Abstract
Background
Rho1 is a small GTPase of the Ras superfamily that serves as the central component in a highly conserved signaling pathway that regulates tissue morphogenesis during development in all animals. Since there is tremendous diversity in the upstream signals that can activate Rho1 as well as the effector molecules that carry out its functions, it is important to define relevant Rho1-interacting genes for each morphogenetic event regulated by this signaling pathway. Previous work from our lab and others has shown that Rho signaling is necessary for the morphogenesis of leg imaginal discs during metamorphosis in Drosophila, although a comprehensive identification of Rho1-interacting genes has not been attempted for this process.
Methodology/Principal Findings
We characterized an amorphic allele of Rho1 that displays a poorly penetrant dominant malformed leg phenotype and is capable of being strongly enhanced by Rho1-interacting heterozygous mutations. We then used this allele in a second-site noncomplementation screen with the Exelixis collection of molecularly defined deficiencies to identify Rho1-interacting genes necessary for leg morphogenesis. In a primary screen of 461 deficiencies collectively uncovering ∼50% of the Drosophila genome, we identified twelve intervals harboring Rho1-interacting genes. Through secondary screening we identified six Rho1-interacting genes including three that were previously identified (RhoGEF2, broad, and stubbloid), thereby validating the screen. In addition, we identified Cdc42, Rheb and Sc2 as novel Rho1-interacting genes involved in adult leg development.
Conclusions/Significance
This screen identified well-known and novel Rho1-interacting genes necessary for leg morphogenesis, thereby increasing our knowledge of this important signaling pathway. We additionally found that Rheb may have a unique function in leg morphogenesis that is independent of its regulation of Tor.
Introduction
Cell shape changes, cell rearrangements, oriented cell divisions and regulated cell death collectively shape tissues and ultimately influence the development of the organism as a whole. These processes, generically referred to as morphogenesis, are the underlying mechanisms for numerous developmental events including gastrulation, neurulation, organogenesis and metamorphosis. Morphogenetic processes are largely driven by regulated changes of the actin cytoskeleton. Members of the highly conserved Rho family of small GTPases are key regulators of the actin cytoskeleton (reviewed in [1], [2]), and genetic and pharmacological studies in a variety of organisms have demonstrated critical roles for Rho, Rac and Cdc42 in regulating specific morphogenetic events during development. For example, Rho signaling has been implicated in neural tube closure [3] and cardiac morphogenesis [4] during vertebrate embryogenesis, whereas Rac signaling is critical for neurite outgrowth in vertebrates, worms and flies [5]-[7], and Cdc42 signaling plays similar roles in neurite outgrowth and axon guidance in vertebrates [8], [9].
Drosophila melanogaster has proven to be an excellent model organism for elucidating the function of Rho proteins during development and for identifying genes that function in concert with Rho proteins to transduce signals through these GTPases (reviewed in [10]). The Drosophila genome encodes one Rho gene (Rho1), one Cdc42 gene (Cdc42), three Rac genes (Rac1, Rac2, and Mtl), and several additional, more divergent, Rho family genes (e.g. RhoBTB and RhoL). In Drosophila, loss of function genetic analysis has demonstrated a role for Rho1 signaling during oogenesis, and for cellularization of the blastoderm embryo, gastrulation, dorsal closure, and head involution during embryogenesis [11]–[13]. Similarly, complete loss of all three Rac genes leads to defects in dorsal closure and neural development [7], whereas zygotic loss of Cdc42 results in defects in germ band retraction during embryogenesis [14].
Like all members of the Rho family, Rho1 functions primarily as a molecular switch, alternating between an inactive, GDP-bound state and an active, GTP-bound form (reviewed in [1]). Guanine nucleotide exchange factors (GEFs) activate Rho1 by removing bound GDP, whereas GTPase activating proteins (GAPs) stimulate the weak GTPase activity of Rho1, thereby inactivating it. In certain contexts, guanine nucleotide dissociation inhibitors (GDIs) bind to and sequester GDP-bound Rho1, reinforcing the inactive Rho1 state. A number of effector proteins can bind activated Rho1 to transduce signals in specific ways. For example, Rho kinase is a serine/threonine kinase that regulates contractile events at the actin cytoskeleton primarily by phosphorylating and thereby inactivating the myosin binding subunit of the myosin phosphatase complex (reviewed in [15]). Since myosin phosphatase normally dephosphorylates the myosin regulatory light chain (MRLC; encoded by spaghetti squash or sqh in Drosophila), the net effect of activated Rho1 signaling through Rho kinase is an increase in the phosphorylation of MRLC. Phospho-MRLC induces a conformational change in the myosin heavy chain (encoded by zipper or zip in Drosophila) that increases the ability of myosin to bind to actin filaments, thereby generating a chemomechanical force on the actin cytoskeleton sufficient for cell shape changes.
Less is known about the upstream events that activate Rho1 signaling during Drosophila development. RhoGEF proteins directly activate Rho1, but the Drosophila genome encodes more than 20 genes that are predicted to have RhoGEF activity, some of which may have preferential specificity for a single Rho family member, whereas others may be more promiscuous. In addition, the mechanisms by which specific RhoGEFs are activated to signal through Rho1 are complex and not fully understood.
The morphogenesis of leg imaginal discs that occurs during Drosophila metamorphosis is a particularly useful genetic model for studying Rho1 signaling. Adult legs are derived from imaginal discs that were specified during embryogenesis and underwent extensive proliferation and patterning during larval development (reviewed in [16]). At the end of the third larval instar each of the leg imaginal discs consists of a single-layered columnar epithelium that is covered and apposed by a squamous peripodial epithelium. In response to the late larval pulse of the steroid hormone ecdysone that triggers puparium formation and initiates metamorphosis, these flat epithelial discs are transformed into rudimentary adult legs in approximately 12 hours (reviewed in [17]). Classical studies by the Fristrom lab and more recent imaging studies have revealed that this morphogenetic process is largely driven by changes in cell shape and by cell rearrangements [18]–[20]. Furthermore, studies by Fristrom and Fristrom [21] demonstrated that the elongation and eversion of the leg imaginal discs could be reversibly inhibited by cytochalasin B, indicating a central role for the actin cytoskeleton in driving leg disc morphogenesis. Not surprisingly, independent genetic modifier screens using zip and an ecdysone-induced transcription factor, broad, have identified genes in the Rho signaling pathway as playing a critical role in regulating leg morphogenesis [22]–[25]. Similar experiments also demonstrated robust genetic interactions between the ecdysone-induced type II transmembrane serine protease stubbloid (sbd) and components of the Rho signaling pathway [26].
