Abstract
Abnormally expanded polyglutamine domains in proteins are associated with several neurodegenerative diseases, of which the best known is Huntington’s. Expansion of the polyglutamine domain facilitates aggregation of the affected protein, and several studies directly link aggregation to neurotoxicity. The age of onset of disease is inversely correlated with the length of the polyglutamine domain; this correlation motivates an examination of the role of the length of the domain on aggregation. In this investigation, peptides containing 8 to 24 glutamines were synthesized, and their conformational and aggregation properties were examined. All peptides lacked secondary structure. Fluorescence resonance energy transfer (FRET) studies revealed that the peptides became increasingly collapsed as the number of glutamine residues increased. The effective persistence length was estimated to decrease from ~11 Å to ~7 Å as the number of glutamines increased from 8 to 24. A comparison of our data with theoretical results suggests that phosphate-buffered saline is a good solvent for Q8 and Q12, a theta solvent for Q16, and a poor solvent for Q20 and Q24. By dynamic light scattering, we observed that Q16, Q20 and Q24, but not Q8 or Q12, immediately formed soluble aggregates upon dilution into phosphate buffered saline at 37°C. Thus, Q16 stands at the transition point between good and poor solvent, and between stable and aggregation-prone peptide. Examination of aggregates by transmission electron microscopy, along with kinetic assays for sedimentation, provided evidence indicating that soluble aggregates mature into sedimentable aggregates. Together, the data support a mechanism of aggregation in which monomer collapse is accompanied by formation of soluble oligomers; these soluble species lack regular secondary structure but appear morphologically similar to the sedimentable aggregates into which they eventually mature.
Keywords: aggregation, dynamic light scattering, fluorescence resonance energy transfer (FRET), peptide conformation, polyglutamine
INTRODUCTION
The “CAG” diseases are a collection of nine known neurodegenerative disorders, including Huntington’s disease.1 These disorders are causally linked to abnormally extended tracts of CAG codons in disease-specific genes. Expression produces a protein with an expanded polyglutamine (polyQ) domain; invariably, aggregates of the affected protein are found in neuronal inclusions.2, 3 Expansion of the polyQ domain beyond a critical length results in a disease phenotype; longer polyQ regions correspond to an earlier age of onset and a greater severity of symptoms.4 The proteins involved in the CAG diseases have no sequence, compositional, or structural homologies, with the exception of the polyQ stretch,5 which varies in both length and placement within the primary sequence of the various disease-associated proteins.6
The expanded polyQ domain facilitates aggregation of the affected protein, and several studies have directly linked aggregation to toxicity. For example, delivery of synthetic polyQ peptide aggregates to the nucleus was shown to cause cell death,7 and the molecular chaperones Hsp40 and Hsp70 effectively decreased aggregation and toxicity in a zebrafish model of polyQ aggregation.8 Onset of disease is more likely due to gain of toxic function rather than loss of normal function of the affected protein.9 As one piece of evidence for this statement, addition of an extended polyQ tract into proteins unrelated to the known polyQ diseases produced polyQ-like aggregates and neurodegeneration in mice.10 Some research suggests that large polyQ aggregates may be toxic,11, 12 but others have argued that a specific monomer conformation,13 or soluble oligomers,14 are responsible for cell death. Alternatively, the polyQ region may interact with other cell components, preventing them from fulfilling their functions and thus causing cell death. In particular, polyQ may interact with transcription factors, thereby disrupting transcriptional regulation,15 as transcription factors have been found within nuclear inclusions.16
PolyQ peptides have been used in many studies as a tool for examining conformation and aggregation.7, 17–19In vitro, longer polyQ domains aggregate faster.20 Aggregation kinetic data were used to propose a nucleation-elongation model of polyQ peptide aggregation, in which the aggregation nucleus is a structured monomer in equilibrium with the bulk pool of disordered monomers; the equilibrium constant, though small, increases with increasing polyQ chain length.20, 21 However, recent work in our group demonstrated that the monomer loss data, used to support the hypothesis of a critical monomeric nucleus,20 is insufficient to firmly establish a specific kinetic model, since a number of alternative kinetic mechanisms can capture the monomer loss data equally well.22 Elucidation of the mechanism of aggregation requires a closer examination of monomer conformation and its relationship to aggregation.
PolyQ monomers lack regular secondary structural elements, as determined by both CD and NMR.17, 23–25 While polyQ monomers are disordered, they may still preferentially adopt collapsed or extended conformations. Theoretical studies have begun to probe the structure and conformation of polyQ peptides. Discontinuous molecular dynamics simulations revealed that, although some α and β backbone–backbone hydrogen bonds form in isolated polyQ monomers, there are not as many of these bonds as there are in a full α-helix or β-sheet.26 Another molecular dynamics study concluded that the driving force for intermolecular associations in polyQ aggregation is governed by spontaneous conformational fluctuations within monomeric polyglutamine, and thus aggregation is unlikely to follow a homogeneous nucleation mechanism with the monomer as the critical nucleus.27
Two experimental results have led to distinctly different conclusions about polyQ peptide conformation. A recent study, utilizing a tryptophan/cysteine contact quenching technique to measure the kinetics of loop closure, concluded that polyQ peptides adopt unusually extended conformations.28 However, other researchers, using florescence correlation spectroscopy29 to determine translational diffusion coefficients, reported that polyQ peptides adopt highly collapsed conformations. The clear discrepancy in these results warrants further investigation.
In this study we systematically varied the length of polyQ peptides from 8 to 24 residues and examined the structure, conformation, and aggregation properties as a function of length. We employed fluorescence resonance energy transfer (FRET), which has previously been successfully applied to the study of the end-to-end distance of disordered peptides,30–32 to measure whether polyQ peptides are extended or collapsed in aqueous solution. We also examined the aggregation kinetics using a combination of size exclusion chromatography, laser light scattering, sedimentation, and electron microscopy.
Using FRET, we observed an increasing propensity for collapsed conformations as the number of glutamine residues is increased. Peptides containing 8 or 12 glutamines were relatively extended in solution and remained monomeric. Peptides containing 20 or 24 glutamines were relatively collapsed in solution and immediately associated into large soluble aggregates, which subsequently formed sedimentable aggregates whose sedimentation kinetics fit a sigmoidal curve. A comparison of the morphology of aggregates formed early and late in the aggregation process suggests that maturation of structure includes realignment within the aggregates themselves. Peptides containing 16 glutamines were intermediate in behavior; these formed large soluble aggregates that did not precipitate. The data support a mechanism of aggregation in which monomer collapse drives the formation of soluble oligomers. These soluble species lack regular secondary structure but appear morphologically similar to the sedimentable aggregates into which they eventually mature.
RESULTS
Peptides of the type K2WQnXK2 were synthesized, with n = 8, 12, 16, 20, and 24. Flanking lysine residues were added to the polyQ core to increase solubility, consistent with previous work,20 while tryptophan served as a FRET donor and for concentration determination. The N-terminus was acetylated and the C-terminus was amidated to eliminate the possibility of charge interactions of the termini. The residue X was either alanine or dansylated lysine. Peptides will be referred to subsequently as Q8, Q12, Q16, Q20, and Q24 when X = alanine, and Q8D, Q12D, Q16D, Q20D, and Q24D when X = dansylated lysine.
