harney JW, Larsen PR. Endocrinology. 1998;139:4895–905. doi: 10.1210/endo.139.12.6334. [DOI] [PubMed] [Google Scholar]
- [36].Kamiya Y, Murakami M, Araki O, Hosoi Y, Ogiwara T, Mizuma H, Mori M. Endocrinology. 1999;140:1272–8. doi: 10.1210/endo.140.3.6594. [DOI] [PubMed] [Google Scholar]
- [37].Tanaka K, Murakami M, Greer MA. Biochem Biophys Res Commun. 1986;137:863–8. doi: 10.1016/0006-291x(86)91159-9. [DOI] [PubMed] [Google Scholar]
- [38].Guadano-Ferraz A, Escamez MJ, Rausell E, Bernal J. J Neurosci. 1999;19:3430–9. doi: 10.1523/JNEUROSCI.19-09-03430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baumgartner A, Hiedra L, Pinna G, Eravci M, Prengel H, Meinhold H. J Neurochem. 1998;71:817–26. doi: 10.1046/j.1471-4159.1998.71020817.x. [DOI] [PubMed] [Google Scholar]
- [40].Zou L, Burmeister LA, Styren SD, Kochanek PM, DeKosky ST. J Neurochem. 1998;71:887–90. doi: 10.1046/j.1471-4159.1998.71020887.x. [DOI] [PubMed] [Google Scholar]
- [41].Dentice M, Luongo C, Huang S, Ambrosio R, Elefante A, Mirebeau-Prunier D, Zavacki AM, Fenzi G, Grachtchouk M, Hutchin M, Dlugosz AA, Bianco AC, Missero C, Larsen PR, Salvatore D. Proc Natl Acad Sci U S A. 2007;104:14466–71. doi: 10.1073/pnas.0706754104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen JK, Taipale J, Cooper MK, Beachy PA. Genes Dev. 2002;16:2743–8. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang G, Wang B, Jiang J. Genes Dev. 1999;13:2828–37. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tu HM, Legradi G, Bartha T, Salvatore D, Lechan RM, Larsen PR. Endocrinology. 1999;140:784–90. doi: 10.1210/endo.140.2.6486. [DOI] [PubMed] [Google Scholar]
- [45].Emerson CH, Bambini G, Alex S, Castro MI, Roti E, Braverman LE. Endocrinology. 1988;122:809–16. doi: 10.1210/endo-122-3-809. [DOI] [PubMed] [Google Scholar]
- [46].Roti E, Braverman LE, Fang SL, Alex S, Emerson CH. Endocrinology. 1982;111:959–63. doi: 10.1210/endo-111-3-959. [DOI] [PubMed] [Google Scholar]
- [47].Esfandiari A, Gagelin C, Gavaret JM, Pavelka S, Lennon AM, Pierre M, Courtin F. Glia. 1994;11:255–61. doi: 10.1002/glia.440110306. [DOI] [PubMed] [Google Scholar]
- [48].Kester MH, Kuiper GG, Versteeg R, Visser TJ. Endocrinology. 2006;147:5845–54. doi: 10.1210/en.2006-0590. [DOI] [PubMed] [Google Scholar]
- [49].Lamirand A, Pallud-Mothre S, Ramauge M, Pierre M, Courtin F. Endocrinology. 2008;149:3713–21. doi: 10.1210/en.2007-1462. [DOI] [PubMed] [Google Scholar]
- [50].Steinert P, Bachner D, Flohe L. Biol Chem. 1998;379:683–91. doi: 10.1515/bchm.1998.379.6.683. [DOI] [PubMed] [Google Scholar]
- [51].Grigo K, Wirsing A, Lucas B, Klein-Hitpass L, Ryffel GU. Biol Chem. 2008;389:179–87. doi: 10.1515/BC.2008.011. [DOI] [PubMed] [Google Scholar]
- [52].Walter PL, Steinbrenner H, Barthel A, Klotz LO. Biochem Biophys Res Commun. 2008;365:316–21. doi: 10.1016/j.bbrc.2007.10.171. [DOI] [PubMed] [Google Scholar]
- [53].Furukawa-Hibi Y, Kobayashi Y, Chen C, Motoyama N. Antioxid Redox Signal. 2005;7:752–60. doi: 10.1089/ars.2005.7.752. [DOI] [PubMed] [Google Scholar]
- [54].Beilstein MA, Vendeland SC, Barofsky E, Jensen ON, Whanger PD. J Inorg Biochem. 1996;61:117–24. doi: 10.1016/0162-0134(95)00045-3. [DOI] [PubMed] [Google Scholar]
- [55].Jeong D, Kim TS, Chung YW, Lee BJ, Kim IY. FEBS Lett. 2002;517:225–8. doi: 10.1016/s0014-5793(02)02628-5. [DOI] [PubMed] [Google Scholar]