As a means to identify genes that function in concert with Rho1 during leg morphogenesis we have conducted a modifier screen using an amorphic allele of Rho1 and the Exelixis collection of molecularly defined deficiencies [27]. Screening through a collection of 461 deficiencies that collectively uncover ∼50% of the Drosophila genome, we identified 12 deficiencies that likely contain Rho1-interacting genes necessary for leg morphogenesis. Included in this set were deficiencies that removed broad, RhoGEF2 and stubbloid, three genes that had previously been identified as interacting with Rho1 during leg imaginal disc morphogenesis. Further, we were able to identify Cdc42, Rheb, and Sc2 as likely Rho1-interacting genes.
Materials and Methods
Drosophila stocks
All Drosophila stocks were maintained on media consisting of corn meal, sugar, yeast, and agar in incubators maintained at a constant temperature of 21°C, or in a room that typically fluctuated between 21°C and 22.5°C. Many of the deficiency stocks used in this study were generated by Exelixis, Inc., and obtained from the Bloomington Drosophila stock center at Indiana University (Bloomington, IN) [27]. Other deficiency stocks and specific mutations used in the screen were also obtained from the Bloomington Drosophila stock center. The Rho1E(br)233 and Rho1E(br)246 stocks used in this study were isolated in a screen for dominant modifiers of broad [25]. The zipE(br), Rho1J3.8, and Rho1E3.10 stocks were obtained from S. Halsell (James Madison University; [23], [28]). The RhoGEF211-3 stock was obtained from L. von Kalm (University of Central Florida; [26]). Dll-Gal4, UAS-Rheb.Pa2, UAS-Rheb.Pa3, and UAS-PI3K92E.CAAX were obtained from the Bloomington Drosophila stock center. Genetic experiments were conducted in incubators controlled at a constant temperature of either 21°C or 25°C, as indicated.
Characterization of Rho1E(br)233 and Rho1E(br)246
Rho1E(br)233 and Rho1E(br)246 were balanced with CyO, P{w+, Dfd-EYFP} [29] to allow for unambiguous identification of homozygous mutant embryos starting at ∼12 hours after egg laying. Genomic DNA isolated from homozygous mutant late embryos was sequenced at the DNA Facility of the Iowa State University Office of Biotechnology (Ames, IA). Lethal phase analyses were performed by collecting homozygous mutant embryos produced through a four hour egg lay of Rho1E(br)246/CyO, P{w+, ActGFP} or Rho1E(br)233/CyO, P{w+, ActGFP} at 25°C, and determining the percentage of unhatched embryos after 48 hours. Non-hatched embryos were then dechorionated in 50% bleach, mounted on microscope slides in Hoyer's medium and subsequently examined for cuticular phenotypes on a Nikon Eclipse 80i compound microscope.
RNA isolation and northern blot analysis
Non YFP-expressing embryos were isolated from 4 hour collections of Rho1E(br)246/CyO, P{w+, Dfd-EYFP}, Rho1E(br)233/CyO, P{w+, Dfd-EYFP}, or w1118 that were aged to be 12–16 hours after egg laying (AEL), 16–20 AEL, or 20–24 hours AEL. ∼100 embryos from each collection were dechorionated and lysed in Tripure isolation reagent (Roche Applied Science, Indianapolis, IN). Total RNA was extracted from these lysates, and approximately 10 µg of total RNA per sample were separated by formaldehyde agarose gel electrophoresis and transferred to a nylon membrane (GeneScreen Plus, PerkinElmer, Waltham, MA). The membrane was hybridized and stripped as described by [30]. Specific probes were labeled by random priming of gel-purified fragments (Stratagene, La Jolla, CA). Generation of probe fragments for Rho1 is described in [25], and for rp49 in [31].
Protein isolation and western blot analysis
Non YFP-expressing embryos were isolated from 4 hour collections of Rho1E(br)246/CyO, P{w+, Dfd-EYFP}, Rho1E(br)233/CyO, P{w+, Dfd-EYFP}, or w1118 that were aged to be 12–16 hours AEL, 16–20 AEL, or 20–24 hours AEL. ∼100 embryos from each collection were dechorionated and lysed in 1X SDS sample buffer [32]. The protein samples were boiled, separated on a 12% SDS-PAGE, and transferred to PVDF membrane (Immun-Blot, Bio-Rad, Hercules, CA) for 1 h at 100 V at 4°C. Blots were blocked in 5% nonfat milk in TBS plus 0.1% Tween-20 for 30 min. at room temperature, and then incubated overnight at 4°C in primary antibody. Anti-Rho1 (p1D9 from the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA) was used at 1:500 and anti-β-tubulin (E7 from the DSHB) was used at 1:3,000. After incubation with horseradish peroxidase–coupled secondary antibodies (Jackson ImmunoReseach Laboratories, West Grove, PA), the immunoreactive proteins were visualized using chemiluminescent detection (Pierce, Rockford, IL).
Deficiency screen
Second-site noncomplementation (SSNC) tests between autosomal deficiencies (or specific mutations) and Rho1E(br)246 were performed by mating eight to ten Rho1E(br)246/CyO, P{w+, ActGFP} virgin females to eight to ten deficiency– or specific mutation–bearing heterozygous males in vials. After 3 days the adults were transferred to fresh vials, and then to a third vial after two additional days. Newly eclosing F1 flies were separated by genotype and examined for malformed legs each day for a total of 10 days per vial. Subsequent secondary screening with Rho1E(br)233 and Rho1E3.10 were performed in the same manner, as were SSNC tests between Rho1-interacting deficiencies or specific Rho1-interacting mutations and mutations in RhoGEF2 and zip. SSNC tests involving X-linked deficiencies or mutations were performed in a similar manner, but we reversed the sexes of the crossed stocks (i.e. Rho1E(br)246/CyO, P{w+, ActGFP} males crossed to deficiency-bearing hemizygous virgin females).
