Abstract
The role of Mre11 phosphorylation in the cellular response to DNA double-strand breaks (DSBs) is not well understood. Here, we show that phosphorylation of Mre11 at SQ/TQ motifs by PIKKs (PI3 Kinase-related Kinases) induces MRN (Mre11-Rad50-Nbs1) complex dissociation from chromatin by reducing Mre11 affinity for DNA. Whereas phosphorylation of Mre11 at these residues is not required for DSB-induced ATM activation, abrogation of Mre11 dephosphorylation impairs ATM signaling. Our study provides a functional characterization of the DNA damage-induced Mre11 phosphorylation, and suggests that MRN inactivation participates in the down-regulation of damage signaling during checkpoint recovery following DSB repair.
1. Introduction
DNA double-strand breaks (DSBs) pose a considerable threat to genomic stability because the DNA loses integrity and information content on both strands, in addition to its physical connection to the centromere [1]. To maintain the integrity of the genome, eukaryotic cells rely on the coordinated action of proteins that sense and transduce the damage signal to ultimately allow repair of the lesion, or to initiate an apoptotic program [1-3]. Initially, discontinuities in the integrity of DNA are sensed, followed by the rapid activation of the DSB checkpoint kinase ATM (Ataxia-Telangiectasia mutated). ATM phosphorylates many DNA damage response components, which in turn trigger the activation of cellular pathways responsible for cell-cycle arrest, DNA repair, and apoptosis. In the final and largely uncharacterized step of the DNA damage response, these pathways are down-regulated following resolution of the breaks to allow for resumption of the normal cell cycle.
Central to the cellular response to DSBs is the MRN (Mre11-Rad50-Nbs1) complex, which is composed of two highly conserved enzymatic subunits, Mre11 and Rad50, and a non-catalytic component, Nbs1 (Xrs2 in budding yeast) [4,5]. MRN are among the first proteins that localize at the site of DSBs [6,7]. The formation of MRN-DNA complex is responsible for the recruitment of ATM to the break site and for its subsequent activation [8-12]. In this regard, the MRN complex acts as a DNA damage sensor and an upstream regulator of ATM activity. The MRN complex also plays a crucial role downstream of ATM activation in DSB checkpoint signaling. DSB-induced phosphorylation of Nbs1 by ATM is crucial for the intra-S phase checkpoint, and is required for ATM-dependent phosphorylation of the checkpoint factors SMC1 and FANCD2 [13-17]. In addition to being involved in the detection of breaks, initiation and amplification of the checkpoint cascade, MRN complex is also required for the repair of DSBs by homologous recombination (HR). Deficiency in MRN components results in a dramatic reduction of targeted integration frequencies [18], gene conversion and sister chromatid exchanges [19,20], and impaired single-strand annealing [21]. Furthermore, inhibition of Mre11 by the small molecule Mirin impairs HR at a I-SceI DSB [22].
The MRN complex has a bipolar architecture characterized by a globular head and two flexible protruding coiled-coils [23-26]. The globular head consists of an Mre11 dimer and two Rad50 ATPase domains, and mediates MRN complex association with chromatin, an ability that is specified by Mre11 intrinsic DNA binding activity. DNA binding induces a parallel alignment of the coiled-coils, which prevents intra-complex apex interaction and favors the inter-molecular association of MRN complexes bound to different DNA molecules [27]. MRN-dependent DNA end-tethering activity has been reported both in vitro and in Xenopus cell-free extracts [9,25,28-31], and is consistent with MRN role in bridging together broken chromosomal parts to prevent loss of genomic DNA by mis-segregation after breakage [32,33].
Mre11 is constitutively Arg-methylated by methyltransferase PRMT1 [34]. This post-translational modification regulates MRN activity during the intra-S-phase checkpoint, as abrogation of Mre11 methylation results in S-phase checkpoint defects, impaired exonuclease activity, and defective recruitment of MRN complex to sites of DNA damage [34,35]. In addition, all three subunits of the complex are phosphorylated following exposure to IR, HU, and UV [36-39]. These phosphorylation events are mediated by PIKK (PI3 Kinase-related Kinases) members ATM and ATR (ATM and Rad3 related), which phosphorylate their substrates preferentially on Ser or Thr that precede Gln residues, the SQ/TQ motifs [40]. Mre11 is hyperphosphorylated in Saccharomyces cerevisiae and human cell lines following treatment with DNA damaging agents, and in Xenopus cell-free egg extracts supplemented with DSBs [41-45]. Furthermore, Mre11 is phosphorylated during DNA replication both under unperturbed conditions and in response to camptothecin treatment [41,46]. However, the biological significance of Mre11 hyperphosphorylation and nature of the DNA damage-regulated phosphorylation sites are still unknown. Mre11 is also constitutively phosphorylated in vitro and in vivo on Ser 649 by casein kinase 2 (CK2) [47].
In this study, we investigated the DSB-induced phosphorylation of Mre11 at SQ/TQ sites, and analysed the consequences on MRN complex functions in Xenopus cell-free egg extracts. We found that Mre11 phosphorylation is extremely rapid and transient. Mutagenesis of Mre11 SQ/TQ phosphorylation sites by Ser/Thr to Ala substitution increased MRN complex loading onto DNA, as well as ATM recruitment. Conversely, treatment of extracts with phosphatase inhibitors, Ser/Thr to Asp mutagenesis of Mre11 SQ/TQ sites, or phosphorylation of MRN by ATM dramatically decreased Mre11 binding to DNA, and consequently ATM loading onto chromatin. Our findings support a model in which Mre11 phosphorylation by PIKKs triggers MRN inactivation via disassembly from chromatin thus down-regulating the DNA damage response and allowing for recovery.
2. Materials and Methods
Xenopus egg extracts and immunodepletion
X. laevis were kept in accordance with Columbia University Institutional Animal Care Usage Committee guidelines. CSF-arrested egg cytosol, prepared according to Murray [48], was supplemented with 100 μg/ml cycloheximide and released into interphase by addition of 0.4 mM CaCl2 for 15 min at 22 °C. Membrane-free egg cytosol was prepared according to Smythe and Newport [49], and stored in aliquots at −80 °C. Immunodepletion of Xenopus Mre11 and ATM proteins was performed according to published protocols [30,50].
Chemicals
Okadaic acid, tautomycin, caffeine, wortmannin and NU7026 were purchased from Sigma-Aldrich. The ATM inhibitor KU55933 was a generous gift from G. Smith (KuDOS Pharmaceuticals). Caffeine stocks were freshly prepared on the day of the experiment in 10 mM PIPES at pH 7.8. Okadaic acid, wortmannin, NU7026 and KU55933 were dissolved in DMSO, and stored in aliquots at −20 °C.
Antibodies
Rabbit polyclonal serum against Xenopus Mre11 was previously described [41]. Antibodies against human Nbs1 were a generous gift of A. Nussenzweig (National Cancer Institute). Anti-hRad50 antibodies were purchased from Bethyl Laboratories. Antibodies to Xenopus ATM were previously described [30,51]. Anti-phospho-H2AX Ser 139 antibodies were purchased from Upstate. Rabbit polyclonal serum against Xenopus MCM6 was previously described [52].
Preparation of DSBs-containing DNA
DSBs-containing DNA was prepared by digesting circular pBS KS II plasmid to completion with HaeIII restriction enzyme (Roche). HaeIII cuts the plasmid 14 times, thus generating 14 fragments containing 2 DSBs each (8.68 × 1012 ends (DSBs) per μg of HaeIII-digested pBS KS II). Digested pBS KS II was diluted and used at the indicated concentrations (8.68×107 to 8.68×1011/ μl of extracts). The range of DSBs for which we start to observe phosphorylation-mediated shift in Mre11 mobility is comparable to the damage resulting from photo-activated laser beam micro-irradiation in mammalian cells. Laser beam irradiation generates between 2,500 and 3,700 DSBs/cell [53,54]. Given that 1 μl of Xenopus extract supports the assembly of 10,000 genomes from maternally stockpiled proteins, laser beam irradiation would be equivalent to 2.5-3.7×107 DSBs per μl of extract.