Circular dichroism
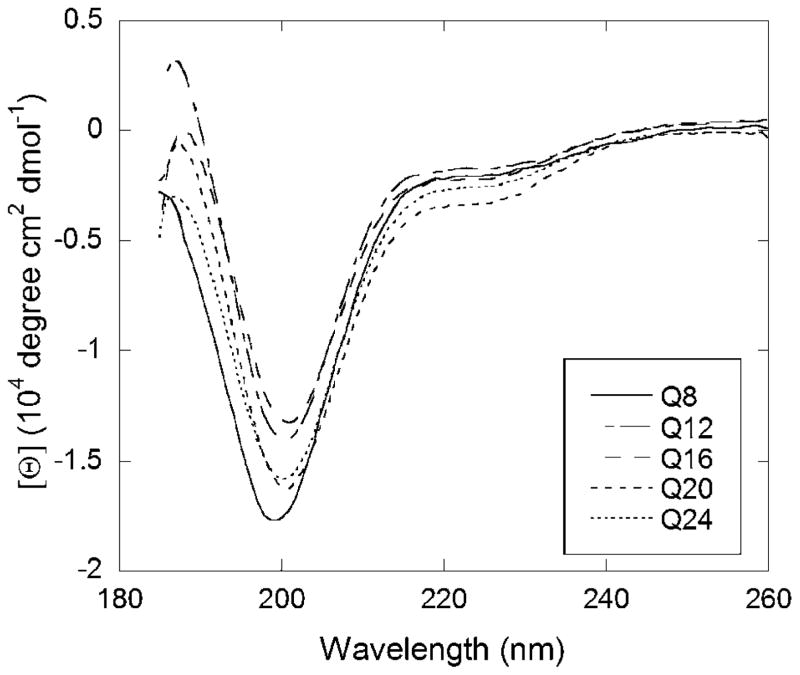
Circular dichroic spectra were collected for solutions of Q8, Q12, Q16, Q20 and Q24. The spectra (Figure 1) are consistent with that of a disordered peptide. Slight variations between the peptides are likely a consequence of tryptophan making up a different percentage of each peptide, as tryptophan can have an impact on CD spectra33 Spectra taken after 13 days (data not shown) are similar to those taken immediately after dilution into buffer. Our data are consistent with other CD results in the literature, indicating monomeric polyQ peptides occupy a disordered state regardless of length.17, 24, 34 NMR data indicated that all Q’s in polyQ regions experience a similar chemical environment,23 again supporting the conclusion that polyQ peptides under these conditions lack regular defined secondary structural elements.
Figure 1. CD spectra of polyQ peptides.
Peptide stock solutions were diluted into a pH 7.4 phosphate buffer, (10 mM buffer salts, 140 mM NaF) to a concentration of 20 μM peptide, and filtered through a 0.45 μm membrane directly into a cuvette. Data was normalized to per residue molar ellipticity.
FRET of polyQ monomers
We next sought to determine the average extension of the polyQ peptide in solution. CD spectra indicated that the peptides are disordered and lack regular secondary structure, but that does not necessarily mean that they are true random coils. We inserted a fluorescent donor (Trp) and acceptor (dansyl) at each end of the polyQ stretch. Dipole-dipole interactions between the fluorescence donor and acceptor allow the transfer of excited state energy from donor to acceptor. The efficiency of energy transfer E is a function of the distance between donor and acceptor.37 For a rigid material, E is related to the distance R between donor and acceptor as
| (1) |
where R0 is the Forster radius for the donor-acceptor pair. We assumed R0 = 21.0 Å for the Trp-dansyl pair based on literature data.30, 35, 36 R0 varies somewhat with peptides and solvents; for instance, Singh measured R0 for dansyl-(AGQ)3-W as 19.3 Å in water and 19.55 Å in 6 M GdmCl.28 We decided to use 21 Å since it is a more established value.
Data were collected with the peptides at 5 μM in buffer at physiological pH (7.4) and salt concentration (10 mM buffer salts plus 140 mM NaCl). The measured E was fit to Eq. (1) to obtain R; results are summarized in Figure 2A. Generally, R increased as the number of glutamines increased. Limited experiments were conducted at lower peptide concentration (1 μM), and over a 2-week incubation time. We observed no change in R with concentration or with time (data not shown), suggesting that the measured R value is for monomers.
Figure 2. Length of polyQ region determined by FRET.
Fluorescence spectra of peptides with and without the fluorescence acceptor dansyl were collected and used to calculate the distance between the fluorophores R. Peptides were diluted to 5 μM in pH 7.4 PBSA (□) or pH 12 buffer (●). Readings were taken in duplicate, and error bars indicate the standard deviation of the measurement. (a): Average length of polyQ region determined by FRET. (b) Persistence length, lp.
Eq. (1) is true for homogeneous rigid materials. Two additional factors must be considered in analyzing FRET for disordered polypeptides: diffusion of the ends during the fluorescence lifetime of the donor, and conformational heterogeneity. The intramolecular diffusion can be ignored if
| (2) |
where D is the intramolecular diffusion coefficient (relative diffusion of donor and acceptor), τF is the fluorescence lifetime of the donor, and s is the mean distance between donor and acceptor.37 For peptides similar to ours containing 16 glutamines, D ≈ 2 × 10−7 cm2/s.28 Fluorescence lifetimes for tryptophan depend somewhat on the peptide and the solvent but are typically less than 3 ns at pH 7.4.38 From our data, we estimate s ≈ 20 Å. Thus, DτF/s2 ≈ 0.02 ≪ 1, so it is reasonable to neglect diffusion during the fluorescence lifetime of the donor.
To address the issue of conformational heterogeneity, we assumed that the peptides populate a conformational ensemble with a probability distribution of donor-acceptor distances P(r). The measured energy transfer is therefore an average <E> over the conformational distribution P(r):
| (3) |
and the measured R in Figure 2A is an average distance between donor and acceptor:
| (4) |
We considered several different models of polypeptide conformation in an effort to interpret the FRET data. If the behavior of the polypeptides could be modeled as freely jointed chains having a Gaussian distribution,39 then
| (5) |
where lb = the bond length and is taken to be 3.8 Å, the distance between repeating atoms in a polypeptide backbone.40 The Gaussian chain model has been used to describe the conformational distribution of unfolded polypeptides in other studies, frequently with the bond length lb as a fitted parameter; the model is better suited for longer chains.41 We inserted the distribution given in Eq. (5) into Eq. (3) to calculate RFJ, the average distance between donor and acceptor one would expect for a freely-jointed chain with a bond length equal to one residue. Alternatively, if the polypeptides were modeled as fully extended rigid rods, then the donor-acceptor distance is simply Lc = (n + 1)lb, where n is the number of glutamines. (We neglect the displacement of the side chains from the backbone in this analysis.) The measured values of R fall between the freely-jointed chain RFJ and the fully extended rod Lc, as shown in Table 1. Thus, the behavior of our polyglutamine peptides appears to fall in-between these two limiting cases.
Table 1.
Measured (R) for polyQ peptides in PBSA, compared to peptide chain distances for a freely jointed chain (RFJ), a fully extended chain (Lc) and a chain with excluded volume in a theta solvent (Rθ), calculated as described in the text.
| No. of Q | R (Å) | RFJ(Å) | Lc(Å) | Rθ(Å) |
|---|---|---|---|---|
| 8 | 19.9 ± 0.5 | 13.3 | 34.2 | 17.1 |
| 12 | 21.2 ± 0.1 | 15.2 | 49.4 | 20.5 |
| 16 | 22.5 ± 0.6 | 16.6 | 64.6 | 23.5 |
| 20 | 22.4 ± 0.3 | 17.7 | 79.8 | 26.1 |
| 24 | 23.9 ± 0.2 | 18.6 | 95.0 | 28.5 |
Vitalis et al. recently completed molecular dynamic simulations of polyglutamine peptides (Ac-Qn-Nm), and calculated average distances in a chain as a function of length and temperature.27 They proposed that the average end-to-end distance for polyglutamine in a theta solvent could be estimated from Rθ = 5.7(n+1)0.5, with Rθ in Å. We calculated Rθ from this equation as another point of comparison with the experimental data. This comparison indicates that Q8 and Q12 are more extended than one would expect for polyglutamine in a theta solvent (R > Rθ), while Q20 and Q24 are more collapsed than one would expect in a theta solvent (R < Rθ). This analysis suggests that PBS becomes less able to solvate polyQ peptides with increasing Q length, and that the peptides become relatively more collapsed as the number of glutamines increases.