Prior to conducting the screen we considered five alleles of Rho1 (Rho1E(br)233, Rho1E(br)246, Rho1k02107b, Rho1J3.8 and Rho1E3.10) and tested them with mutations in two known Rho1-interacting genes, zip and RhoGEF2, at 21°C and 25°C. We considered an animal to be malformed if it displayed even a single malformed leg, and defined a leg as malformed if any femur, tibia or tarsal segment was bent or twisted or was excessively short and fat (examples are shown in Figure 1B–D). As shown in Table 1, only Rho1E(br)246/+ and Rho1E3.10/+ showed a background penetrance of malformed legs less than 2% at either temperature (Rho1E(br)246/+ at 21°C and Rho1E3.10/+ at both temperatures). Both of these Rho1 alleles are also capable of being strongly enhanced by heterozygous mutations in RhoGEF2 and zip, with Rho1E(br)246 showing a modestly better interaction. Given the fact that Rho1E(br)246 is an amorphic allele, whereas Rho1E3.10 is likely an antimorphic allele [23], we decided to use Rho1E(br)246 for the screen. In addition, although we observed stronger interactions at 25°C than 21°C for all the Rho1 alleles, we decided to conduct the screen at 21°C since the background level of malformations was lower at this temperature, and thus would likely maximize our ability to identify Rho1-interacting loci. Using these conditions, we established our threshold for interaction at 10% malformed legs in animals doubly heterozygous for Rho1E(br)246 and any deficiency or specific mutation.
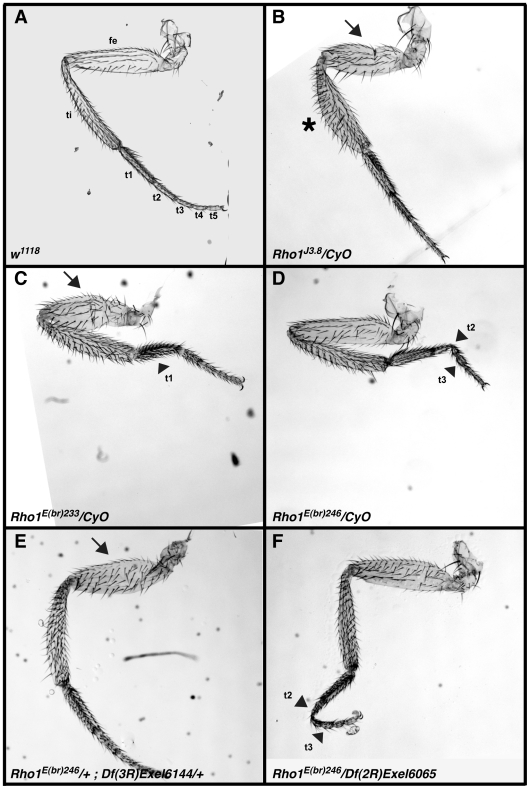
Figure 1. Representative malformed leg phenotypes in animals heterozygous for mutations in Rho1, or doubly heterozygous for mutations in Rho1E(br)246 and specific Rho1-interacting deficiencies.
Brightfield photomicrographs of representative adult legs from the third thoracic segment of w1118 (A), Rho1J3.8/Cyo (B), Rho1E(br)233/CyO (C), Rho1E(br)246/CyO (D), Rho1E(br)246/+;Df(3R)Exel6144/+ (E), and Rho1E(br)246/Df(2R)Exel6065 (F). Femur (fe), tibia (ti), and the five tarsal segments (t1-t5) are labeled in (A). Some animals show short, fat femurs (arrows in B, C and E) or tibias (asterisk in B), whereas others show tarsal segments that are short and fat, or are long and thin and occasionally show severe bends (arrowheads in C, D and F). Note that malformations in all leg segments are seen with each Rho1 allele, and in the interaction between Rho1 alleles and specific Rho1-interacting deficiencies.
Table 1. SSNC control data with select Rho1 alleles.
| Genotypea | Temperature (°C) | %Malformed (n)b |
| Rho1E(br)246/+ | 21 | 1.6 (689) |
| 25 | 3.1 (878) | |
| Rho1E(br)233/+ | 21 | 3.1 (709) |
| 25 | 4.9 (733) | |
| Rho1E3.10/+ | 21 | 1.0 (295) |
| 25 | 0.7 (277) | |
| Rho1k02107b/+ | 21 | 4.7 (172) |
| 25 | 4.1 (195) | |
| Rho1J3.8/+ | 21 | 2.0 (406) |
| 25 | 3.1 (425) | |
| Rho1E(br)246 +/+ RhoGEF211-3b | 21 | 37 (252) |
| 25 | 86 (43) | |
| Rho1E(br)233 +/+ RhoGEF211-3b | 21 | 31 (239) |
| 25 | 75 (61) | |
| Rho1E3.10 +/+ RhoGEF211-3b | 21 | 20 (143) |
| 25 | 20 (30) | |
| Rho1k02107b +/+ RhoGEF211-3b | 21 | 80 (5) |
| 25 | 100 (9) | |
| Rho1J3.8 +/+ RhoGEF211-3b | 21 | 55 (62) |
| 25 | 91 (22) | |
| Rho1E(br)246 +/+ zipE(br) | 21 | 50 (82) |
| 25 | 66 (44) | |
| Rho1E(br)233 +/+ zipE(br) | 21 | 20 (102) |
| 25 | 66 (29) | |
| Rho1E3.10 +/+ zipE(br) | 21 | ND |
| 25 | 33 (12) | |
| Rho1k02107b +/+ zipE(br) | 21 | ND |
| 25 | ND | |
| Rho1J3.8 +/+ zipE(br) | 21 | 95 (20) |
| 25 | 97 (30) |
Balanced, Rho heterozygous mutant virgin females were crossed to either w1118 males, or males bearing RhoGEF2 or zip mutations over a second chromosome balancer at 21°C and 25°C. b% malformed indicates the percentage of animals of the indicated genotype showing a malformed leg phenotype in at least one leg. n, total number of flies of the indicated genotype that were scored. ND, not determined.
For the primary screen we used the Exelixis collection of deficiencies maintained by the Bloomington Drosophila Stock Center. It has been reported that these deficiencies collectively uncover ∼56% of the Drosophila genome (predicted genes; [27]), although complementation tests conducted by the Drosophila stock center have shown that at least 10% are not completely deficient for the indicated intervals (http://flystocks.bio.indiana.edu/Browse/df-dp/dfextract.php?num=all&symbol=exeldef), and thus the collection provides closer to 50% coverage. It should be noted that two of the stocks that passed our primary screen, Df(1)Exel8196 and Df(1)Exel6253, were not tested by the stock center, but all of the remaining deficiencies that we describe in the text have been confirmed by the stock center.