Mre11 SQ/TQ site mutagenesis and baculovirus generation
The pTP17 plasmid used for the initial mutagenesis reactions contains the cDNA for a C-terminal His6-tagged version of human Mre11 cloned into pFastBac1 vector [55]. pFastBac1-SAI was generated by sequential Ser/Thr to Ala mutagenesis of S264, T329, and S382. pFastBac1-SAII was produced by sequential Ser/Thr to Ala mutagenesis of T481, S531, S590, S676, and S678. pFastBac1-SDI was generated by sequential Ser/Thr to Asp mutagenesis of S264, T329, and S382. pFastBac1-SDII was produced by sequential Ser/Thr to Asp mutagenesis of T481, S531, S590, S676, and S678. The oligonucleotides used for site-directed mutagenesis are listed in Supplementary Table 1. pFastBac1-SA was generated by sub-cloning the BsgI-KpnI fragment of the SAII cDNA from pFastBac1-SAII into BsgI-KpnI sites of pFastBac1-SAI plasmid. pFastBac1-SD was produced by sub-cloning the BsgI-KpnI fragment of the SDII cDNA from pFastBac1-SDII into BsgI-KpnI sites of pFastBac1-SDI plasmid. All site-directed mutageneses were confirmed by sequencing. pTP17, pFastBac1-SA, pFastBac1-SAI, pFastBac1-SAII, and pFastBac1-SD plasmids were used to generate recombinant baculoviruses using the BAC-TO-BAC Baculovirus Expression System (Invitrogen). High-titer baculoviruses were obtained by sequential rounds of viral amplification in Sf9 insect cells. Viral titer was estimated for each baculovirus by end-point dilution protocol.
Expression and purification of recombinant human wild type and mutant MRN complexes
Sf9 cells (9 × 106 cells/75 cm2 flask) were co-infected with baculoviruses expressing human Rad50, Nbs1, and Mre11 (WT, SA, SAI, SAII, or SD) proteins at an MOI of ∼10, and harvested 48-72 h post-infection. Purification was performed by Nickel chromatography according to published protocols [56].
Co-immunoprecipitation analysis
For the co-immunoprecipitation analysis in Fig. 2B, purified recombinant MRN-WT, MRN-SA, and MRN-SD complex preparations (5 μg) were incubated with preimmune or anti-XMre11 serum for 45 min at 4 °C, followed by incubation with protein A-Sepharose beads for 45 min at 4 °C. Immunoprecipitates were washed six times with 1× PBS supplemented with 0.2% NP-40, and electrophoresed on a NuPAGE® Novex 3-8% Tris-Acetate Gel (Invitrogen). For the co-immunoprecipitation analysis in SFig. 1C, MRN-SD complex (500 nM) was incubated in Mre11-depleted extract for the indicated time points, and immunoprecipitated with either preimmune or anti-XMre11 serum following the protocol described above.
Figure 2. Mre11 is phosphorylated at SQ/TQ motifs in response to DSBs.
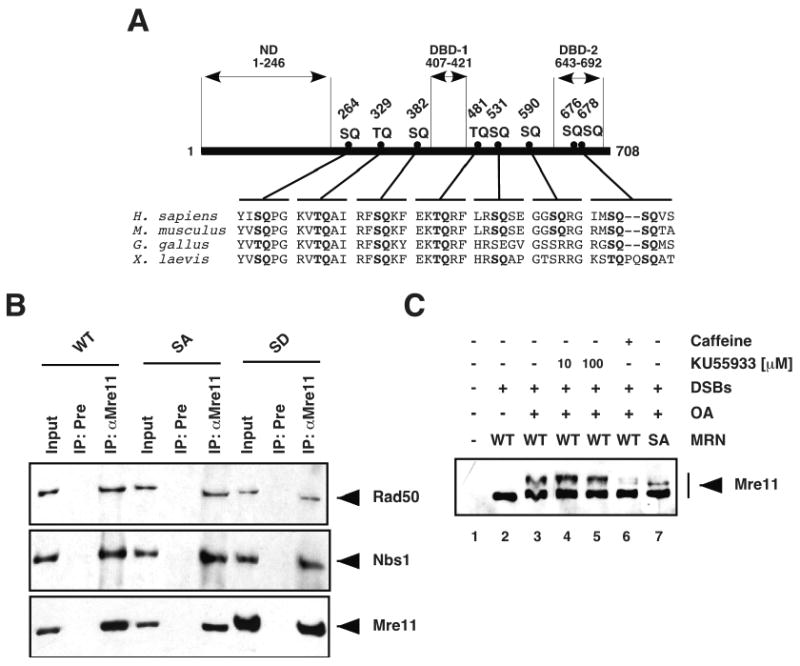
(A) The amino acid sequences of Mre11 proteins from Homo sapiens (ACcession number: P49959), Mus musculus (AC: Q61216), Gallus gallus (AC: Q9IAM7), and Xenopus laevis (AC: Q9W6K1) were aligned using the CLUSTAL W (1.83) multiple sequence alignment program. SQ/TQ motifs are highlighted in bold. The amino acid positions of the Ser/Thr residues in the eight SQ/TQ motifs of the human protein are indicated. ND: Nuclease domain; DBD-1/2: DNA-binding domain 1/2. (B) Purified MRN-WT, MRN-SA, and MRN-SD complexes were immunoprecipitated with preimmune (Pre) or anti-XMre11 (αMre11) serum, and the immunoprecipitates were immunoblotted with anti-hRad50 serum (upper panel), anti-hNbs1 serum (middle panel), and anti-XMre11 serum (lower panel). The inputs shown are equivalent to 15% of the purified MRN complex used in the co-immunoprecipitation reactions. (C) Mre11-depleted membrane-free extracts supplemented with recombinant MRN-WT or MRN-SA complex were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl), okadaic acid (OA; 4 μM), and KU55933 (10 or 100 μM) or caffeine (10 mM), and analysed by Western blotting with anti-XMre11 serum.
Analysis of Mre11 phosphorylation status
Untreated, mock-, ATM-, or Mre11-depleted extracts (10 μl) supplemented with recombinant MRN-WT, -SA, -SAI, -SAII, or -SD complex were incubated with DSBs at 22 °C for 15 min unless otherwise indicated (Fig. 1C). Reactions were stopped with Laemmli buffer, electrophoresed on 8% SDS-PAGE (sample volume equivalent to 0.5-1 μl of undiluted extract), and analysed by Western blotting with anti-XMre11 serum. Where indicated, tautomycin, okadaic acid, caffeine, wortmannin, NU7026 or KU55933 were added to the extracts 10 min before incubation with DSBs. Lambda Protein Phosphatase (New England BioLabs) treatment was performed at the end of the incubation period with DSBs for 1 hour at 30 °C, and products were analysed on 8% SDS-PAGE.
Figure 1. DSBs induce rapid and transient phosphorylation of Mre11 in Xenopus cell-free egg extracts.