As an alternative analysis, we postulated that our polyQ peptides could be modeled as semiflexible (wormlike) chains.28, 42, 43 The wormlike chain model has been used with reasonable success to describe the conformational distributions of unfolded polypeptides and flexible linkers within folded proteins.41 We used an approximate analytical expression derived by Hyeon and Thirumalai42 for P(r) as a function of Lc (the fully-extended distance between donor and acceptor), L (the total length of the peptide from N to C terminus), and lp, the persistence length, which is a measure of the extent to which the chain persists in the same direction. In calculating L, we neglected the fact that the donor and acceptor are on side chains; we estimated that this neglect would affect the calculated value of lp by less than 10% and would not change our conclusions. The wormlike chain model can account for polymer chains anywhere from a stiff rigid rod (high lp) to a collapsed chain (low lp). This distribution does not account for excluded volume effects, which become more important as L increases and as lp decreases but should be negligible for peptides of the length studied here.41 We numerically integrated Eq. (3) using P(r) from Hyeon and Thirumalai42 for each peptide length, as a function of lp, calculated R per Eq. (4) and regressed the measured R value from the FRET data to find the least-squares best fit of lp. Results of these calculations are presented in Figure 2B. There is a smooth decrease in the effective persistence length as the number of glutamines increases. Thus, the peptides become progressively more collapsed as the number of glutamines increases. This conclusion is similar to that obtained from the analysis summarized in Table 1.
Aggregation kinetics
We next used laser light scattering to test for the presence of aggregates in peptide solutions. No scattering above background was detected from solutions of Q24 at pH 3 over 24 hours (data not shown), which we interpreted to indicate that the stock solution was unaggregated. Peptides (Q8, Q12, Q16, Q20, and Q24) were diluted from stock solutions into PBSA to a concentration of 20 μM and held at 37°C. Both the average apparent hydrodynamic radius, Rhz, and the normalized scattered intensity at 90°, Inorm were measured. With Q8 and Q12, we did not detect scattering intensities above background over the course of 24 hours (not shown), therefore Inorm ~ 0 and Rhz was not measurable. Thus Q8 and Q12 did not aggregate to any measurable extent when diluted into PBSA.
For Q16, Q20, and Q24, scattered light was detectable, and soluble aggregates formed essentially immediately after sample preparation. For these samples, the average apparent Rhzincreased continuously over time and showed little variation with the length of the peptides, as shown in Figure 3. Since Rhz is heavily weighted toward larger particles, this indicates that aggregate sizes for all 3 peptides are similar. Rhz reached ~100 nm after approximately 1 h; particles grew to an apparent radius of several hundred nanometers within 4 h (Figure 3). For these 3 peptides, the total normalized scattering intensity, Inorm, increased over time for the first ~50 minutes (Figure 4A). All samples extrapolate to zero intensity at time zero, supporting the notion that the peptides are not aggregated in the low pH stock solution and that dilution into PBSA at 37°C initiates the aggregation process. Inorm increased with Q length: Inorm for Q20 was about 30% greater than that for Q16, and Inorm for Q24 was about 2 times that of Q20.
Figure 3. Dynamic light scattering hydrodynamic radius of polyQ peptides.
Disaggregated peptide stock solution was diluted to a concentration of 20μM in PBSA, then immediately filtered directly into a light scattering cuvette and held at 37°C. Apparent z-averaged hydrodynamic radius Rhz, as determined from cumulants analysis of autocorrelation data collected at 90° scattering angle. Data shown for Q16 (■), Q20 (○), and Q24 (▲). Q8 and Q12 did not scatter above background.
Figure 4. Scattered light intensity of polyQ peptides.
Data shown for Q16 (■), Q20 (○), and Q24 (▲). Q8 and Q12 did not scatter above background. (a) Intensity of scattered light at 90°. Scattering due to the solvent was subtracted and results were normalized to the scattering intensity of toluene to account for changes in laser strength and aperture and to the mass concentration of peptide. (b) Total intensity of scattered light over a longer period of time.
Note that Inorm ∝ 〈M〉w P(90°), where 〈M〉 w =Σwi Mi is the weight-averaged molecular weight when wi is the weight fraction of species with molecular weight Mi, and P(90°) is a particle scattering factor that depends on particle shape and size. Aggregate sizes (as measured by Rhz) are roughly equal for Q16, Q20, and Q24 (Figure 3). Assuming that particle shapes are equivalent, then P(90°) is roughly equal for all 3 peptides, and the differences in Inorm among the peptides likely indicate differences in <M>w. We assumed that there are only monomer and soluble aggregate populations during this time (see SEC data, below), and that the molecular weight of the aggregates is much greater than that of the monomers, or Magg ≫ Mmon. With these assumptions, 〈M〉w = wmonMmon + Σwagg Magg, and increases in Inorm reflect an increase in the Σ wagg Magg term. Because the average size Rhz is not much different for Q16, Q20, and Q24, as long as aggregate shapes are similar then Magg is similar for all 3 peptides, and therefore the increase in Inorm with increasing Q length is primarily due to an increase in wagg, the mass fraction of peptide that is incorporated into soluble aggregates.
At times longer than 1 h, Inorm plateaued for Q16 and Q20, and decreased for Q24. There are two possible explanations for these observations. First, as Rhz increases, P(90°) decreases sharply. For example, at our conditions P(90°) for spheres = 0.9 if Rh = 30 nm but 0.27 if Rh = 100 nm. Thus, an increase in <M>w could be offset by an equivalent decrease in P(90°), leading to little change in Inorm. Under some circumstances the decrease in P(90°) could more than offset an increase in <M>w. Second, any loss of material due to precipitation could cause a decrease in Inorm.34 Since we did not detect any visible precipitates at this early time in any of the samples, we suspect that the first explanation is the most likely.
TEM
We prepared solutions of all 5 peptides in PBSA and examined them by TEM 3 h after preparation. In agreement with the light scattering data, we failed to see aggregates in the Q8 and Q12 samples. Aggregates were observed with both Q20 (Figure 5A) and Q24 (Figure 5B), although they were sparse, particularly in Q20. No aggregates were found in the Q16 sample. The gross morphology of Q20 and Q24 aggregates were similar, as both appeared as loose clusters of linear structures of similar size. However, there were some differences at a finer resolution. Q20 clusters contained many short, poorly defined rod-like structures. In contrast, Q24 aggregates showed a higher degree of order, with more clearly defined, longer rod-like structures. However, the rods themselves were not highly aligned.
Figure 5. TEM image of polyQ peptide aggregates.
Samples were prepared at 20 μM in PBSA, and then incubated at 37°C prior to imaging. Images are representative of a large number of fields examined. The length of the white bar is 200 nm. (a) Q20, 3 hours incubation. (b) Q24, 3 hours incubation. (c) Q20, 40 days incubation. (d) Q16, 40 days incubation.
Solutions were incubated for 40 days and then imaged. Again, no aggregates were detected in the Q8 and Q12 samples. Aggregates were observed in Q16, Q20 and Q24; these were more plentiful than in the early-time samples. Figure 5C shows a representative Q20 aggregate while Figure 5D shows a Q16 aggregate. Morphologically similar aggregates were observed in Q24 samples (not shown). The mature aggregates shown in Figure 5C and 5D are of similar cluster size, but exhibit a more ordered appearance than the immature aggregates of Figure 5A. For example, the aged Q20 aggregates are clearly fibrillar, with laterally aligned rods that are very well defined. Frequently, the ends of bundled fibrils contact the sides of a single rod, but it was rare to observe rods piled on top of one another, in contrast to the early aggregates. The aged aggregates of Q16 shown in Figure 5D exhibit a high degree of ordered bundling of fibrils.
Sedimentation assay
A sedimentation assay was used to study kinetics of growth of insoluble aggregates. Results are shown in Figure 6. The sedimentation assay utilizes the increased density of insoluble aggregates to determine the mass of peptide left in the supernatant, which includes both monomer and soluble aggregates. No sediment was observed in Q8 or Q12 samples even after 35 days of aggregation. This result is consistent with light scattering and TEM data, as we were unable to detect aggregation in these peptides. With Q16, we did not measure any sedimentable aggregates even after 35 days, despite evidence from light scattering and TEM that aggregates exist. This indicates that either the aggregates present in Q16 remain soluble, or the mass fraction of sedimentable aggregates in the sample is smaller than the error in this measurement (< ~5%). Sedimentable aggregates clearly form in the case of Q20 and Q24, with Q24 aggregating more quickly and to a greater extent than Q20. Both Q20 and Q24 undergo a lag phase before sedimentable aggregates are detected. For Q20, the lag phase lasts approximately 5 days, while Q24 aggregates after 3 days.
Figure 6. Sedimentation kinetics of polyQ peptides.