Adult specimen preparations
Adult leg cuticles were prepared by dissecting legs from the third thoracic segment of w1118, Rho1E(br)246/+, Rho1E(br)233/+, Rho1J3.8/+, Rho1E(br)246/+;Df(3R)Exel6144, or Rho1E(br)246/Df(2R)Exel6065 in PBS, clearing them overnight in 10% KOH, and mounting them in Euporal (Bioquip, Gardena, CA) on microscope slides. Images of adult leg cuticles were captured on a Photometrics CoolSNAP ES high performance digital CCD camera mounted on a Nikon Eclipse 80i microscope. Images of adult legs from live wild type flies and flies overexpressing Tor signaling pathway genes were captured on Photometrics CoolSNAP cf color digital CCD camera mounted on a Leica MZFLIII stereomicroscope. All digital images were cropped and adjusted for brightness and contrast in Adobe Photoshop (version CS3, San Jose, CA).
Results
Characterization of newly isolated Rho1 alleles
Molecular characterizations of two new EMS-induced mutations of Rho1 that we recovered from a modifier screen of br1 [25] demonstrate that they are null alleles. Sequence analysis of genomic DNA from Rho1E(br)246 embryos revealed a G/C to A/T transition in the start codon (nucleotide 9130 from genomic clone AF177871; Figure 2A). Consistent with this observation, we did not detect any Rho1 protein by western blot or by indirect immunofluorescence of fixed embryos in Rho1E(br)246 mutant animals from 12 hr after egg laying (the earliest time point we could unambiguously identify mutant embryos; Figure 2C and data not shown). We also observed reduced Rho1 transcript levels in mutant embryos (Figure 2B), raising the possibility that this mutation engages a nonsense-mediated RNA decay pathway. Similarly, genomic sequencing of Rho1E(br)233 revealed a G/C to A/T transition at the invariant G in the splice donor site of the first exon (nucleotide 9284 from AF177871; Figure 2A). Northern blot analysis of total RNA isolated from 12–24 hour Rho1E(br)233 mutant embryos revealed nearly wild type levels of expression, but altered Rho1 transcript sizes, consistent with a defect in splicing (Figure 2B). We have not determined the nature of the altered transcript since there are several potential splice donor sites in the 1.2 kb intron. The protein predicted from this allele would encode the amino terminal 52 amino acids of Rho1 followed by 12 novel (non-Rho1) amino acids before a premature stop codon. Consistent with this result we do not detect any Rho1 protein by western blot or by indirect immunofluorescence of fixed embryos in Rho1E(br)233 mutant animals (Figure 2C and data not shown), although the 1D4 anti-Rho1 monoclonal antibody recognizes an epitope within the carboxyl-terminal 55 amino acids [33], and therefore would not detect the mutant protein. Together, these molecular characterizations suggest that Rho1E(br)246 and Rho1E(br)233 are likely null alleles.
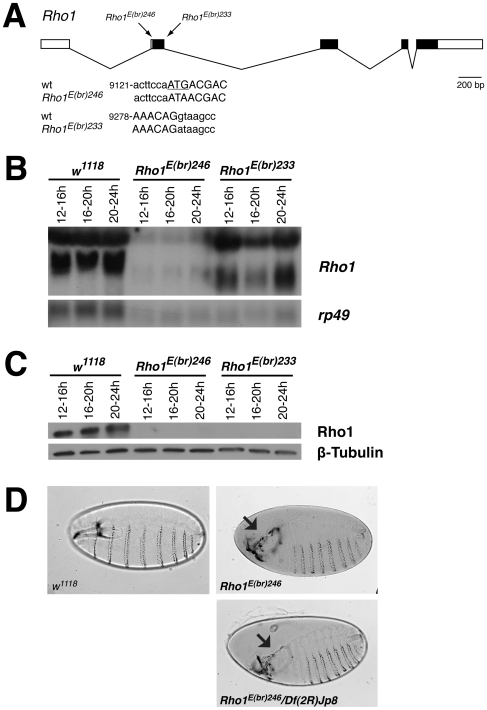
Figure 2. Molecular and genetic characterization of Rho1E(br)246 and Rho1E(br)233.
(A) Schematic diagram of the Rho1 transcript. Exons are indicated by boxes with coding regions filled in black. The locations of the Rho1E(br)246 and Rho1E(br)233 mutations are indicated above the diagram and the molecular lesions are shown in the sequences below. 5′ untranslated sequence and intronic sequences are written in lowercase letters while coding sequence is written in capital letters. The ATG start codon is underlined. The number refers to the sequence from genomic clone AF177871. (B) Northern blot analysis of total RNA isolated from w1118, Rho1E(br)246, and Rho1E(br)233 mutant embryos at 12–16 hrs, 16–20 hrs, and 20–24 hrs after egg laying. Hybridization to rp49 was used as a control for loading and transfer, and indicates that the Rho1 samples are underloaded relative to w1118. Hybridization of the blot with Rho1 indicates that Rho1E(br)246 produces substantially less Rho1 transcripts, whereas Rho1E(br)233 produces transcripts at ∼ wild type levels but with altered size. The two prominent bands in the w1118 samples are the 2.1 kb and 1.3 kb transcripts described by [12]. (C) Western blot analysis of total protein lysate isolated from w1118, Rho1E(br)246, and Rho1E(br)233 mutant embryos at 12–16 hrs, 16–20 hrs, and 20–24 hrs after egg laying. No Rho protein is observed in Rho1E(br)246 or Rho1E(br)233 mutant embryos. β-Tubulin was used as a control for loading and transfer. (D) Brightfield photomicrographs of cuticle preparations of w1118, Rho1E(br)246 and Rho1E(br)246/Df(2R)Jp8 mutant embryos. All animals are shown with anterior to the left and the dorsal surface up. All Rho1E(br)246 and Rho1E(br)246/Df(2R)Jp8 mutant animals die as embryos, with nearly completely penetrant defects in head involution (arrows point to dorsal anterior holes in the cuticle).
Genetic experiments on Rho1E(br)246 and Rho1E(br)233 support the conclusions of the molecular data and demonstrate that they are amorphic alleles. Rho1E(br)246 mutant embryos show complete embryonic lethality with a nearly completely penetrant defect in head involution (135/136 mutant embryos showed anterior open defects). The phenotype is identical in penetrance and expressivity to Rho1E(br)246/Df(2R)Jp8 (Figure 2D), and is consistent with the zygotic loss of function phenotype reported for the strong loss of function Rho1 allele reported by Magie et al. [12]. Similarly, Rho1E(br)233 mutant embryos show complete embryonic lethality with nearly completely penentrant head involution defects (data not shown).