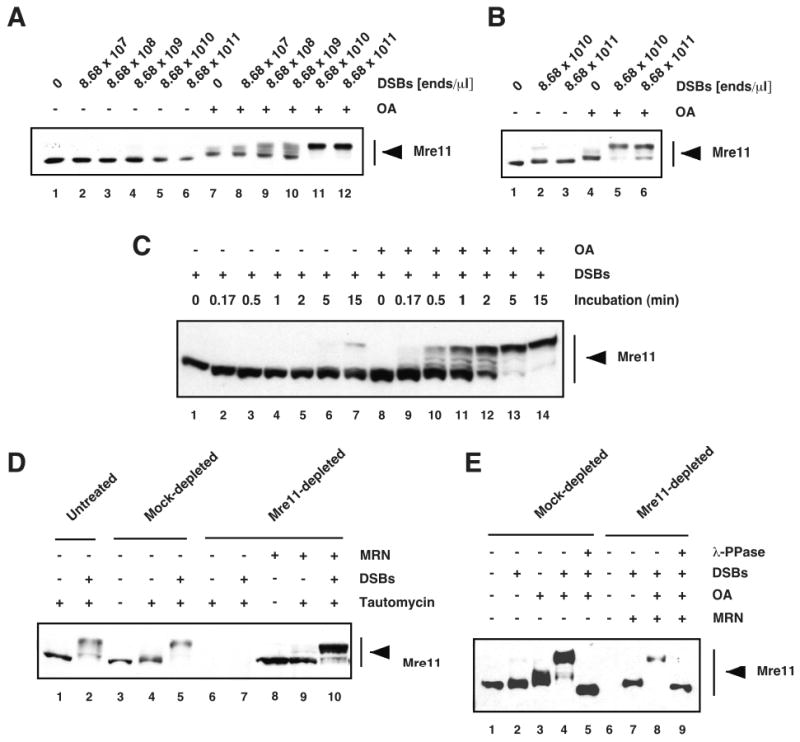
Interphase egg cytosol (A) and membrane-free egg cytosol (B) extracts were incubated for 15 min with DSBs-containing DNA (DSBs) at the indicated concentrations in the absence or presence of 4 μM of okadaic acid (OA), and analyzed by Western blotting with anti-XMre11 serum. (C) Membrane-free egg cytosol was incubated for the indicated times with DSBs (4.34 × 1010 ends/μl) in the absence or presence of 4 μM of okadaic acid (OA), and analyzed by Western blotting with anti-XMre11 serum. (D) Untreated interphase egg cytosol, mock-, and Mre11-depleted extracts supplemented with recombinant MRN complex were incubated with DSBs (4.34 × 1011 ends/μl) in the presence or absence of tautomycin (4 μM), and analyzed by Western blotting with anti-XMre11 serum. (E) Mock- and Mre11-depleted membrane-free extracts supplemented with purified MRN complex were incubated in the presence or absence of DSBs (4.34 × 1010 ends/μl) and OA (4 μM), treated with Lambda Protein Phosphatase (λ-PPase) (lanes 5 and 9), and analyzed by Western blotting with anti-XMre11 serum.
Biotinylated DNA pull-down assay
Biotinylated linear double-stranded DNA molecules of 150 bp and 1 Kb were generated by PCR using M13 single-stranded DNA as a template and either a forward or a reverse biotinylated primer as previously described [30]. Biotinylated DNA was coupled to streptavidin-coated magnetic beads (M280, Dynal Biotechnology) overnight at 4 °C. DNA-streptavidin beads were washed six times in ELB buffer (10 mM HEPES pH 7.7, 2.5 mM MgCl2, 0.05 mM KCl, and 250 mM sucrose), and incubated with extracts at 21 °C at a final concentration of 1.2 × 1011 ends/μl. When indicated, okadaic acid was added to the extracts at a final concentration of 4 μM 10 min before incubation with biotinylated DNA. DNA-bound fraction was separated from unbound supernatant according to the manufacturer's instruction, washed six times with ELB buffer supplemented with 0.1% Triton X-100, and electrophoresed on NuPAGE® Novex 3-8% Tris-Acetate Gels (Invitrogen).
Chromatin binding assay on demembranated sperm nuclei
Demembranated Xenopus sperm nuclei were prepared as described by Murray [48], and used as chromosomal templates for chromatin binding assays. Mre11-depleted interphase egg extracts (65 μl) were supplemented with purified MRN-WT, MRN-SA, or MRN-SD complex (200 nM), and incubated with 10,000 sperm nuclei/μl for 60 min at 22 °C. Following incubation, extracts were diluted in 800 μl of Chromatin isolation buffer (100 mM KCl, 2.5 mM MgCl2, 50 mM HEPES pH 7.8) supplemented with 0.125% Triton X-100, and chromatin was isolated through a 30% sucrose cushion in Chromatin isolation buffer (320 μl) at 6,000 rpm for 30 min at 4 °C. Chromatin pellets were resuspended in Laemmli buffer and electrophoresed on NuPAGE® Novex 3-8% Tris-Acetate Gels (Invitrogen).
In vitro kinase assay
Mre11-depleted membrane-free egg extracts were supplemented with recombinant MRN-WT or MRN-SA complex (500 nM), and incubated for 5 min at 22 °C with streptavidin beads coated with biotinylated 150 bp DNA (6.1 × 1010 ends/μl). The beads were separated from extracts according to the manufacturer's instruction, washed six times with ELB buffer, and incubated with monomeric ATM (T. Paull, University of Texas, Austin) in the presence of 1 mM ATP in Kinase buffer (50 mM HEPES pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 10% glycerol, 10 mM MnCl2) for 30 min at 30 °C. Streptavidin beads were washed six times with ELB buffer, and DNA-bound proteins were analysed by Western Blotting.
DNA end-tethering assay
32P-labeled double-stranded 1 Kb DNA molecules were generated by PCR using M13 single-stranded DNA as a template and α-32P dCTP in the reaction mix as previously described [9]. Mock- or Mre11-depleted membrane-free egg extracts (10 μl) supplemented with recombinant MRN-WT, MRN-SA, or MRN-SD complex (125 nM, 250 nM, or 500 nM) were incubated with streptavidin beads coated with biotinylated 1 Kb DNA and free unbiotinylated 32P-1 Kb DNA (total free ends: 1.2 × 1011 ends/μl) for 25 min at 21 °C. Following pull-down from extracts, beads were washed six times with ELB buffer supplemented with 0.1% Triton X-100, and the associated radioactivity was measured in a scintillation counter.
3. Results
DSBs induce rapid and transient phosphorylation of Mre11 in Xenopus egg extracts
To characterize the DNA damage-induced phosphorylation of Mre11, we first monitored Mre11 mobility on PAGE as an indication of its phosphorylation in egg extracts incubated with DSBs-containing DNA. We used two types of cell-free egg extracts for this study, egg cytosol and membrane-free egg cytosol, which recapitulate faithfully MRN/ATM- and ATR-dependent DNA damage signaling pathways [9,30,41,57-59]. To trap phosphorylated intermediates, extracts were supplemented with either okadaic acid or tautomycin, which inhibit the serine and/or threonine protein phosphatases PP1 and PP2A-like (PP2A, PP4, PP5, and PP6). Treatment of egg extracts with increasing concentrations of DSBs for 15 minutes induced a dose-dependent phosphorylation of total (soluble and DNA-bound) Mre11 protein (Fig. 1, A and B), as seen by the appearance of slower migrating isoforms that were eliminated by treatment with Lambda phosphatase (Fig. 1E, lane 5). Mre11 mobility shift becomes detectable at doses of DSBs comparable to DNA damage triggered by laser microbeam irradiation in mammalian cells (See Materials and Methods). Phosphorylation of Mre11 was extremely rapid as seen by the appearance of shifted polypeptides as early as after 10 seconds post-incubation (Fig. 1C, lane 9). The slower migrating forms were hardly detectable in the absence of okadaic acid (Fig. 1, A, B and C) indicating that PP1/PP2A-like phosphatase activities rapidly dephosphorylate Mre11 in the extracts. The presence of multiple isoforms indicates that Mre11 is modified at several sites. Of note, phosphorylation of the DNA-bound fraction of Mre11 is detectable in the absence of okadaic acid (data not show). As previously reported [41], addition of phosphatase to extracts results in the appearance of a polypeptide of higher electrophoretic mobility (Fig. 1E, compare lane 5 with lanes 1 and 3). This suggests that Mre11 is constitutively phosphorylated in the absence of DNA damage (hypo-phosphorylated form). Interestingly, DSB treatment resulted in the formation of slower migrating forms of the Nbs1 protein (data not shown and [22]), suggesting that Nbs1 is also phosphorylated in Xenopus extracts in response to DSBs. A low level of Mre11 hyper-phosphorylation was detected in okadaic acid-supplemented extracts in the absence of DNA damage (Fig. 1A, lane 7, Fig, 1B, lane 4, and Fig. 1E, lane 3). At concentration of DSBs higher than 1010 ends/μl, Mre11 was detected predominantly as a single discrete slower migrating band (Fig. 1A, lanes 11-12, and Fig. 1B, lanes 5-6) suggesting that at high concentrations of DSBs the majority of Mre11 molecules, soluble and DNA-bound, is phosphorylated.