Peptides [Q8 (△), Q12 (□), Q16 (■), Q20 (○), and Q24 (▲)] were diluted into PBSA at 20 μM, aliquoted into tubes, and incubated at 37°C. At the times indicated, a tube was centrifuged and the peptide concentration in the supernatant was determined using a BCA assay. Data is reported as the wt% peptide soluble, calculated from the ratio of the supernatant to the total concentration in an uncentrifuged sample. Lines are drawn as an aid to the eye.
SEC analysis
Freshly prepared peptide samples were injected onto a calibrated size exclusion column (3–70 kDa range) and elution peaks were monitored by absorbance. In all cases, a single peak was observed, corresponding to a molecular weight slightly larger than expected based on column calibration. Q8 eluted at a molecular weight of 2420 Da (1854 expected), Q12 at 2939 (2366 expected), Q16 at 3181 (2879 expected), Q20 at 3892 (3391 expected), and Q24 at 5000 (3904 expected). No peaks eluting at higher molecular weights were detected. Identical samples were injected without the column in place, and the peak area of the eluting material was measured. The percent monomer recovered was calculated as the area of the monomer peak divided by the total peak area without the column. The % recovered decreased from ~90% for Q8 and Q12, to 83% for Q16, to ~76–78% for Q20 and Q24 on the fresh samples. The material that was not recovered in the monomer peak could include monomers that are adsorbed or otherwise lost elsewhere in the system, soluble oligomers that elute as a broad band rather than a peak and therefore are not detected above baseline, and/or aggregates that adsorb to, or are trapped by, the column or filter and do not elute.
Repeated injections of Q16, Q20 and Q24 were made over a 35-day period (Table 2). The elution time of the monomer peak did not change (data not shown). The monomer peak area for each peptide and each day was normalized by the peak area without the column in place (% recovered), or the monomer peak area on day 0 (% initial). These data mirrored the trends observed with the sedimentation assay. Specifically, there was a lag time during which the monomer peak area did not change, and when the peak area did decrease, the change was detected earliest for Q24 and latest for Q16. Numerical values for monomer recovery at 35 days are consistent with the results from the sedimentation assay (Figure 6).
Table 2.
Mass fraction of PolyQ peptide monomer measured by SEC. Peptides were diluted into PBSA at 20 μM, incubated at 37°C, and solutions were analyzed by size exclusion chromatography. % recovered equals the peak area of the monomer divided by the peak area of the peptide injected without the column, times 100%. % initial equals the peak area of the monomer divided by the peak area of the monomer at 0 days, times 100%. ND = not determined.
| Time (days) | Q8 | Q12 | Q16 | Q20 | Q24 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| %rec | %initial | %rec | %initial | %rec | %initial | %rec | %initial | %rec | %initial | |
| 0 | 88 | 100 | 90 | 100 | 83 | 100 | 76 | 100 | 78 | 100 |
| 2 | 93 | 105 | 88 | 97 | 83 | 100 | 76 | 99 | 75 | 97 |
| 5 | 95 | 107 | 95 | 105 | 86 | 104 | 75 | 99 | 80 | 102 |
| 10 | ND | ND | ND | ND | 86 | 104 | 77 | 102 | 72 | 92 |
| 35 | ND | ND | ND | ND | 67 | 81 | 47 | 61 | 19 | 24 |
Effect of pH on mean end-to-end distance and aggregation
As described earlier, we designed our peptides with two lysines flanking both ends of the polyglutamine stretch. Thus, at pH 7.4, the peptides have a net +4 charge. These lysines facilitate solubility, but likely also influence the conformational and aggregation properties through electrostatic repulsion. We diluted disaggregated peptide into buffer at pH 12, above the pKa of the lysine side chain, where the lysines should be uncharged. FRET data were collected and analyzed to obtain values of R at pH 12. Neutralization of the lysine charges led to a small (~5%) decrease in R for Q8 relative to the results in PBS. The effect of pH was dramatically greater for the longer peptides, with R for Q24 decreasing by ~25% with the change from pH 7.4 to pH 12. In fact, at pH 12, R was essentially independent of peptide length (Figure 2A).
We fitted the data to P(r) for a wormlike chain as described earlier, and found that the effective persistence length decreased from ~9 Å for Q8 to less than 3 Å for Q24 (Figure 2B). Thus, the electrostatic repulsion provided by the charged lysines at neutral pH masks the intrinsic tendency for polyQ peptides to collapse, and the influence of the flanking residues becomes increasingly important as the Q length increases.
We examined peptide solutions at pH 12 by light scattering. For Q8 and Q12, although there was some scattering above background, the signal was weak and there was no growth in aggregate size or scattering intensity over a 4-hour measuring period. For Q16 at pH 12, scattered intensity and growth kinetics were similar to that obtained in PBS. For Q20, scattered intensity was about 4 times stronger at pH 12 than in PBS and the growth kinetics were similar. We were unable to collect light scattering data for Q24 because the peptide immediately formed insoluble precipitates upon dilution into pH 12 buffer. Thus, the extent of aggregation was accentuated by neutralization of lysines, but the Q16 still sat at the transition point separating non-aggregating and aggregating lengths of polyglutamine peptides.
DISCUSSION
Two groups who have examined the conformation of polyQ peptides in aqueous solution came up with surprisingly different conclusions. Tryptophan-cysteine triplet quenching experiments led one group to conclude that polyQ peptides are unusually extended.28 Based on fluorescence correlation spectroscopy, another group concluded that polyQ peptides are collapsed and that water is a surprisingly poor solvent.29 There are two key differences between the peptides used by the two groups that may explain these discrepant results: length and charge. Singh and Lapidus worked with peptides of 4 to 16 Q’s, while Crick et al. used peptides containing 15 to 53 Q’s. Second, Singh and Lapidus worked with peptides with two flanking lysines at either end of the polyglutamine domain (KKWQnCKK), similar to our peptides, whereas Crick et al. used peptides of the form GQnC*KK, where C* indicates a cysteine coupled to a fluorescent dye that carries two negative charges. Thus, the Singh peptide has a net charge of +4, whereas the Crick peptide contains both positive and negative charges but is overall neutral.
As an alternative approach to the conformational question, we used FRET to measure the mean end-to-end distance of monomeric polyQ peptides of Q lengths varying from 8 to 24 residues. We observed that the average R increased only slightly as the number of Q residues increased (Figure 2). We interpreted the measured R by comparing to calculated values for freely-jointed chains with a bond length of one residue (RFJ), fully extended (rigid) chains (Lc), or polyQ peptides in a theta solvent (Rθ) (Table 1). In PBS, R for all 5 peptides was greater than that for the freely-jointed chain but significantly less than that for a fully extended chain. Q8 is relatively extended (R/Lc = 0.58, R/Rθ = 1.16), whereas Q24 is relatively collapsed (R/Lc = 0.25, R/Rθ = 0.84). This comparison led us to conclude that PBS is a good solvent for our shorter peptides (e.g., Q8) but a poorer solvent for longer peptides (e.g., Q24). The transition (R ~ R θ) occurs at Q16.
To look at the data in a different way, we also used a wormlike chain model to calculate an effective persistence length. The persistence lengths shown in Figure 2B clearly trend downward with increasing polyQ length, from 11 Å for Q8 to 7 Å for Q24. Singh and Lapidus reported a persistence length of 13.5 Å for their series of short polyQ peptides.28 Their value is similar to, but somewhat greater than, our value of 11 Å for Q8 in PBS; the difference could be explained by the fact that triplet quenching and FRET sample different parts of the conformational distribution, and/or inaccuracies in the wormlike chain model. Thus, our data are consistent with that of Singh and Lapidus: short polyQ peptides with flanking charged lysines are quite extended in aqueous buffer.
Using results from fluorescence correlation spectroscopy measures of translational diffusion times, Crick et al.29 reported that Rh of polyQ peptides varies as N0.32 and therefore concluded that the peptides were highly collapsed, and essentially globular. We calculated the hydrodynamic radius for wormlike chains as a function of lp and N, assuming a chain thickness of 4 Å, and using analytical expressions derived by Yamakawa and Fujii44. We then fit the calculated values to Rh = aNν and found that ν = 0.33 (compact sphere) only when the persistence length is very short (lp = 2 Å). We observed lp < 3Å for Q24 at pH 12. Thus, our data are consistent with that of Crick et al: longer polyQ peptides with no flanking charged residues are highly collapsed in aqueous buffer.