A second-site noncomplementation (SSNC) screen for modifiers of Rho signaling during leg morphogenesis
In order to identify genes that function as part of a Rho signaling pathway required for leg imaginal disc morphogenesis during metamorphosis, we screened the Exelixis deficiency collection [27] for deficiencies that increased the penetrance of malformed legs in animals doubly heterozygous for the deficiency and a loss of function allele of Rho1 (Examples of malformed legs in animals doubly heterozygous for Rho1E(br)246 and specific deficiencies are shown in Figure 1E and F). The Exelixis collection consists of ∼500 molecularly-defined deficiencies generated by FLP-mediated recombination of FRT-bearing P-element stocks. The key advantages to this collection of deficiencies are that they were generated in a near isogenic background, the deletions are small (∼140 kb on average), and the breakpoints are known. Prior to the screen we conducted a series of control experiments to determine which Rho allele and what conditions were best for conducting the large-scale screen (details are presented in Materials and Methods). It should be noted that all of the potential Rho1 alleles show a dominant, partially penetrant malformed leg phentoype that varies between 1 and 5 percent depending upon the allele and temperature of development (Table 1; Figure 1B–D). From these experiments we chose to use Rho1E(br)246 at 21°C for the screen, and established a threshold for interaction at 10% malformed legs in animals doubly heterozygous for Rho1E(br)246 and any deficiency or specific mutation. Of the 461 deficiency stocks tested in the primary screen, 18 reached this threshold (there were two exceptions in which we observed an overall penetrance of 8%, but at least one of the three vials tested showed greater than 10% and we had additional evidence supporting the interval; see below). The entire dataset is presented in Table S1.
Through extensive secondary screening we confirmed 12 regions as containing putative Rho1-interacting genes and refined the intervals containing these genes (Table 2; the entire dataset including all SSNC tests with deficiencies and specific mutations for intervals that passed the primary screen is presented in Table S2). To accomplish this we retested the 18 deficiencies that passed the primary screen, along with additional overlapping deficiencies, for SSNC with Rho1E(br)246, and then tested the primary deficiency with two additional Rho1 alleles (Rho1E(br)233 and Rho1E3,10). After these secondary tests, we considered that a region contains a Rho1-interacting gene if the primary deficiency interacted with at least two alleles of Rho1 and at least two overlapping deficiencies interacted with an allele of Rho1 (one exception to this rule was Df(1)Exel6253 in which overlapping deficiencies were not available, see below). It should be noted that for the twelve deficiencies that passed these secondary tests there was low variability in the penetrance of malformed legs between the primary screen and the subsequent retest. We first determined the mean penetrance of malformed legs and the standard error of measurement. We then calculated the standard error as a percentage of the mean. Overall there was 18% variance around the mean for all of these deficiencies, with a range from 0% of the mean for Df(3R)Exel6144 to 34% of the mean for Df(3R)Exel7328.
Table 2. Summary of Rho1-interacting deficiencies and specific mutations.
| % malformed (n)c | |||||
| Primary screen Dfa | Secondary screen Df or specific mutation | Cytologyb | Rho1E(br)246 | Rho1E(br)233 | Rho1E3.10 |
| Df(1)Exel8196 | 2B1; 2B5 | 9 (78) | 31 (42) | 50 (28) | |
| Df(1)A94 | 1E3; 2B12 | 48 (23) | |||
| br5 | 2B3-5 | 9 (33)d | 8 (49)d | ||
| br1 | 2B3-5 | 18 (57)d | 21 (38)d | 62 (53)d | |
| dor8 | 2B5 | 11 (61) | |||
| Df(1)Exel6245 | 11E11; 11F4 | 11 (46) | 11 (47) | 0 (84) | |
| Df(1)N12 | 11D; 11F2 | 13 (82) | |||
| Df(1)C246 | 11D1; 12A1 | 13 (38) | |||
| Df(1)Exel6253 | 18D13; 18F2 | 11 (85) | 25 (51) | 18 (105) | |
| Cdc421 | 18E1 | 52 (40) | 47 (34) | 47 (38) | |
| Cdc423 | 18E1 | 9 (99) | 13 (68) | 27 (30) | |
| Df(2L)Exel6017 | 27E4; 27F5 | 19 (54) | 3 (124) | 1 (107) | |
| Df(2L)spdj2 | 27B2; 27F2 | 13 (122) | |||
| Df(2L)ED489 | 27E4; 28B1 | 12 (78) | |||
| Df(2L)Exel7055 | 34A2; 34A7 | 15 (65) | 10 (52) | 11 (105) | |
| Df(2L)prd1.7 | 33B3; 34A2 | 0 (172) | |||
| Df(2L)ED776, | 33E4; 34A3 | 1 (136) | |||
| Df(2L)ED777 | 33E7; 34A3 | 0 (72) | |||
| Df(2L)ED773 | 33E9; 34A3 | 0 (177) | |||
| Df(2L)ED778 | 33E9; 34A7 | 18 (89) | |||
| Df(2L)Exel8028 | 34A1; 34A2 | 0 (135) | |||
| Df(2L)ED774 | 34A3; 34A3 | 0 (121) | |||
| Df(2L)BSC30 | 34A3; 34B9 | 15 (89) | |||
| Df(2L)ED784 | 34A4; 34B6 | 32 (31) | |||
| Df(2L)Exel9023 | 34A6; 34A7 | 0 (211) | |||
| TorDeltaP | 34A4 | 5 (354) | 5 (195) | 5 (129) | |
| P{lacW}TorK17004 | 34A4 | 4 (161) | |||
| Df(2R)Exel7098 | 44D5; 44E3 | 10 (71) | 14 (125) | 9 (183) | |
| Df(2R)ED1742 | 44B9; 44E3 | 11 (149) | |||
| Df(2R)H3D3 | 44D1; 44F5 | 8 (77) | |||