Next, we analyzed the ability of Xenopus egg extract to induce phosphorylation of recombinant human Mre11 (hMre11) in response to DSBs. Recombinant hMRN complex can rescue all known phenotypes associated with XMRN-depletion in Xenopus extracts [9,30,41]. Extracts were treated with anti-XMre11 serum to quantitatively immunodeplete endogenous Mre11 protein, and supplemented with purified recombinant human MRN complex. Note that XMre11-depletion results in the co-depletion of XNbs1 and XRad50 [30]. Incubation of hMRN-supplemented extracts with DSBs in the presence of phosphatase inhibitors induced a significant shift in the electrophoretic mobility of hMre11 (Fig. 1D, lane 10, and Fig. 1E, lane 8), which was abrogated by treatment with Lambda Phosphatase (Fig. 1E, lanes 9). Similar to endogenous Mre11, hMre11 exhibited some level of hyper-phosphorylation in the absence of DNA damage in tautomycin-treated extracts (Fig. 1D, lane 9). hMre11 hyper-phosphorylation was not detectable in extracts in the absence of phosphatase inhibitors (Fig. 1E, lane 7), indicating that hMre11 was rapidly dephosphorylated in extracts as observed for endogenous Mre11 (Fig 1, A, B and C). These findings indicate that DSBs induce rapid and transient phosphorylation of endogenous Mre11 and recombinant hMre11 in Xenopus extracts.
Mre11 is phosphorylated at SQ/TQ motifs in response to DSBs
DSBs activate primarily ATM and subsequently ATR [57,60]. Mre11 phosphorylation is mediated at least in part by these PIKKs [36-39,41,44,59], which phosphorylate their substrates preferentially at Ser/Thr residues in SQ/TQ motifs [40]. SQ/TQ motifs in Mre11 are highly conserved in higher eukaryotes (Fig. 2A). Therefore, we sought to analyze the consequences of site-directed mutagenesis of Ser/Thr residues in SQ/TQ motifs on Mre11 phosphorylation and functions.
hMre11 contains eight such motifs (Fig. 2A and SFig.1A). The first 6 SQ/TQ motifs are located within spacer domains, whereas the C-terminal doublet S676Q/S678Q is within the second DNA binding domain of Mre11 [4,61] (Fig. 2A). The Ser/Thr residues were substituted for Ala to generate hMre11 protein lacking all eight SQ/TQ consensus sequences (SFig. 1A, SA mutant), or hMre11 lacking specific subsets of SQ/TQ motifs (SFig. 1A, SAI and SAII mutants). In addition, we generated a form of hMre11 in which the Ser/Thr residues of all eight SQ/TQ motifs were mutated to Asp (SFig. 1A, SD mutant) in an attempt to generate a phosphomimic Mre11 mutant. The expression of mutant Mre11 proteins was verified by Western blot and Coomassie staining analysis of recombinant MRN complexes purified from insect cells co-infected with baculoviruses for hRad50, hNbs1, and each of the hMre11 SQ/TQ site mutant proteins (SFig. 1B and data not shown).
We then performed co-immunoprecipitation analysis of purified recombinant MRN complexes to assess stable complex formation between Mre11, Rad50 and Nbs1. We used anti-XMre11 serum to immunoprecipitate hMre11 from purified MRN-WT, MRN-SA, and MRN-SD complexes, followed by Western blot analysis with antibodies specific for each MRN subunit. hRad50 and hNbs1 were detected at similar levels in Mre11-immunoprecipitates from MRN-WT, MRN-SA, and MRN-SD complexes (Fig. 2B) indicating that mutagenesis of Mre11 SQ/TQ motifs does not affect the ability of Mre11 to interact with Rad50 and Nbs1, and therefore the integrity of the MRN complex. We observed that Nbs1 was less abundant in all three recombinant complexes (data not shown), a phenomenon previously reported for WT-MRN [10].
Next, we assessed the phosphorylation of Mre11 SQ/TQ site mutant proteins in Mre11-depleted extracts supplemented with recombinant MRN complexes. We found that mutagenesis of all eight SQ/TQ motifs to Ala reduced but not abrogated DSB-induced Mre11 phosphorylation (Fig. 2C, lane 7). A similar behavior was observed with Mre11 protein lacking the five most C-terminal consensus sites (SFig. 2A, lane 4), whereas mutagenesis of the three most N-terminal SQ/TQ motifs had no effect on Mre11 electrophoretic mobility (SFig. 2B, lane 7). These findings establish that one or more Ser/Thr residues among the five C-terminal Mre11 SQ/TQ motifs are phosphorylated in a DNA damage-dependent manner, and that additional sites other than SQ/TQ motifs contribute to Mre11 phosphorylation as seen by electrophoretic mobility shift.
Next, we investigated the contribution of PIKKs to Mre11 phosphorylation following DNA damage by assessing the consequence of PIKK inhibition on Mre11 electrophoretic mobility. We found that treatment with caffeine, which inhibits both ATM and ATR, or wortmannin, which inhibits ATM, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and possibly ATR, resulted in the partial disappearance of hyperphosphorylated Mre11 (Fig. 2C and SFig. 2C). Treatment with the ATM specific inhibitor KU55933 [62] or ATM-depletion had no appreciable effect on Mre11 electrophoretic mobility (Fig. 2C and SFig. 2C). This finding is consistent with data in mammalian cells that showed that DNA damage-induced Mre11 mobility shift was normal in A-T cells deficient for ATM protein kinase [43,45]. However, several studies have implicated ATM as one of the kinases responsible for Mre11 phosphorylation in response to DSBs [36-39,44,59], and in vitro kinase reaction assays with catalytically active ATM provide evidence for a functional role of Mre11 phosphorylation by ATM (see below). This suggests that either both ATR and ATM can phosphorylate Mre11 in a redundant manner following DNA damage and/or Mre11-phosphorylation by ATM is not associated with a detectable shift in gel mobility. Finally, treatment with NU7026, which inhibits the catalytic subunit of DNA-PK [63], did not affect Mre11 mobility at concentrations that inhibit specifically DNA-PKcs (SFig. 2, C and D). We have previously established that NU7026 significantly reduces repair of DSBs by non-homologous end joining in Xenopus egg extracts at concentrations as low as 0.5 μM [50]. We also tested caffeine and wortmannin treatment in combination with ATM-depletion and, similar to what we observed with undepleted extracts (Fig. 2C and SFig. 2C), we found a significant reduction but not abrogation of Mre11 phosphorylation (data not shown). Together, these findings suggest that Mre11 phosphorylation following DNA damage is mediated by ATR/ATM as well as by additional protein kinases that are insensitive to PIKKs inhibitors.
Phosphorylation of Mre11 SQ/TQ motifs modulates the affinity of MRN for DNA
We investigated whether damage-induced phosphorylation regulates Mre11 binding to DNA. We monitored DNA binding to biotinylated linear DNA molecules or chromatin from demembranated sperm nuclei. Xenopus egg extracts support chromatin assembly on purified DNA [64-66], and biotinylated linear DNA molecules have been successfully used to analyse the kinetics of chromatin and pre-replicative complex assembly [52,67-70]. We first examined loading of endogenous Mre11 protein onto biotinylated DNA molecules in extracts by streptavidin-mediated pull-down and Western blot analysis of bound proteins with anti-XMre11 serum. Samples were analyzed under electrophoretic conditions that limit the resolution of phosphorylation-mediated gel shifts, in order to better estimate the total amount of DNA-bound Mre11. Time course analysis of untreated extracts showed that Mre11 bound to DNA with an extremely rapid kinetics, being detectable in the DNA-bound fraction as soon as DNA was incubated in extracts (Fig. 3A, lower panel, lanes 2 and 6). Mre11 binding to DNA peaked at 2 minutes, and declined afterwards (Fig. 3A, lanes 3-5). We then compared Mre11 binding to DNA in the absence and presence of the phosphatase inhibitor okadaic acid, which prevents Mre11 dephosphorylation in egg extracts (Fig. 1). Binding of Mre11 to DNA was reduced in presence of okadaic acid compared to untreated extracts at each time point (Fig. 3A, lower panel, compare lanes 2 to 5 with lanes 6 to 9). This suggests that Mre11 binding to DNA is regulated in part by phosphorylation of Mre11 that can be reversed by okadaic acid-sensitive phosphatases. Interestingly, when we analyzed Mre11 binding to DNA under electrophoretic conditions that allowed for resolution of the phosphorylated Mre11 isoforms (data not shown), we found that both hypo- and hyper-phosphorylated forms of Mre11 were readily detectable on DNA in the absence of okadaic acid. This suggests that hyper-phosphorylated Mre11 can transiently associate with DNA, and that DNA-bound Mre11 is more resistant to dephosphorylation than Mre11 in the cytosol.