Our data indicated that lp decreases with increasing peptide length. For further comparison with the Crick data, we fit a simple power law expression to lp versus the number of Q’s, NQ, and obtained lp ~ 22 NQ −0.36. If we then use the variable lp to calculate Rh as a function of polyQ peptide length, and plot Rh vs NQ, we find an apparent v ~ 0.30 (Figure 7). In other words, a Q-length-dependent lp flattens a plot of Rh on NQ and leads to an estimate of a smaller v than what one would obtain if one used a constant lp. Without taking into account the variable persistence length, one would conclude that the polypeptides are more collapsed than they actually are. This is one additional possible contribution to the discrepancy between the Singh and Lapidus conclusion that polyQ is extended, and the Crick et al. conclusion that polyQ is collapsed.
Figure 7. Hydrodynamic radius of polyQ peptides from wormlike chain model.
Rh was calculated from the Yamakawa-Fujii equations for a wormlike chain, 44 assuming a chain diameter of 4 Å and a contour chain length (Å) of 3.8N = 3.8(NQ + 6), where N = NQ + 6 accounts for the number of glutamines in the peptide plus 6 flanking residues. Results are shown for lp = 5 Å (□, ν= 0.37), lp = 12 Å (○, ν= 0.44), and lp = 22 NQ −0.36 (●, ν= 0.30).
Our data show that charge (or lack thereof) of the flanking residues strongly influences the conformational properties of polyQ peptides, as illustrated by a comparison of FRET data taken at pH 7.4, where our peptides have a net +4 charge, and at pH 12, where there is no net charge. When the charge on the flanking lysine residues is neutralized by raising the pH to 12, Q8 remains relatively extended, with R > Rθ and lp ~ 9 Å. Thus, even in the absence of electrostatic repulsion, we surmise that Q8 remains relatively well solvated. However, the effect of pH becomes dramatically stronger as the Q length increases. In the absence of electrostatic repulsion from the flanking lysines, R for Q24 approaches the freely-jointed chain limit (RFJ), and lp decreases dramatically, to <3 Å. These results demonstrate the strong intrinsic tendency of longer polyQ peptides towards collapse, as well as the major role that repulsive interactions from flanking residues play in mitigating collapse.
It is useful to compare our estimated lp values to other data in the literature. A short persistence length, 3 Å, was reported for loops in folded proteins.43 From FRET data reported by Tucker et al. for a 14-mer amphipathic peptide, Mastoparan X.45, we also calculated a persistence length of 3 Å More typically, unfolded proteins and polypeptides have persistence lengths ranging from 5 to 9 Å. For example, using single-molecule pulling experiments lp was found to be 6 – 7 Å for unfolded titin.46 Similarly, Buscaglia et al. found lp = 6 – 7 Å for C(AGQ)nW, with n=1–9.47 We applied the wormlike chain analysis to FRET data reported in the literature and calculated lp = 6.3 Å, 6.5 Å, and 9.9 Å for 14-mers of repeating AGQ, AGE, and AQE units, respectively.32 A similar analysis of data from a study of prion repeats30 containing 11, 21, and 31 total residues, yielded estimates of lp = 5.6, 6.8, and 5.1 Å respectively, showing no dependence on the length of the segment, unlike our polyQ peptides. Schuler et al.48 analyzed single-molecule FRET data for polyproline and found lp = 44 Å, with a slight increase with increasing number of Pro residues. This result is not surprising given that polyproline is very stiff and is used as a “molecular ruler”. In comparing these literature values to our results, we conclude that short polyQ peptides in relatively good solvents (e.g., Q8 in PBS) are more extended than typical unfolded peptides, while longer polyQ peptides in the absence of repulsive interactions in flanking residues (e.g., Q24 in pH 12) are highly collapsed.
We propose the following simple hypothesis to explain why polyQ peptides become more collapsed as the Q length increases. Glutamine is considered a hydrophilic polar side chain;40 the amide side chain participates in hydrogen bonding with water as both a donor and acceptor. With short polyQ peptides, we hypothesize that hydrogen bonding between side chain amides and water is prevalent, the peptide is well solvated, and is therefore somewhat more extended than the typical polypeptide. As the number of Q residues and the length increases, random contacts between distant parts of the chain become more likely, and the probability of forming intramolecular hydrogen bonds (either side chain-side chain, side chain-backbone, or backbone-backbone) increases. Formation of multiple intramolecular hydrogen bonds traps the collapsed conformation, leading to a decrease in the effective persistence length. Although the peptides become more collapsed as the number of glutamines increases, they do not develop any regular secondary structure (Figure 1). Our data and hypothesis are consistent with several theoretical studies. For example, Marchut et al. describe polyglutamine monomers at lower temperatures as collapsed random coils.26 Vitalis et al. showed by atomistic simulations that polyglutamine has no preference for a specific, unique conformation.27 A very recent theoretical study proposes that the total number of intramolecular hydrogen bonds, especially those involving side-chain groups, increases as Q length increases from 5 to 30, and that these collapsed peptides remain disordered rather than forming a regular β-sheet structure.49
It is important to note here that the FRET data could be skewed if the peptides were aggregated to a significant extent. Aggregation could place the fluorescence donors/acceptors of multiple peptides in close proximity to each other, causing significant error in the measurement of FRET efficiency and length of the polyQ region. This issue was controlled for by using a low (5 μM) concentration of polyQ peptides and taking measurements at room temperature. Further, R was found to be independent of peptide concentration or incubation time, indicating that aggregation is unlikely to influence the FRET data.
We next examined the aggregation properties as a function of Q length, using a variety of techniques. Previous work in our lab showed that early, soluble aggregates were detected in a polyQ peptide by laser light scattering long before sedimentable aggregates were found using other methods.34 These results were confirmed in the present study. No soluble aggregates were detected for Q8 or Q12 in PBS by light scattering, and none were observed in TEM or by sedimentation assay. Q16 appears to straddle a transition point: we detected immediate formation of soluble aggregates by light scattering and found aggregates in aged samples but not fresh samples by TEM. SEC analysis suggested some loss of monomer for Q16 after 35 days, although there were no sedimentable aggregates. In PBS, Q20 and Q24 both immediately formed soluble aggregates, detectable by both light scattering and TEM. The higher scattered intensity for Q24 compared to Q20, coupled with the similar average size, suggests that the primary difference is the fraction of material incorporated into the soluble aggregates. Sedimentable aggregates, and loss of monomer, were detected in both samples, but only after several days’ incubation. The lag in appearance of insoluble aggregates is consistent with the lag phase observed by other groups for polyQ aggregation.20, 50 This lag phase has been cited as evidence of a nucleation-elongation type mechanism for aggregate formation.19–21 However, the sedimentation assay is only sensitive to insoluble aggregates; the light scattering data of Figures 3 and 4 shows no lag time in formation of soluble aggregates. This result, along with previous work in our lab,34 indicates that the lag phase is a lag in the accumulation of sedimentable aggregates, not a lag in the formation of aggregates themselves.
Our experimental results are summarized in Table 3. Interestingly, the polyglutamine length at which the transition between non-aggregating and aggregating peptides is observed, Q16, is also the length at which the mean end-to-end distance transitions from a good to a poor solvent (R ~ R θ). This result suggests that collapse is strongly correlated with aggregation.
Table 3.