| Df(2R)ED1770 | 44D8; 45B4 | 55 (60) | 60 (58) | 23 (64) | |
| Df(2R)Exel6065 | 53D14; 53F9 | 17 (163) | 34 (127) | 50 (70) | |
| Df(2R)ED2751 | 53D14; 53F9 | 38 (128) | |||
| Df(2R)ED1 | 53E4; 53F9 | 31 (80) | |||
| PBac{RB}RhoGEF2e03784 | 53E4-F1 | 57 (28) | |||
| P{EPgy2}RhoGEF2EY08391 | 53E4-F1 | 17 (92) | |||
| RhoGEF211-3b | 53E4-F1 | 37 (252) | |||
| Df(2R)Exel6098 | 63F2; 63F7 | 8 (120) | 8 (76) | 10 (72) | |
| Df(3L)ED208 | 63C1; 63F5 | 23 (59) | 35 (62) | 56 (19) | |
| Df(3L)ED4341 | 63F6; 64B9 | 10 (63) | |||
| Sc21 | 63F5 | 14 (133) | 33 (131) | 11 (80) | |
| P{PZ}Sc205634 | 63F5 | 3 (139) | |||
| Sc2A4 | 63F5 | 4 (92) | |||
| Sc2F9 | 63F5 | 0 (95) | |||
| P{UASp-YFP.Rab8.Q67L}Sc210 | 63F5 | 0 (126) | |||
| Df(3R)Exel6144 | 83A6; 83B6 | 10 (249) | 25 (87) | 5 (111) | |
| Df(3R)ED5177 | 83B4; 83B6 | 0 (61) | |||
| P{Mae-UAS.6.11}RhebAV4 | 83B2 | 8 (130) | 14 (77) | 4 (113) | |
| P{SUPor-P}RhebKG02006 | 83B2 | 3 (149) | |||
| P{EPgy2}RhebEY08085 | 83B2 | 2 (133) | |||
| P{Mae-UAS.6.11}RhebLA01053 | 83B2 | 4 (162) | |||
| Df(3R)Exel7328 | 89B1; 89B9 | 8 (227) | 37 (84) | 55 (116) | |
| Df(3R)Exel7327 | 89A8; 89B3 | 3 (95) | |||
| Df(3R)bxd100 | 89B6; 89E2 | 18 (89) | |||
| SbdE(br)536 | 89B4-6 | 14 (72)d | |||
| Df(3R)Exel6178 | 90E7; 91A5 | 18 (212) | 42 (85) | 18 (152) | |
| Df(3R)P14 | 90C2; 91B2 | 9 (68) | 6 (82) | 14 (59) | |
| Df(3R)Cha7 | 90F1; 91F5 | 11 (76) | |||
| Df(3R)ED5815 | 90F4; 91B8 | 8 (75) | |||
| Df(3R)Exel6179 | 91A5; 91B5 | 12 (272) | 21 (112) | 11 (159) | |
| Df(3R)Cha1a | 91A2; 92A1 | 28 (46) | |||
| Df(3R)ED2 | 91A5; 91F1 | 15 (127) | 28 (25) | ||
| Df(3R)BX5 | 91B1; 91D2 | 4 (134) | |||
| Df(3R)07280 | 91B2; 91C1 | 13 (125) | |||
Exelixis deficiencies identified in the primary screen that interact with more than one Rho1 allele and have been confirmed by the identification of overlapping Rho1-interacting deficiencies. bCytology is based upon flybase annotations as of January 2009 (reflects release 5 of the Drosophila genome). c% malformed indicates the percentage of animals heterozygous for the indicated Rho1 allele and heterozygous for the indicated deficiency or specific mutation showing the malformed leg phenotype in at least one leg. n, total number of flies of the indicated genotype that were scored. dData from [25].
Three of these interacting regions contain the previously identified Rho1-interacting genes broad, RhoGEF2, and stubbloid, and we have strong genetic evidence that another of the interacting intervals is due to an interaction between Rho1 and Cdc42. Further, our results indicate that Rheb and Sc2 interact genetically with Rho1 during imaginal disc morphogenesis, and that the Target of rapamycin (Tor) may also interact with Rho1 during this process. The details of these interactions are presented below.
broad (br)
Df(1)Exel8196 is predicted to uncover 18 genes from cytological region 2B1 to 2B5, including br. Although the deficiency stock was sick and we consistently obtained only small numbers of Df/+;Rho1E(br)246/+ animals, we did observe malformed legs at a frequency that placed this interval above the threshold in the majority of the vials tested. Consistent with this observation, Df(1)Exel8196 also showed SSNC with Rho1E(br)233 and Rho1E3.10 (Table 2). Furthermore, a larger overlapping deficiency, Df(1)A94, also showed SSNC with Rho1E(br)246. Since we previously identified Rho1E(br)246 and Rho1E(br)233 as dominant modifiers of the malformed leg phenotype associated with br1, and demonstrated that both Rho alleles show SSNC with the amorphic allele br5 [25], we are confident that the genetic interaction observed with this interval is due to an interaction between Rho1 and br. It should be noted that dor8 also showed SSNC with Rho1E(br)246 (Table 2), raising the possibility that deep orange may also be a Rho1-interacting gene, although thus far we have not followed up on this observation.
RhoGEF2
Df(2R)Exel6065 is a large molecularly defined deficiency that uncovers 47 predicted genes in the cytological interval 53D14 to 53F9. This deficiency displayed SSNC with all three alleles of Rho1 tested (Table 2). In addition, Df(2R)ED2751, a molecularly defined deficiency whose breakpoints are each within 200 bp of those for Df(2R)Exel6065, also shows SSNC with Rho1E(br)246. In order to refine this interval we tested Df(2R)ED1 and observed a robust genetic interaction with Rho1E(br)246. This molecularly defined deficiency has a left breakpoint within the first intron of RhoGEF2 and a right breakpoint identical to Df(2R)Exel6065. In total this deficiency is predicted to uncover 14 genes. There were loss of function alleles for four of these genes and we tested all of them for interaction with Rho1E(br)246. We observed SSNC with multiple RhoGEF2 alleles, but with none of the other mutations (Table 2 and Table S2). The identification of RhoGEF2 as a modifier for Rho1 during imaginal disc morphogenesis supports earlier observations from Halsell and Kiehart [23] and Bayer et al. [26].