Figure 3. Phosphorylation of Mre11 SQ/TQ motifs modulates MRN complex association with DNA.
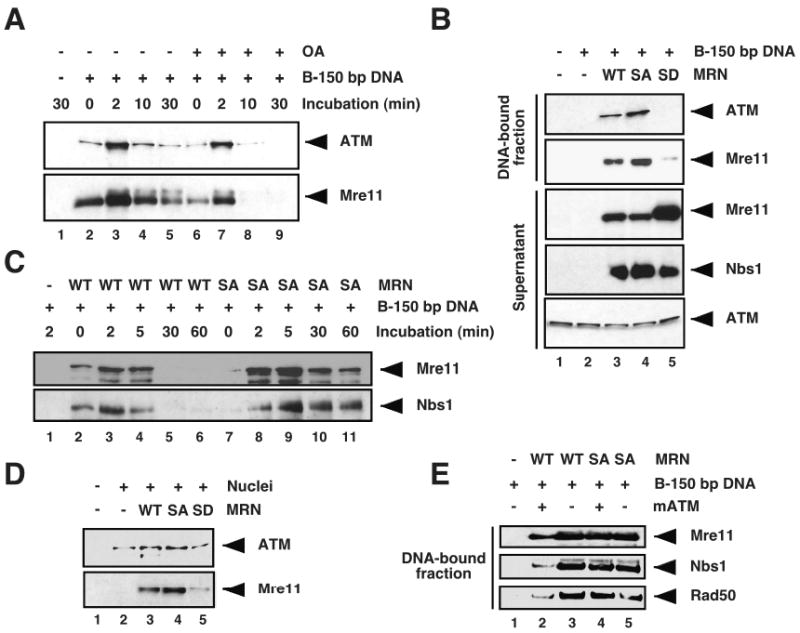
(A) Untreated membrane-free egg extracts were incubated with biotinylated 150 bp linear double-stranded DNA (B-150 bp DNA) bound to streptavidin beads for the indicated times in the absence (lanes 1-5) or presence (lanes 6-9) of 4 μM okadaic acid (OA). The DNA-bound fractions were pulled down, electrophoresed on NuPAGE® Novex 3-8% Tris-Acetate gel and analyzed by Western blot with anti-XATM serum (upper panel) or anti-XMre11 serum (lower panel). (B) Mre11-depleted membrane-free egg extracts supplemented with MRN-WT, -SA, or -SD recombinant complex were incubated for 30 min with B-150 bp DNA bound to streptavidin beads. The resulting supernatants were separated from the DNA-bound fractions, and both fractions were analyzed by Western blotting with antibodies against the indicated proteins. (C) Mre11-depleted membrane-free egg cytosol supplemented with MRN-WT or MRN-SA complex was incubated with B-150 bp DNA bound to streptavidin beads for the indicated time points. The DNA-bound fractions were separated from the supernatants, and analyzed by Western blotting with anti-XMre11 serum (upper panel) and anti-hNbs1 serum (lower panel). (D) Mre11-depleted interphase egg cytosols supplemented with MRN-WT, MRN-SA, or MRN-SD complex were incubated for 1 h with demembranated sperm nuclei (10,000 nuclei/μl), and the chromatin fractions were isolated and analyzed by Western blotting with anti-XATM serum (upper panel) and anti-XMre11 serum (lower panel). (E) Mre11-depleted membrane-free egg extracts supplemented with either MRN-WT or MRN-SA complex were incubated for 5 min at 22 °C with biotinylated 150 bp linear double-stranded DNA (B-150 bp DNA) bound to streptavidin beads. The DNA-bound fractions were separated from the supernatants and used as a substrate for an in vitro kinase reaction in the presence or absence of purified monomeric ATM kinase (mATM) and dATP. DNA-bound proteins were then separated on SDS-PAGE, and analyzed by Western blotting with anti-XMre11 serum (upper panel), anti-hNbs1 serum (middle panel), and anti-hRad50 serum (lower panel).
ATM is recruited, at least initially, through its binding with the C-terminus of Nbs1 [71]. Accordingly, we found that decreased Mre11 binding to DNA in okadaic acid-treated extracts correlated with reduced ATM recruitment to DNA at each time point (Fig. 3A, upper panel).
Taken together, these findings strongly suggest that Mre11 phosphorylation modulates its DNA binding activity. We sought to test the possibility that phosphorylation of Mre11 at SQ/TQ sites could modulate Mre11 ability to load onto DNA. We analyzed DNA binding of recombinant MRN-WT, MRN-SA and MRN-SD complexes following incubation of extracts with biotinylated linear DNA. Western blot analysis of DNA-bound fractions with anti-XMre11 serum revealed that substitution of Ser/Thr for Ala in the MRN-SA mutant complex results in a significant increase in Mre11 and ATM binding to DNA (Fig. 3B, top two panels, compare lanes 3 and 4). Notably, Mre11 and ATM binding were dramatically reduced in DNA-bound fraction purified from extracts supplemented with MRN-SD complex (Fig. 3B, lane 5). To rule out the possibility that MRN-SD reduced binding could result from an unstable complex, we monitored Mre11-SD, Rad50 and Nbs1 protein levels and MRN-SD complex stability following incubation in extracts (SFig. 1C). We found that MRN-SD complex was stable, ruling out the possibility that MRN-SD might be specifically unstable in extracts. We then compared the kinetics of MRN-WT and MRN-SA complex binding to biotinylated DNA, and found that the amount of Mre11 and Nbs1 proteins in the DNA-bound fractions purified from extracts supplemented with MRN-SA was higher than in extracts supplemented with WT complex at all time points (Fig. 3C).
Next, we monitored the ability of MRN SQ/TQ site mutant complexes to bind chromatin from demembranated sperm nuclei. Extracts supplemented with MRN complex were incubated for 1 hour with sperm nuclei, and Mre11 chromatin binding was monitored by Western blot analysis of purified chromatin fractions. Consistent with the results obtained with biotinylated DNA templates, Mre11-SA mutant displayed enhanced loading onto sperm chromatin compared to the WT protein, whereas Mre11-SD binding was dramatically reduced (Fig. 3D, lower panel). As expected, ATM loading correlated with MRN binding to chromatin (Fig. 3D, upper panel) Taken together, these results suggest that phosphorylation of Mre11 at SQ/TQ motifs regulates MRN complex affinity for DNA. These experiments do not exclude the possibility that phosphorylation at non-SQ/TQ sites could contribute to this regulation as MRN-SA binding to biotinylated DNA in extracts supplemented with okadaic acid was reduced compared to untreated extracts (data not shown).