- [56].Wimmer U, Wang Y, Georgiev O, Schaffner W. Nucleic Acids Res. 2005;33:5715–27. doi: 10.1093/nar/gki881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Amantana A, Vorachek WR, Butler JA, Ream W, Whanger PD. J Inorg Biochem. 2004;98:1513–20. doi: 10.1016/j.jinorgbio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- [58].Amantana A, Vorachek WR, Butler JA, Costa ND, Whanger PD. J Inorg Biochem. 2002;91:356–62. doi: 10.1016/s0162-0134(02)00453-1. [DOI] [PubMed] [Google Scholar]
- [59].Aso T, Conaway JW, Conaway RC. J Biol Chem. 1994;269:26575–83. [PubMed] [Google Scholar]
- [60].Gross P, Oelgeschlager T. Biochem Soc Symp. 2006:225–36. doi: 10.1042/bss0730225. [DOI] [PubMed] [Google Scholar]
- [61].Arkhipova IR. Genetics. 1995;139:1359–69. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dreher I, Jakobs TC, Kohrle J. J Biol Chem. 1997;272:29364–71. doi: 10.1074/jbc.272.46.29364. [DOI] [PubMed] [Google Scholar]
- [63].Hernandez A, Lyon GJ, Schneider MJ, St Germain DL. Endocrinology. 1999;140:124–30. doi: 10.1210/endo.140.1.6423. [DOI] [PubMed] [Google Scholar]
- [64].Hatfield MBD, Gladyshev V. Selenium its Molecular Biology and Role in Human Health Springer Science+business Media. 2006. [Google Scholar]
- [65].Orozco A, Jeziorski MC, Linser PJ, Greenberg RM, Valverde RC. Gen Comp Endocrinol. 2002;128:162–7. doi: 10.1016/s0016-6480(02)00071-0. [DOI] [PubMed] [Google Scholar]
- [66].Colgan J, Manley JL. Proc Natl Acad Sci U S A. 1995;92:1955–9. doi: 10.1073/pnas.92.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Black DL. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- [68].Kim E, Goren A, Ast G. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- [69].Lopez AJ. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- [70].Thierry-Mieg D, Thierry-Mieg J. Genome Biol. 2006;7(Suppl 1):S12 1–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bartha T, Kim SW, Salvatore D, Gereben B, Tu HM, Harney JW, Rudas P, Larsen PR. Endocrinology. 2000;141:229–37. doi: 10.1210/endo.141.1.7282. [DOI] [PubMed] [Google Scholar]
- [72].Ohba K, Yoshioka T, Muraki T. Mol Cell Endocrinol. 2001;172:169–75. doi: 10.1016/s0303-7207(00)00368-3. [DOI] [PubMed] [Google Scholar]
- [73].Pfeifer H, Conrad M, Roethlein D, Kyriakopoulos A, Brielmeier M, Bornkamm GW, Behne D. FASEB J. 2001;15:1236–8. [PubMed] [Google Scholar]
- [74].Puglisi R, Tramer F, Panfili E, Micali F, Sandri G, Boitani C. Biol Reprod. 2003;68:405–11. doi: 10.1095/biolreprod.102.006544. [DOI] [PubMed] [Google Scholar]
- [75].Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, Muntoni F, Topaloglu H, Guicheney P. Nat Genet. 2001;29:17–8. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- [76].Osborne SA, Tonissen KF. BMC Genomics. 2001;2:10. doi: 10.1186/1471-2164-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sun QA, Zappacosta F, Factor VM, Wirth PJ, Hatfield DL, Gladyshev VN. J Biol Chem. 2001;276:3106–14. doi: 10.1074/jbc.M004750200. [DOI] [PubMed] [Google Scholar]
- [78].Rundlof AK, Carlsten M, Giacobini MM, Arner ES. Biochem J. 2000;347(Pt 3):661–8. [PMC free article] [PubMed] [Google Scholar]
- [79].Lescure A, Gautheret D, Carbon P, Krol A. J Biol Chem. 1999;274:38147–54. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- [80].Pushpa-Rekha TR, Burdsall AL, Oleksa LM, Chisolm GM, Driscoll DM. J Biol Chem. 1995;270:26993–9. doi: 10.1074/jbc.270.45.26993. [DOI] [PubMed] [Google Scholar]
- [81].Suske G. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- [82].Dynan WS, Tjian R. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- [83].Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Adv Cancer Res. 1998;72:141–96. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.