Stubbloid (sbd)
The final previously identified Rho1-interacting gene we identified through the primary screen was sbd. Df(3R)Exel7328 showed 8% malformed legs in the primary screen, but several of the individual vials showed greater than 10% malformations. We therefore tested this deficiency with Rho1E(br)233 and Rho1E3.10 and observed much stronger interactions (Table 2). This deficiency is predicted to uncover 29 genes between 89B1 and 89B9. From the primary screen, we determined that an overlapping deficiency, Df(3R)Exel7327, does not show SSNC with Rho1E(br)246. Df(3R)Exel7327 has a left breakpoint in 89A8 and a right breakpoint in 89B3, thereby limiting the interacting interval to 19 genes including sbd. An overlapping deficiency, Df(3R)bxd100 (predicted interval: 89B6-89E2), also shows SSNC with Rho1E(br)246, strongly supporting this interval. Although we did not test any of the other genes in this interval, we had previously shown a genetic interaction between sbd and Rho1 [25], as had Bayer et al. [26].
Cdc42
We observed a SSNC between Df(1)Exel6253 (predicted interval: 18D13-18F2) and Rho1E(br)246, Rho1E(br)233 and Rho1E3.10. Although we were not able to confirm this finding with an overlapping deficiency that also showed SSNC with Rho1E(br)246, we were able to test loss of function alleles for 6 of the 32 genes predicted for this interval (all that had loss of function alleles available from the Bloomington Drosophila stock center), and found SSNC between two alleles of Cdc42 and all three tested alleles of Rho1. The interaction was stronger with Cdc421 than with Cdc423. In fact, Cdc421 showed a greater interaction with Rho1E(br)246 than did Df(1)Exel6253, and produced very few Cdc421/+; Rho1E(br)246/+ animals, although the other classes of offspring were well represented. This observation is consistent with a report that Cdc421 is an antimorphic allele [14].
Rheb
During the primary screen we observed SSNC between Df(3R)Exel6144 (predicted interval 83A6-83B6) and both Rho1E(br)246 and Rho1E(br)233. The Rho1-interacting region was further refined by the lack of interaction between Rho1E(br)246 and the molecularly defined deficiency Df(3R)ED5177 (predicted interval 83B4-83B6), thereby limiting the region to 17 predicted protein-coding genes and 16 small nucleolar RNA genes in 83A6-83B4. We were able to test lethal alleles for eight of these genes, seven of which showed no SSNC with Rho1E(br)246 (Table S2). The final mutation, P{Mae-UAS.6.11}RhebAV4, showed SSNC with both Rho1E(br)246 and Rho1E(br)233 (Table 2). This allele of Rheb is a P-element insertion that has an UAS element to allow for overexpression of Rheb, but the insertion also acts as a recessive lethal allele of Rheb, resulting in slow growth and eventual death as delayed first instar larvae [34]. Two additional P element alleles of Rheb, P{Mae-UAS.6.11}RhebLA01053 and P{EPgy2}RhebEY08085, failed to show SSNC with Rho1E(br)246 (Table 2).
Sc2
We observed a weak SSNC between Df(3L)Exel6098 (predicted interval 63F2-63F7) and Rho1E(br)246 during the primary screen. We observed a similar interaction between this deficiency and Rho1E(br)233 and Rho1E3.10. During the secondary screen we observed a very strong SSNC between Rho1E(br)246 and Df(3L)ED208, a molecularly defined deficiency with breakpoints in 63C1 and 63F5. We also observed SSNC between Rho1E(br)246 and Df(3L)ED4341, another molecularly defined deficiency (predicted interval: 63F6-64B9) that overlaps with Df(3L)Exel6098, but not with Df(3L)ED208, suggesting that there are likely two genes responsible for these interactions (Table 2). One of these genes is predicted to map to interval 63F6-7 that contains only four genes, Sc2, ida, mge, and Eip63F-1. We therefore tested loss of function alleles for Sc2, ida and mge (all of the Eip63F-1 alleles are viable) and found SSNC between Sc21 and Rho1E(br)246, Rho1E(br)233 and Rho1E3.10 (Table 2 and Table S2). Sc21 is an EMS generated hypomorphic allele resulting in pupal lethality [35]. The other three alleles of Sc2, however, showed 3-4% malformed legs when heterozygous with Rho1E(br)246/+, and thus were deemed to not interact. To identify the other potential Rho1-interacting gene we tested mutations in 13 genes that mapped to the interval 63C1 to 63F5 for SSNC with Rho1E(br)246 (representing all of the genes that had loss of function mutations available from the stock center). None of these mutations, however, showed an interaction with Rho1E(br)246 (Table S2).
Tor
We identified a SSNC between Rho1E(br)246 and Df(2L)Exel7055 (predicted interval 34A2-34A7). Df(2L)Exel7055 also showed SSNC with Rho1E(br)233 and Rho1E3.10. To refine this interval we tested ten additional deficiencies with three of them interacting and seven failing to interact (Table 2). Using the molecularly defined deficiencies, we were able to limit the Rho1-interacting region to 13 potential genes between Target of rapamycin (Tor) and Sir2 in 34A4-34A7. We tested loss of function alleles for five of these genes (all that were available), but did not observe a genetic interaction with any of them (Table S2). We did, however, observe an interaction between TorΔP and Rho1E(br)246 that, although below our threshold, consistently gave 5% malformed legs with all three tested Rho1 alleles. TorΔP is a deletion resulting from an imprecise excision of a P-element that removes the start codon and the amino terminal 902 codons of Tor and thus is likely an amorphic allele [36]. We tested a weaker loss of function allele of Tor, P{lacW}Tork17004, but observed less than 5% malformed legs with Rho1E(br)246 (Table 2).
Other Rho1-interacting loci
There are five additional intervals that are predicted to contain Rho1-interacting genes based upon the criteria outlined above: 11E11-11F2, 27E4-27F2, 44D8-44E3, 90F4-91A5, and 91A5-91B1. We have narrowed these intervals as far as possible using all available molecularly defined deficiencies, and in each case have tested putative loss of function mutations for all the genes for which stocks are available, but have not found any additional potential Rho1-interacting genes (Table S2). Although we have carefully examined the lists of genes for these remaining intervals, there are no obvious candidates for Rho1-interacting genes, and thus the cloning and characterization of these genes should provide novel insights into Rho1 signaling during leg development.