To test whether Mre11 phosphorylation at SQ/TQ sites regulates MRN complex release from DNA, we monitored the binding of MRN-WT and MRN-SA complexes to DNA before and after phosphorylation by ATM. In vitro kinase reaction with catalytically active monomeric ATM and purified MRN-WT complex in presence of 32P-ATP yielded three major bands with apparent molecular weights expected for Rad50, Nbs1, and Mre11 (data not shown), consistent with recent proteomic-based studies showing that all three subunits of the MRN complex are phosphorylated on the ATM/ATR substrate SQ/TQ consensus motifs following DNA damage [36,37,39]. Mre11-depleted extracts were incubated with biotinylated DNA in the presence of MRN-WT or mutant SA complex to trigger MRN binding to DNA, and the purified DNA-bound fractions were phosphorylated in vitro with purified monomeric ATM. MRN-WT binding to DNA was reduced following phosphorylation by monomeric ATM (Fig. 3E, lanes 2-3). In contrast, MRN-SA binding to DNA was not affected by incubation with active ATM protein kinase (Fig. 3E, lanes 4-5). Altogether, these findings strongly suggest that ATM/PIKKs-mediated phosphorylation of Mre11 SQ/TQ motifs facilitates the release of the MRN complex from DNA.
Phosphorylation of Mre11 SQ/TQ motif impairs DNA tethering and subsequent ATM activation
We showed that Mre11 phosphorylation reduces MRN DNA binding activity and in turn, ATM recruitment. Next, we wanted to test if this could affect ATM activation. We first characterized the effects of SQ/TQ mutagenesis on MRN complex DNA-end tethering activity, which is strictly dependent upon MRN complex ability to bind to DNA, and is required for proper ATM activation in response to DSBs [9,30]. Mre11-depleted extracts were incubated with biotinylated linear DNA molecules and radioactive non-biotinylated DNA in the presence of MRN-WT, -SA, or -SD recombinant complexes. MRN DNA end-tethering activity was monitored by measuring the amount of radioactivity associated with biotinylated DNA following pull-down. As previously reported [9,30], Mre11-depletion reduced, but did not eliminate DNA tethering activity in extracts, suggesting that additional factors contribute to this activity (Fig. 4A). MRN complex is rate-limiting in extract for DNA end-bridging activity as addition of increasing amounts of MRN-WT complex results in a proportional increase in the radioactivity associated with biotinylated DNA (SFig. 3 and [9]). At concentration of 500 nM, MRN-WT complex rescued the DNA tethering defect of Mre11-depleted extracts above the levels observed for mock-depleted extracts (Fig. 4A and SFig. 3). Addition of MRN-SA complex to Mre11-depleted extracts rescued the DNA end-tethering defect (Fig. 4A) indicating that phosphorylation of SQ/TQ sites was not required for DNA-bridging activity. However, the DNA tethering activity of MRN-SA did not correlate with its affinity for DNA, which is higher than that of the WT complex. Notably, the MRN-SD complex failed to correct the DNA tethering defect in Mre11-depleted extracts (Fig. 4A), which is consistent with Mre11-SD impaired ability to load onto DNA.
Figure 4. Phosphorylation of Mre11 SQ/TQ motif impairs DNA tethering and subsequent ATM activation.
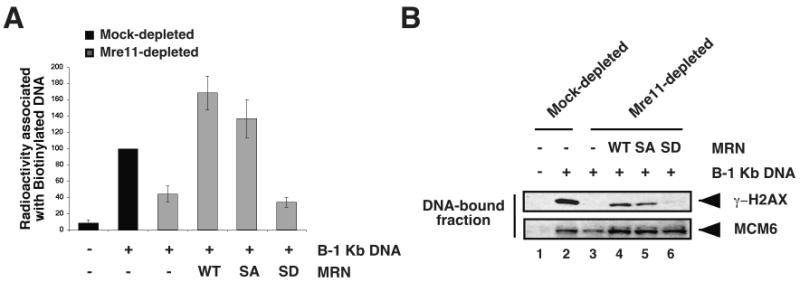
(A) Mock- and Mre11-depleted membrane-free egg extracts supplemented with MRN-WT, -SA, or -SD (500 nM) were incubated with biotinylated 1 Kb linear double-stranded DNA (B-1 Kb DNA) bound to streptavidin beads and free radioactive non-biotinylated 1 Kb DNA. Streptavidin-immobilized DNA fractions were isolated, and the associated radioactivity (cpm) was measured. Values are expressed as percentages relative to B-1 Kb DNA-supplemented Mock-depleted sample, which is set to 100%, and error bars represent standard deviations from three independent experiments. (B) Mock- and Mre11-depleted membrane-free egg extracts supplemented with MRN-WT, MRN-SA, or MRN-SD (500 nM) were incubated with B-1 Kb DNA (1.1 × 1011 ends/μl) bound to streptavidin beads for 10 min at 22°C. The DNA-bound fractions were separated from the supernatants and analyzed by Western blotting with antibodies against γ-H2AX (upper panel) and MCM6 (lower panel; loading control).
An early consequence of the MRN-dependent recruitment of ATM to DSBs is the phosphorylation of histone H2AX in the immediate vicinity of the break [72,73]. To determine the consequences of substituting Mre11 SQ/TQ phosphorylation sites, we monitored the ability of mutant MRN complexes to support ATM activation in response to DSBs as measured by histone H2AX phosphorylation. Mre11-depleted extracts supplemented with MRN-WT, -SA, or -SD complexes were incubated with biotinylated 1 Kb DNA. DNA was purified and phosphorylation of histone H2AX was monitored on immobilized DNA by Western blot analysis with antibodies against γ-H2AX. As expected, histone H2AX phosphorylation was dependent upon MRN, and Mre11-depletion abrogated H2AX phosphorylation (Fig. 4B, lane 3). Addition of recombinant MRN-WT complex rescued H2AX phosphorylation (Fig. 4B, lane 4). MRN-SA complex also restored H2AX phosphorylation, albeit to a slightly lower extent than the WT complex (Fig. 4B, lane 5). In contrast, no γ-H2AX signal was detected in the DNA-bound fraction from extracts supplemented with MRN-SD (Fig. 4B, lane 6) indicating that this mutant complex fails to support DSB-induced ATM activation, even though it was stable in extracts (SFig.1C). Altogether, these data demonstrate that DNA damage-induced phosphorylation of Mre11 is not required for ATM activation, however, abrogation of Mre11 dephosphorylation dramatically impairs ATM signaling.
4. Discussion
The MRN complex is required upstream of ATM activation [8-12,30], and all three Mre11, Rad50 and Nbs1 proteins are targets of ATM or ATR [13,14,17,36,37,39,44,59]. This suggests that feedback loops regulate the activity of the MRN complex through phosphorylation. To test this hypothesis in the case of Mre11, we have assessed the consequence of substituting all PIKK consensus phosphorylation sites (SQ and TQ) by non-phosphorylatable alanines or phosphomimic aspartates. Alanine substitution of the eight putative phosphorylation sites yields functional MRN complexes: 1) Mre11-SA assembles into MRN complexes (Fig. 2B); 2) MRN-SA complexes bind to chromatin and DNA (Fig. 3); 3) recombinant MRN-SA supports DNA tethering in MRN-depleted extracts (Fig. 4A); and 4) MRN-SA supports ATM activation in MRN-depleted extracts (Fig. 4B).
While we provide evidence that phosphorylation of Mre11 by ATM alone has significant functional consequences by decreasing MRN binding to DNA, we also provide evidence that ATM is not responsible for the bulk of phosphorylation following DNA damage. Indeed, substitution of all SQ/TQ sites to AQ did not abrogate Mre11 mobility shift, strongly suggesting that additional DNA damage-dependent phosphorylations are taking place. Consistent with this, caffeine or wortmannin only partially inhibits wild type Mre11 phosphorylation. Finally, specific inhibition of ATM or DNA-PKcs has minimal effect on Mre11 mobility, indicating that SQ/TQ sites in Mre11 might be phosphorylated by both ATM and ATR, possibly in a redundant manner. This hypothesis is consistent with previous observations in mammalian cells and in cell-free extracts [43,45,59].