Summary of experimental results. R/Rθ is the ratio of the measured average distance spanned between donor and acceptor R compared to that calculated for a polyQ peptide in a theta solvent, from Vitalis et al.27
| Peptide | R/Rθ (FRET) | Soluble aggregates (LS)? | Aggregates at 3 hr (TEM)? | Aggregates at 40 days (TEM)? | Monomer loss at 35 days (SEC)? | Sedimentation at 35 days? |
|---|---|---|---|---|---|---|
| Q8 | 1.16 | No | No | No | No | No |
| Q12 | 1.03 | No | No | No | No | No |
| Q16 | 0.96 | Yes | No | Yes | Yes (19%) | No |
| Q20 | 0.86 | Yes | Yes | Yes | Yes (39%) | Yes (37%) |
| Q24 | 0.84 | Yes | Yes | Yes | Yes (76%) | Yes (76%) |
Neutralization of the lysine charges by adjusting the pH to 12 affected both conformation and aggregation, with a greater effect as the Q length increased. Interestingly, R became essentially independent of Q length under these conditions. This result is consistent with a recent single-molecule study that showed that polyQ domains inserted into an unfolded protein were remarkably resistant to mechanical stretching and that the length of the polypeptide under stretching forces was independent of Q length.51 For the shortest polyQ peptides, neutralization of the charge on the flanking residues has minor impact on R. This result suggests that short polyQ peptides are normally well solvated and prefer extended conformations, independent of the flanking residues. In contrast, in the absence of repulsive interactions, longer polyQ peptides have a strong intrinsic tendency towards collapse. With neutralization of the charge on the flanking residues, both the extent of collapse (as determined by FRET) and the degree of aggregation (as determined by light scattering and observation of precipitates) becomes more strongly dependent on Q length. In other words, repulsive interactions of the flanking residues modulates the otherwise strong Q length-dependence of both conformation and aggregation. Interestingly, the Q length at which a transition from minimal to strong aggregation occurs remains at Q16 at both neutral and basic pH.
Close examination of the TEM images (Figure 5) leads to a number of observations that may shed further light on the mechanism of polyglutamine aggregation. First, the Q20 and Q24 aggregates formed after 3 h are roughly similar in size and shape – both appear as loose clusters of linear aggregates. However, the Q20 aggregates have a more amorphous appearance than Q24, with less sharply defined surfaces. The mature aggregates of Q20 take on a much more defined and highly ordered appearance compared to the early Q20 aggregates, while preserving the general shape and size of the clusters. We did not observe any of the more amorphous aggregates in the aged sample. These observations lead us to propose that the immature soluble aggregates are not “off-pathway”, but undergo a slow internal conformational re-arrangement over time that leads to the highly ordered mature aggregates. The fact that the early Q24 aggregates look more defined than the early Q20 aggregates could indicate that the Q24 peptide matures more quickly than the Q20 aggregate. We hypothesize that the slow internal conformational rearrangement involves making and breaking of multiple hydrogen bonds, and dehydration, with the net effect being an increase in regular β-sheet structure, an increase in packing density and decrease in solubility of the aggregates. If true, then sedimentation is an indication of this conformational re-arrangement within the pre-formed large aggregates. Our interpretation of the data is conceptually consistent with conclusions from a recent theoretical study.49
In the mature aggregates, the degree of order is striking, as the rod-like aggregates seem to have a clear preference for lateral alignment. Interestingly, the ends of the rods may have a similar property. In Figure 5C, there are a number of locations where the ends of multiple rods contact the sides of another rod, and seem to hold it into place. Thus, it is possible that once a certain number of contacts are made, be it through lateral alignment with another rod or with end-to-side contact with multiple rods, the relative positions of the rods become fixed. Additionally, it appears as though most of the aggregates lie in a single plane, with only a few examples of rods lying on top of each other. In other words, the conformational rearrangement leads to increased asymmetry of the rod-like structures, resulting in different properties on the top and bottom faces compared to those of the sides and ends.
We propose a schematic (Figure 8) for Q-length-dependent aggregation that is consistent with our results. Short polyQ peptides occupy extended conformations while longer polyQ peptides adopt collapsed coil conformations; all lack regular secondary structure. Collapse is facilitated by a switch from peptide-solvent hydrogen bonding to intramolecular hydrogen bonding as Q length grows (phase 1). The amide side chain of glutamine allows for side chain-side chain and side-chain backbone hydrogen bonding, as well as backbone-backbone hydrogen bonding. Based on molecular dynamic simulations, all three types of hydrogen bonds are likely to occur in polyglutamine aggregates.26, 49 Water becomes a poorer solvent, and the collapsed polypeptides tend to self-associate (phase 2). The transition occurs at Q16. In phase 3, the aggregation-competent peptides rapidly associate into soluble oligomers that grow over time. The mass fraction of peptide incorporated into soluble oligomers is higher for Q24 than for Q16 or Q20, based on the increased scattering intensity (Figure 4). These soluble aggregates lack regular secondary structure. However, at lower resolution they possess some degree of order: by TEM the early aggregates appear as loose clusters of rodlike particles. In phase 4, aggregates transition to fully mature, insoluble aggregates, characterized by a higher degree of order. The gross morphological similarities between the aggregates imaged after 3 h and those imaged after 40 days suggests that the soluble aggregates are precursors to the insoluble aggregates rather than “off-pathway”. We hypothesize that maturation of the aggregates involves dehydration and an increase in density, thus leading to sedimentation. Over the time frame we investigated, Q20 and Q24 but not Q16 aggregates make this final transition.
Figure 8. Schematic illustrating proposed kinetic pathway.
Phase 1 consists of disordered peptides, with longer peptides showing more collapse. In phase 2, peptides with Q ≥ 16 associate in a nonspecific manner, leading to the amorphous aggregates present in phase 3. The amount of aggregated material in phase 3 is proportional to the number of glutamines. In phase 4, Q24 forms fully mature, structured, insoluble aggregates, while Q16 failed to sediment in 35 days.
MATERIALS AND METHODS
Peptide synthesis and purification
All materials were from Fisher Scientific (Pittsburgh, PA) except where indicated. Peptides were synthesized using standard Fmoc solid phase methods on a PerSeptive Biosystems Pioneer synthesizer. Alanine, glutamines with a Trityl side chain protecting group, and lysines and tryptophan with a Boc side chain protecting group were purchased from Novabiochem (Gibbstown, NJ). Dansylated lysine was purchased from Anaspec (San Jose, CA). The resin used was Fmoc-PAL-PEG-PS from Applied Biosystems (Foster City, CA). Resin sites were partially blocked with lysine-Boc to reduce on-bead aggregation. Extended cycles and double couplings were used to improve yield. The peptides were cleaved from the resin using a solution of 95% trifluoracetic acid (TFA), 2.5% ethanedithiol (Fluka, Buchs, Switzerland) and 2.5% H2O. Peptides were precipitated into cold t-butylmethyl ether and digested in aqueous solvent for 1 hour before lyophilization. Crude peptide was pre-solubilized in a 1:1 solution of TFA and hexafluoroisopropanol (HFIP) (Fluka). This solution was evaporated under gentle N2 flow and the peptide was resuspended in water adjusted to pH 3 before purifying by reverse-phase HPLC on a Vydac C18 column. Peptide was eluted from the column with a linear gradient of acetonitrile and water with 0.1% TFA, starting at 5% and ending at 30% acetonitrile. The peak corresponding to the correct sequence was collected and lyophilized. Peptide identity was confirmed by MALDI-TOF mass spectrometry, which yielded molecular weights of 1854.0 (Q8, 1854.2 theoretical), 2366.2 (Q12, 2366.7 theoretical), 2879.5 (Q16, 2879.2 theoretical), 3390.7 (Q20, 3391.8 theoretical), and 3903.9 (Q24, 3904.3 theoretical). For the dansylated peptides, molecular weights were 2144.0 (Q8D, 2144.2 theoretical), 2656.0 (Q12D, 2656.7 theoretical), 3168.8 (Q16D, 2169.2 theoretical), 3680.8 (Q20D, 3681.7 theoretical), and 4194.0 (Q24D, 4194.2 theoretical).