Our data are consistent with a model in which DSBs trigger the rapid association coupled with the hyper-phosphorylation of Mre11 on DNA. Phosphorylation of Mre11 at SQ/TQ sites inactivates the MRN complex by facilitating its dissociation from chromatin, thus allowing down-regulation of DNA damage signaling and recovery from the checkpoint response. Furthermore, our data strongly suggest that phosphorylation of Mre11 within the MRN complex is the most critical event. Whereas phosphorylation of MRN by ATM significantly decreases MRN-WT DNA binding, it does not affect the binding of MRN-SA (Fig. 3E). This shows that PIKK-dependent modulation of MRN DNA binding and DNA tethering is mediated primarily by phosphorylation of Mre11 SQ/TQ sites. However, we cannot exclude the possibility that non-consensus sites on Mre11 or additional targets of okadaic acid-sensitive phosphatases might contribute to this regulation, since okadaic acid treatment reduced the binding of MRN-SA to DNA (data not shown). Upon DNA damage, MRN localizes rapidly to DSBs [6,7] and recruits ATM to the sites of damage through the interactions of Nbs1 C-terminus with ATM [71]. We show that both Mre11 phosphorylation and binding to DNA are extremely rapid and concomitant, supporting the idea that MRN localization to sites of damage is tightly coupled with ATM/PIKKs recruitment. Dissociation of MRN from damage sites should not precede the successful completion of the DNA damage response: phosphorylation of ATM downstream targets and DSB repair. We propose that a mechanism to prevent the precocious inactivation of the MRN complex by phosphorylation of Mre11 is the rapid turnover of phosphate on Mre11. Indeed, we observe rapid dephosphorylation of Mre11 by okadaic acid-sensitive phosphatase(s) in extracts supplemented with short DSB-containing DNA templates that are not repaired during the course of the experiment. As anticipated from its weaker chromatin binding, MRN-SD is defective in ATM activation (Fig. 4B). Candidate phosphatases for such Mre11-dephosphorylating activity are PP2A, which is associated with ATM and released upon ATM activation [74], and PP1, which is activated in an ATM-dependent manner following IR-induced DNA damage [75]. Conversely, extensive spreading of an activated MRN complex harboring both endo- and exo-nuclease activities around damage sites could be harmful. In fact, MRN chromatin spreading surrounding a DSB is limited to few kilobases in yeast [76] and in mammalian cells [77,78]. Dissociation of the complex by phosphorylation would thus prevent extensive chromatin spreading of the complex. Failure to phosphorylate and inactivate Mre11 could then result in a persistent DNA damage response associated with cell-cycle arrest.
DSBs initiate a signaling cascade with dramatic effects on cell cycle and viability, and therefore multiple levels of regulation might have evolved to promptly inactivate checkpoint factors all along the cascade once DNA is repaired. Dephosphorylation is a critical modification for checkpoint down-regulation. In S. cerevisiae, the PP2C-like phosphatases Ptc2 and Ptc3 contribute to adaptation and recovery after HO-induced DSBs by promoting dephosphorylation and inactivation of the checkpoint kinase Rad53 [79]. Pph3-dependent dephosphorylation of γH2A has also been shown to be required for efficient recovery from DNA damage checkpoint in budding yeast [80]. In mammalian cell lines, PP2A dephosphorylates γH2AX during later stages of the DNA damage response, and this allows for DSB repair to be efficiently completed [81]. Our study unveils a potential role for the phosphorylation of the MRN complex subunit Mre11 in the regulation of checkpoint inactivation. Mre11 phosphorylation and γH2AX dephosphorylation could represent two complementary mechanisms that synergistically prevent the recruitment of ATM molecules to the original DSB site following resolution of the break, thus allowing for down-regulation of the DSB checkpoint signaling and resumption of normal cell cycle.
Supplementary Material
(A) Schematic representation of wild type (WT) hMre11 and SQ/TQ site mutant proteins. The amino acid number of the Ser/Thr residues in the eight SQ/TQ motifs of the WT protein is indicated. SA: all eight SQ/TQ sites mutated to AQ; SAI: S264Q, T329Q, S382Q motifs mutated to AQ; SAII: T481Q, S531Q, S590Q, S676Q, S678Q motifs mutated to AQ. SD: all eight SQ/TQ sites mutated to DQ. (B) Recombinant mutant human MRN complexes were purified from Sf9 cells following co-infection with baculoviruses for hRad50, hNbs1, and each of the hMre11 SQ/TQ site mutants shown in panel A, and analysed by Western blotting with anti-hRad50 serum (upper panel), anti-hNbs1 serum (middle panel), and anti-XMre11 serum (lower panel). (C) MRN-SD complex was incubated in Mre11-depleted extracts for the indicated time points, and either directly electrophoresed (left panels) or immunoprecipitated with either preimmune (Pre) or anti-XMre11 (αMre11) serum (right panels). Inputs and immunoprecipitates were analysed by Western blotting with anti-hRad50 serum (upper panel), anti-hNbs1 serum (middle panel), and anti-XMre11 serum (lower panel).
(A) Mre11-depleted extracts supplemented with WT, or mutant recombinant MRN complex (SA, SAII, or SD) were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl) and okadaic acid (OA; 4 μM), and analysed by Western blotting with anti-XMre11 serum. (B) Mock- and Mre11-depleted extracts supplemented with MRN-WT, or mutant recombinant MRN complex (SA, or SAI) were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl) and okadaic acid (OA; 4 μM), followed by Western blot analysis with anti-XMre11 serum. (C) Mock- and ATM-depleted extracts were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl), okadaic acid (OA; 4 μM), and caffeine (10 mM), wortmannin (100 μM) or NU7026 (25 μM), and analysed by Western blotting with anti-XATM serum (upper panel) and anti-XMre11 serum (lower panel). (D) Untreated membrane-free extracts were incubated for 15 min with DSBs (4.34 × 1011 ends/μl) in the presence of okadaic acid (OA; 4 μM) and the indicated concentrations of NU7026, and analysed by Western blotting with anti-XMre11 serum.
Mock- and Mre11-depleted membrane-free egg extracts supplemented with 125 nM, 250 nM, or 500 nM MRN-WT complex were incubated with biotinylated 1 Kb linear double-stranded DNA (B-1 Kb DNA) bound to streptavidin beads and free radioactive unbiotinylated 1 Kb DNA. Streptavidin-immobilized DNA fractions were isolated, and the associated radioactivity (cpm) was measured.
Acknowledgments
We are extremely grateful to T. Paull (University of Texas, Austin) for monomeric ATM protein and for baculoviruses encoding human wild type Rad50 and Nbs1, to A. Nussenzweig (National Cancer Institute) for antibodies against human Nbs1, and to G. Smith (KuDOS Pharmaceuticals) for the ATM inhibitor KU55933.
Funding: This work was supported by the National Institutes of Health [grant CA92245 to J.G.].
Footnotes
Conflict of Interest statement: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 2.O'Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 4.D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 5.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerosaletti K, Concannon P. Independent roles for nibrin and Mre11-Rad50 in the activation and function of Atm. J Biol Chem. 2004;279:38813–38819. doi: 10.1074/jbc.M404294200. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 12.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 14.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, Stotsky M, Mathew CG, Kastan MB, Weaver DT, D'Andrea AD. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- 16.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, Shay JW, Ziv Y, Shiloh Y, Lee EY. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, Kakazu N, Takeda S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. Embo J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, Takata M, Takeda S, Matsuura S, Komatsu K. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 21.Howlett NG, Scuric Z, D'Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair (Amst) 2006;5:251–257. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson DE, Trujillo KM, Sung P, Erickson HP. Structure of the Rad50 × Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J Biol Chem. 2001;276:37027–37033. doi: 10.1074/jbc.M106179200. [DOI] [PubMed] [Google Scholar]
- 24.Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci U S A. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 26.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 27.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem. 2005;280:2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- 30.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 31.Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- 32.Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 33.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 34.Boisvert FM, Dery U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boisvert FM, Hendzel MJ, Masson JY, Richard S. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle. 2005;4:981–989. doi: 10.4161/cc.4.7.1830. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 37.Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y, Dodson GE, Mukhopadhyay PS, Shanware NP, Trinh AT, Tibbetts RS. Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies. J Biol Chem. 2007;282:9236–9243. doi: 10.1074/jbc.M609256200. [DOI] [PubMed] [Google Scholar]
- 39.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 42.D'Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Z, Zhong Q, Chen PL. The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J Biol Chem. 1999;274:19513–19516. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- 44.Yuan SS, Chang HL, Hou MF, Chan TF, Kao YH, Wu YC, Su JH. Neocarzinostatin induces Mre11 phosphorylation and focus formation through an ATM- and NBS1-dependent mechanism. Toxicology. 2002;177:123–130. doi: 10.1016/s0300-483x(02)00220-2. [DOI] [PubMed] [Google Scholar]
- 45.Yuan SS, Su JH, Hou MF, Yang FW, Zhao S, Lee EY. Arsenic-induced Mre11 phosphorylation is cell cycle-dependent and defective in NBS cells. DNA Repair (Amst) 2002;1:137–142. doi: 10.1016/s1568-7864(01)00009-x. [DOI] [PubMed] [Google Scholar]
- 46.Takemura H, Rao VA, Sordet O, Furuta T, Miao ZH, Meng L, Zhang H, Pommier Y. Defective Mre11-dependent Activation of Chk2 by Ataxia Telangiectasia Mutated in Colorectal Carcinoma Cells in Response to Replication-dependent DNA Double Strand Breaks. J Biol Chem. 2006;281:30814–30823. doi: 10.1074/jbc.M603747200. [DOI] [PubMed] [Google Scholar]
- 47.Kim ST. Protein kinase CK2 interacts with Chk2 and phosphorylates Mre11 on serine 649. Biochem Biophys Res Commun. 2005;331:247–252. doi: 10.1016/j.bbrc.2005.03.162. [DOI] [PubMed] [Google Scholar]
- 48.Murray AW. Cell Cycle Extracts. In: Kay BK, Peng HB, editors. Xenopus laevis: practical uses in cell and molecular biology. Academic Press, Inc.; San Diego: 1991. pp. 581–605. [Google Scholar]
- 49.Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. In: Hamkalo BA, Elgin SCR, editors. Functional organization of the nucleus: a laboratory guide. Academic Press, Inc.; San Diego: 1991. pp. 449–468. [DOI] [PubMed] [Google Scholar]
- 50.Di Virgilio M, Gautier J. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J Cell Biol. 2005;171:765–771. doi: 10.1083/jcb.200506029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson K, Hensey C, Gautier J. Isolation and characterization of Xenopus ATM (X-ATM): expression, localization, and complex formation during oogenesis and early development. Oncogene. 1999;18:7070–7079. doi: 10.1038/sj.onc.1203194. [DOI] [PubMed] [Google Scholar]
- 52.Ying CY, Gautier J. The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding. Embo J. 2005;24:4334–4344. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bradshaw PS, Stavropoulos DJ, Meyn MS. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- 55.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–539. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
- 57.Costanzo V, Robertson K, Ying CY, Kim E, Avvedimento E, Gottesman M, Grieco D, Gautier J. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol Cell. 2000;6:649–659. doi: 10.1016/s1097-2765(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 58.Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 59.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. Embo J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 61.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 62.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 63.Hollick JJ, Golding BT, Hardcastle IR, Martin N, Richardson C, Rigoreau LJ, Smith GC, Griffin RJ. 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-Dependent protein kinase (DNA-PK) Bioorg Med Chem Lett. 2003;13:3083–3086. doi: 10.1016/s0960-894x(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 64.Almouzni G, Mechali M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. Embo J. 1988;7:665–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laskey RA, Mills AD, Morris NR. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- 66.Wolffe AP. Chromatin Assembly. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical Uses in Cell and Molecular Biology. Academic Press Inc; 1991. [Google Scholar]
- 67.Crowe AJ, Barton MC. Functional analysis of chromatin assembled in synthetic nuclei. Methods. 1999;17:173–187. doi: 10.1006/meth.1998.0728. [DOI] [PubMed] [Google Scholar]
- 68.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 69.Ladoux B, Quivy JP, Doyle P, du Roure O, Almouzni G, Viovy JL. Fast kinetics of chromatin assembly revealed by single-molecule videomicroscopy and scanning force microscopy. Proc Natl Acad Sci U S A. 2000;97:14251–14256. doi: 10.1073/pnas.250471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner G, Bancaud A, Quivy JP, Clapier C, Almouzni G, Viovy JL. Compaction kinetics on single DNAs: purified nucleosome reconstitution systems versus crude extract. Biophys J. 2005;89:3647–3659. doi: 10.1529/biophysj.105.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 72.Kim JE, Minter-Dykhouse K, Chen J. Signaling networks controlled by the MRN complex and MDC1 during early DNA damage responses. Mol Carcinog. 2006;45:403–408. doi: 10.1002/mc.20221. [DOI] [PubMed] [Google Scholar]
- 73.Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 74.Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. Embo J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang X, Hui ZG, Cui XL, Garg R, Kastan MB, Xu B. A novel ATM-dependent pathway regulates protein phosphatase 1 in response to DNA damage. Mol Cell Biol. 2008;28:2559–2566. doi: 10.1128/MCB.01711-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. Embo J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 78.Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY. Interplay between human DNA repair proteins at a unique double-strand break in vivo. Embo J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leroy C, Lee SE, Vaze MB, Ochsenbien F, Guerois R, Haber JE, Marsolier-Kergoat MC. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 80.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, Lieberman J, Shen X, Buratowski S, Haber JE, Durocher D, Greenblatt JF, Krogan NJ. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 81.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of wild type (WT) hMre11 and SQ/TQ site mutant proteins. The amino acid number of the Ser/Thr residues in the eight SQ/TQ motifs of the WT protein is indicated. SA: all eight SQ/TQ sites mutated to AQ; SAI: S264Q, T329Q, S382Q motifs mutated to AQ; SAII: T481Q, S531Q, S590Q, S676Q, S678Q motifs mutated to AQ. SD: all eight SQ/TQ sites mutated to DQ. (B) Recombinant mutant human MRN complexes were purified from Sf9 cells following co-infection with baculoviruses for hRad50, hNbs1, and each of the hMre11 SQ/TQ site mutants shown in panel A, and analysed by Western blotting with anti-hRad50 serum (upper panel), anti-hNbs1 serum (middle panel), and anti-XMre11 serum (lower panel). (C) MRN-SD complex was incubated in Mre11-depleted extracts for the indicated time points, and either directly electrophoresed (left panels) or immunoprecipitated with either preimmune (Pre) or anti-XMre11 (αMre11) serum (right panels). Inputs and immunoprecipitates were analysed by Western blotting with anti-hRad50 serum (upper panel), anti-hNbs1 serum (middle panel), and anti-XMre11 serum (lower panel).
(A) Mre11-depleted extracts supplemented with WT, or mutant recombinant MRN complex (SA, SAII, or SD) were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl) and okadaic acid (OA; 4 μM), and analysed by Western blotting with anti-XMre11 serum. (B) Mock- and Mre11-depleted extracts supplemented with MRN-WT, or mutant recombinant MRN complex (SA, or SAI) were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl) and okadaic acid (OA; 4 μM), followed by Western blot analysis with anti-XMre11 serum. (C) Mock- and ATM-depleted extracts were incubated for 15 min in the presence or absence of DSBs (4.34 × 1010 ends/μl), okadaic acid (OA; 4 μM), and caffeine (10 mM), wortmannin (100 μM) or NU7026 (25 μM), and analysed by Western blotting with anti-XATM serum (upper panel) and anti-XMre11 serum (lower panel). (D) Untreated membrane-free extracts were incubated for 15 min with DSBs (4.34 × 1011 ends/μl) in the presence of okadaic acid (OA; 4 μM) and the indicated concentrations of NU7026, and analysed by Western blotting with anti-XMre11 serum.
Mock- and Mre11-depleted membrane-free egg extracts supplemented with 125 nM, 250 nM, or 500 nM MRN-WT complex were incubated with biotinylated 1 Kb linear double-stranded DNA (B-1 Kb DNA) bound to streptavidin beads and free radioactive unbiotinylated 1 Kb DNA. Streptavidin-immobilized DNA fractions were isolated, and the associated radioactivity (cpm) was measured.