Sample preparation
Lyophilized peptides were disaggregated using a protocol similar to that developed by others.52 Briefly, peptide was incubated overnight in a 1:1 solution of TFA and HFIP. Solvent was evaporated under a gentle flow of N2 and the peptides were redissolved in water adjusted to pH 3 with TFA. Peptide stock solutions were filtered through a 0.45 μm filter, aliquoted, snap frozen in dry ice and ethanol and stored at −80°C. Prior to each experiment, a vial was thawed and centrifuged at 19 500 rcf for 30 min, and then the supernatant (top 75%) was removed and used immediately. Although the TFA used was the purest available (99.5% TFA for biochemistry, Acros Organics), evaporation of TFA was found to leave a residue that interfered with the absorbance of the peptide at 280nm. Therefore, the concentration of peptide was determined using the fluorescence intensity of standard solutions of tryptophan in 6 M guanidine hydrochloride. The concentration of tryptophan was determined by absorbance at 280 nm using an extinction coefficient of 5500 cm−1M−1. Tryptophan standards of 5 to 50 μM were used to produce a linear standard curve using the emission intensity at 350 nm with an excitation wavelength of 295 nm. Peptide concentrations were determined from the fluorescence intensity of a peptide stock sample diluted into 6 M guanidine hydrochloride fit to the standard curve; the peptide concentration in the stock solution was typically ~200 μM. The concentration of the dansyl peptides was determined using a similar method, with the standard curve based on dansyl lysine (Anaspec), using an extinction coefficent at 330 nm of 4600 cm−1 M−1 to determine the stock concentration,53 and an excitation wavelength of 350 nm and an emission wavelength of 549 nm. Dansyl was used as the standard rather than tryptophan, as tryptophan fluorescence could be quenched by the dansyl group in these peptides.
Fluorescence resonance energy transfer (FRET)
FRET requires both a fluorescence donor and acceptor, where the emission spectrum of the donor overlaps the absorbance spectrum of the acceptor. Excitation of the fluorescence donor results in emission from the donor group. If the acceptor group is located in close proximity to the donor group, excited state energy is transferred from the donor to the acceptor. The decrease in measured emission from the donor is used to calculate the distance between the donor and acceptor groups.54
Peptide samples for FRET experiments were diluted to 5 μM in pH 7.4 PBSA (0.01 M buffer salts, 0.14 M NaCl, and 3 mM sodium azide), or pH 12 potassium chloride buffer (0.01 M KCl and NaOH to adjust to pH 12). Limited additional data were obtained at 1 μM peptide to ascertain the effect of concentration. Fluorescence spectra for solutions of both Qn and QnD peptides were obtained with a QuantaMaster Series spectrofluorometer (PTI, Inc., Birmingham, NJ), with excitation at 295 nm and emission spectra recorded from 310 to 550 nm. Measurements were made in duplicate and at steady state. An excitation wavelength of 295 nm was used to avoid energy transfer between two Trp groups that occurs with excitation at 280 nm.55 FRET efficiency E was calculated as54
| (6) |
where FDA is the donor (tryptophan) fluorescence with the acceptor (dansyl) group present, and FD is the donor fluorescence with the acceptor group absent. The values of FDA and FD in Eq. (6) were determined by integrating the peak area of the emission scan, with background subtracted, from 330–370 nm.
Circular dichroism (CD)
Peptide stock solutions were diluted into phosphate buffer (10 mM K2HPO4/KH2PO4, 140 mM NaF, pH 7.4) to a concentration of 20 μM peptide. Samples were filtered through a 0.45 μm membrane immediately before transfer to a 1 mm cell. CD spectra were collected using an Aviv 202SF circular dichroism spectrometer at 37°C immediately after filtration. Solvent spectra were collected and subtracted. An identical set of samples was produced as described above and incubated in sample tubes for 13 days at 37 °C before collecting CD spectra.
Transmission electron microscopy (TEM)
Peptides at 20 μM were prepared in PBSA and incubated for 3 h or 40 days at 37°C. A drop of sample was applied to a pioloform coated grid and stained with methylamine tungstate stain, then imaged with a Philips CM120 scanning transmission electron microscope.
Sedimentation assay
Peptides were diluted into PBSA to a concentration of 20 μM, distributed into sample tubes in 100 μL aliquots, and incubated at 37°C. At regular time intervals, a sample tube was withdrawn and centrifuged at 19 500 rcf for 30 min. The supernatant (top 75%) was removed and the peptide concentration was determined in triplicate with a BCA (Pierce, Rockford IL) assay in the microplate format, using a BioTek EL 800 Universal Microplate Reader and following the manufacture’s protocol. All concentrations are reported as a percentage of the concentration of an uncentrifuged aliquot.
Laser light scattering
Samples were diluted into PBSA that had been filtered twice with a 0.22 μm filter, then immediately filtered again through a 0.45 μm filter directly into a clean light scattering cuvette. The cuvette was placed in a 37°C bath of the index matching solvent decahydronaphthalene. Data were collected using a Malvern 4700c system and a Coherent Innova 90C argon ion laser operating at 488nm wavelength. The autocorrelation function at 90° scattering angle was measured repeatedly over several hours. The z-averaged diffusivity was determined from the autocorrelation function by the method of cumulants. Rhz, the apparent z-averaged hydrodynamic radius, was calculated from <D>z using the Stokes equation.56 On the same samples and at the same time points, the total intensity of scattered light at 90°, Is(90°), was measured. Scattering of the solvent, Ib(90°), was subtracted from Is(90°) and the result was normalized to the scattered intensity of toluene Itol(90°), to adjust for changes in laser power or aperture, and to the mass concentration of peptide (ctot, mg/mL). Data are reported as a normalized intensity Inorm:
| (7) |
Inorm is proportional to <M>w, the weight-averaged molecular weight, and P(90°), the particle scattering factor at 90° scattering angle:
| (8) |
For small Rg, P(90°) is independent of shape and
| (9) |
where Rg is the radius of gyration and q is the scattering vector. With our system, q = 0.0242 nm−1, so this approximation is valid if Rg < ~40 nm. At larger particle size, P(90°) depends on the particle shape and deviates from Eq. (9); analytical expressions for many particle shapes are available.57
Size exclusion chromatography (SEC)
Samples were prepared as described and incubated at 37°C. Aliquots were removed at regular time intervals and analyzed by SEC on a Superdex 75 PC 3.2/30 column (Pharmacia), with a molecular weight range of 3–70 kDa, using a Waters 625LC system. The mobile phase (PBS, pH 7.4) flow rate was set to 0.1 mL/min, an overfilled 50 μL sample loop was used to apply a standard volume of sample, and peaks were detected by absorbance at 280 nm. The column was calibrated with a set of molecular weight protein standards. The weight fraction of a species eluting from the column at a specific elution time was determined by dividing the area of that peak by the total area of an identical injection without the column in place.
Acknowledgments
This work was supported by grant BES-0330537 from the National Science Foundation and grant 5T32 GM-08349 (RHW) from the National Institute of Health. We gratefully acknowledge the technical assistance of Dr. Gary Case, Dr. Randy Massey, and the UW Biotechnology Center mass spectrometry staff. CD data were obtained at the University of Wisconsin - Madison Biophysics Instrumentation Facility, which was established with support from the University of Wisconsin - Madison and grants BIR-9512577 (NSF) and S10 RR13790 (NIH).
Abbreviations used
- BCA
bicinchoninic assay
- CD
circular dichroism
- FRET
fluorescence resonance energy transfer
- HFIP
hexafluoroisopropanol
- HPLC
high-pressure liquid chromatography
- PBS(A)
phosphate-buffered saline (with azide)
- SEC
size exclusion chromatography
- TEM
transmission electron microscopy
- TFA
trifluoroacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bates G. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT. Beyond the Qs in the polyglutamine diseases. Genes Dev. 2001;15:925–932. doi: 10.1101/gad.888401. [DOI] [PubMed] [Google Scholar]
- 3.Wanker EE. Protein aggregation and pathogenesis of Huntington’s disease: mechanisms and correlations. Biol Chem. 2000;381:937–942. doi: 10.1515/BC.2000.114. [DOI] [PubMed] [Google Scholar]
- 4.Gusella JF, MacDonald ME. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 5.Albrecht M, Golatta M, Wullner U, Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur J Biochem. 2004;271:3155–3170. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- 6.Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Dunlap JR, Andrews RB, Wetzel R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet. 2002;11:2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 8.Schiffer NW, Broadley SA, Hirschberger T, Tavan P, Kretzschmar HA, Giese A, Haass C, Hartl FU, Schmid B. Identification of anti-prion compounds as efficient inhibitors of polyglutamine protein aggregation in a zebrafish model. J Biol Chem. 2007;282:9195–9203. doi: 10.1074/jbc.M607865200. [DOI] [PubMed] [Google Scholar]
- 9.Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 10.Ordway JM, TallaksenGreene S, Gutekunst CA, Bernstein EM, Cearley JA, Wiener HW, Dure LS, Lindsey R, Hersch SM, Jope RS, Albin RL, Detloff PJ. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XQ, Smith DL, Merlin AB, Engemann S, Russel DE, Roark M, Washington SL, Maxwell MM, Marsh JL, Thompson LM, Wanker EE, Young AB, Housman DE, Bates GP, Sherman MY, Kazantsev AG. A potent small molecule inhibits polyglutamine aggregation in Huntington’s disease neurons and suppresses neurodegeneration in vivo. Proc Natl Acad Sci U S A. 2005;102:892–897. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss A, Klein C, Woodman B, Sathasivam K, Bibel M, Regulier E, Bates GP, Paganetti P. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104:846–858. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 14.Shao J, Diamond MI. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum Mol Genet. 2007;16:R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 15.Truant R, Atwal RS, Burtnik A. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington’s disease. Prog Neurobiol. 2007;83:211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Freiman RN, Tjian R. Perspectives: neurodegeneration - a glutamine-rich trail leads to transcription factors. Science. 2002;296:2149–2150. doi: 10.1126/science.1073845. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J Mol Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 18.Slepko N, Bhattacharyya AM, Jackson GR, Steffan JS, Marsh JL, Thompson LM, Wetzel R. Normal-repeat-length polyglutamine peptides accelerate aggregation nucleation and cytotoxicity of expanded polyglutamine proteins. Proc Natl Acad Sci U S A. 2006;103:14367–14372. doi: 10.1073/pnas.0602348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SM, Berthelier V, Hamilton JB, O’Nuallain B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry (N Y) 2002;41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- 20.Chen SM, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci U S A. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharyya AM, Thakur AK, Wetzel R. Polyglutamine aggregation nucleation: thermodynamics of a highly unfavorable protein folding reaction. Proc Natl Acad Sci U S A. 2005;102:15400–15405. doi: 10.1073/pnas.0501651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernacki JP, Murphy RM. Model discrimination and mechanistic interpretation of kinetic data in protein aggregation studies. Biophys J. 2009;96:2871–2887. doi: 10.1016/j.bpj.2008.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masino L, Kelly G, Leonard K, Trottier Y, Pastore A. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett. 2002;513:267–272. doi: 10.1016/s0014-5793(02)02335-9. [DOI] [PubMed] [Google Scholar]
- 24.Altschuler EL, Hud NV, Mazrimas JA, Rupp B. Random coil conformation for extended polyglutamine stretches in aqueous soluble monomeric peptides. J Pept Res. 1997;50:73–75. doi: 10.1111/j.1399-3011.1997.tb00622.x. [DOI] [PubMed] [Google Scholar]
- 25.Klein FAC, Pastore A, Masino L, Zeder-Lutz G, Nierengarten H, Oulad-Abdeighani M, Altschuh D, Mandel J, Trottier Y. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: towards a length-dependent toxicity gradient. J Mol Biol. 2007;371:235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Marchut AJ, Hall CK. Effects of chain length on the aggregation of model polyglutamine peptides: molecular dynamics simulations. Proteins. 2007;66:96–109. doi: 10.1002/prot.21132. [DOI] [PubMed] [Google Scholar]
- 27.Vitalis A, Wang X, Pappu RV. Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization. J Mol Biol. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh VR, Lapidus LJ. The intrinsic stiffness of polyglutamine peptides. J Phys Chem B. 2008;112:13172–13176. doi: 10.1021/jp805636p. [DOI] [PubMed] [Google Scholar]
- 29.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci U S A. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gustiananda M, Liggins JR, Cummins PL, Gready JE. Conformation of prion protein repeat peptides probed by FRET measurements and molecular dynamics simulations. Biophys J. 2004;86:2467–2483. doi: 10.1016/S0006-3495(04)74303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moglich A, Joder K, Kiefhaber T. End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc Natl Acad Sci U S A. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soranno A, Longhi R, Bellini T, Buscaglia M. Kinetics of contact formation and end-to-end distance distributions of swollen disordered peptides. Biophys J. 2009;96:1515–1528. doi: 10.1016/j.bpj.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fasman GD. Circular Dichroism and the Conformational Analysis of Biomolecules. Plenum Press; NY: 1996. p. 737. [Google Scholar]
- 34.Lee CC, Walters RH, Murphy RM. Reconsidering the mechanism of polyglutamine peptide aggregation. Biochemistry. 2007;46:12810–12820. doi: 10.1021/bi700806c. [DOI] [PubMed] [Google Scholar]
- 35.Dunn BM, Pham C, Raney L, Abayasekara D, Gillespie W, Hsu A. Interaction of alpha-dansylated peptide inhibitors with porcine pepsin - detection of complex-formation by fluorescence energy-transfer and chromatography and evidence for a 2-step binding scheme. Biochemistry. 1981;20:7206–7211. doi: 10.1021/bi00528a023. [DOI] [PubMed] [Google Scholar]
- 36.Wu PG, Brand L. Resonance energy-transfer - methods and applications. Anal Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 37.Lakowicz JR. Principles of fluorescence spectroscopy. 2. Springer; NY: 1999. pp. 1–20. [Google Scholar]
- 38.Gudgin E, Lopez-Delgado R, Ware WR. The tryptophan fluorescence lifetime puzzle. A study of decay times in aqueous solution as a function of pH and buffer composition. Canadian Journal of Chemistry. 1981;59:1037. [Google Scholar]
- 39.Flory PJ. Principles of polymer chemistry. Cornell University Press; Ithica, NY: 1953. pp. 404–407. [Google Scholar]
- 40.Creighton TE. Proteins: structures and molecular principles. WH Freeman and Company; NY: 1983. pp. 4–10. [Google Scholar]
- 41.Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 42.Hyeon C, Thirumalai D. Kinetics of interior loop formation in semiflexible chains. J Chem Phys. 2006;124:104905. doi: 10.1063/1.2178805. [DOI] [PubMed] [Google Scholar]
- 43.Zhou HX. Loops in proteins can be modeled as worm-like chains. J Phys Chem B. 2001;105:6763–6766. [Google Scholar]
- 44.Yamakawa H, Fujii M. Translational friction coefficient of wormlike chains. Macromolecules. 1973;6:407–415. [Google Scholar]
- 45.Tucker MJ, Oyola R, Gai F. Conformational distribution of a 14-residue peptide in solution: a fluorescence resonance energy transfer study. J Phys Chem B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 46.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 47.Buscaglia M, Lapidus LJ, Eaton WA, Hofrichter J. Effects of denaturants on the dynamics of loop formation in polypeptides. Biophys J. 2006;91:276–288. doi: 10.1529/biophysj.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proc Natl Acad Sci U S A. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitalis A, Lyle N, Pappu RV. Thermodynamics of β-sheet formation in polyglutamine. Biophy J. 2009;97:303–311. doi: 10.1016/j.bpj.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 51.Dougan L, Li J, Badilla CL, Berne BJ, Fernandez JM. Single homopolypeptide chains collapse into mechanically rigid conformations. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0900678106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen SM, Wetzel R. Solubilization and disaggregation of polyglutamine peptides. Protein Sci. 2001;10:887–891. doi: 10.1110/ps.42301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 54.dos Remedios CG, Moens PDJ. Resonance energy transfer in proteins. In: Andrews DL, Demidov AA, editors. Resonance Energy Transfer. Wiley; Chichester: 1999. pp. 1–64. [Google Scholar]
- 55.Weber G, Shinitzk M. Failure of energy transfer between identical aromatic molecules on excitation at long wave edge of absorption spectrum. Proc Natl Acad Sci U S A. 1970;65:823–830. doi: 10.1073/pnas.65.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen CL, Fitzgerald MC, Murphy RM. Effect of acid predissolution on fibril size and fibril flexibility of synthetic beta-amyloid peptide. Biophys J. 1994;67:1238–1246. doi: 10.1016/S0006-3495(94)80593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burchard W. Static and dynamic light-scattering from branched polymers and bio-polymers. Advances in Polymer Science. 1983;48:1–124. [Google Scholar]