Genetic interactions between Rho1-interacting deficiencies and components of the Rho signaling pathway
In order to further characterize the Rho1-interacting deficiencies, we crossed all of them (and several of the putative Rho1-interacting specific mutations) to zipE(br) and RhoGEF211-3b, and found that nine of the twelve deficiencies showed SSNC with at least one of these mutations. This experiment was predicated on previous studies reporting strong genetic interactions between Rho1 and several components in the Rho signaling pathway, including RhoGEF2 and zip, during leg imaginal disc development [23]–[26]. We first determined the background level of malformed legs in RhoGEF211-3b/+ and zipE(br)/+ adults. RhoGEF211-3b/+ adults showed malformed legs with a frequency of 1% (n = 398) regardless of the direction of the cross (for example, w1118 males crossed to RhoGEF211-3b/CyO females), whereas the frequency of malformations in zipE(br)/+ varied according to the sex of the parents in the cross, with a higher frequency of malformed progeny resulting from a cross in which the mother provided the zipE(br) allele (2%, n = 126 with zipE(br)/SM5 males and 6%, n = 116 with zipE(br)/SM5 females). Since all of the autosomal deficiency crosses were set up with zipE(br)/SM5 mothers, we considered an interaction to be significant when the progeny showed 20% or greater malformations, whereas we retained the 10% threshold for all other crosses. As shown in Table 3, seven of the twelve Rho1-interacting deficiencies showed SSNC with both zipE(br) and RhoGEF211-3b (or failed to complement RhoGEF2 in the case of Df(2R)Exel6065), strongly implicating genes uncovered by these deficiencies in Rho signaling. It should be noted that Df(2R)Exel7328, which removes sbd and showed the strongest SSNC with RhoGEF211-3b, was not tested for interactions with zipE(br), since genetic interactions between sbd and zip had been well documented [25], [26]. Two of the five remaining deficiencies showed SSNC with zipE(br), but not with RhoGEF211-3b.
Table 3. SSNC tests between Rho1-interacting deficiencies and zip and RhoGEF2.
| % malformed (n)b | ||||
| Primary screen Df | Specific mutation | Cytologya | zipE(br) | RhoGEF211-3b |
| Df(1)Exel8196 | 2B1; 2B5 | 59 (39) | 26 (14) | |
| br1 | 2B5 | 24 (88) | ND | |
| Df(1)Exel6245 | 11E11; 11F4 | 13 (45) | 3 (61) | |
| Df(1)Exel6253 | 18D13; 18F2 | 13 (45) | 24 (38) | |
| Cdc421 | 18 E1 | 64 (36) | 34 (47) | |
| Cdc423 | 18 E1 | 8 (114) | 4 (112) | |
| Df(2L)Exel6017 | 27E4; 27F5 | 8 (107) | 1 (150) | |
| Df(2L)Exel7055 | 34A2; 34A7 | 33 (138) | 10 (173) | |
| TorDeltaP | 34A4 | 9 (91) | 3 (119) | |
| Df(2R)Exel7098 | 44D5; 44E3 | 41 (86) | 12 (135) | |
| Df(2R)Exel6065 | 53D14; 53F9 | 77 (99) | Failed to complement | |
| Df(3L)Exel6098 | 63F2; 63F7 | 11 (88) | 1 (127) | |
| Sc21 | 63F5-6 | 17 (46) | 1 (173) | |
| Df(3R)Exel6144 | 83A6; 83B6 | 52 (61) | 5 (148) | |
| RhebAV4 | 83B2 | 19 (69) | 9 (126) | |
| Df(3R)Exel7328 | 89B1; 89B9 | ND | 38 (162) | |
| sbdE(br)536 | 89B4-6 | 33 (73)c | 2 (119)c | |
| Df(3R)Exel6178 | 90E7; 91A5 | 51 (45) | 14 (115) | |
| Df(3R)Exel7179 | 91A5; 91B5 | 11 (111) | 5 (174) | |
Cytology is based upon flybase annotations as of January 2009 (reflects release 5 of the Drosophila genome). b% malformed indicates the percentage of animals heterozygous for the indicated Exelixis deficiency or specific mutation and heterozygous for zipE(br) or RhoGEF211-3b showing the malformed leg phenotype in at least one leg. n, total number of flies of the indicated genotype that were scored. Background penetrance of malformed legs: RhoGEF211-3b/+ 1% (n = 398), zipE(br)/+ 2% (n = 126) with zipE(br)/SM5 fathers and 6% (n = 116) with zipE(br)/SM5 mothers. cData from [25].
We next tested specific Rho1-interacting mutations and found that Cdc421 strongly interacted with both zipE(br) and RhoGEF211-3b, whereas Cdc423 did not interact with these mutations. We also found some indication of an interaction between Sc21 and zipE(br), although it was below the threshold we established. Similarly, we observed a trend suggesting an interaction between RhebAV4 and both zip and RhoGEF2 alleles, although in both cases the results were just below the threshold. These results were encouraging, however, and more strongly suggested an interaction between Rheb and the Rho signaling pathway.
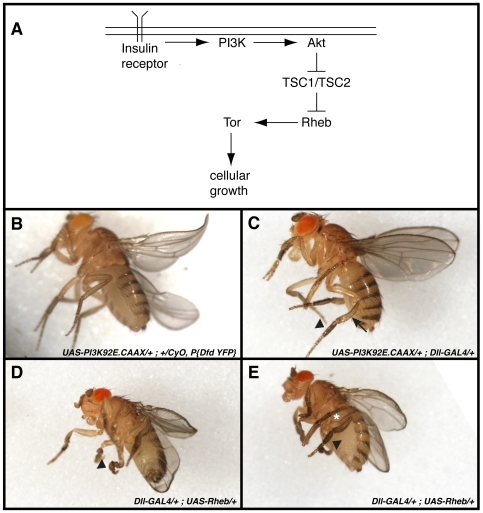
To further address the role of Rheb in leg morphogenesis, we overexpressed Rheb in distal leg segments using a UAS-GAL4 approach [37] and observed severely malformed legs. P{GawB}Dllmd23 flies express the yeast transcription factor GAL4 in the distal half of the tibia and in all the tarsal segments throughout imaginal disc development, as well as in the wing margin, the antennae, and mouth segments (see [38] for the expression pattern of dll-GAL4 in prepupae and pupae). We crossed two different UAS-Rheb transgenic lines to the P{GawB}Dllmd23 stock, and in both cases observed offspring with severely malformed legs and wings (100% malformed, n = 45 with P{UAS-Rheb.Pa}2 and 96% malformed, n = 170 with P{UAS-Rheb.Pa}3; Figure 3D, E). The malformations were characterized by fat tarsal segments that were often nonuniform in diameter and in many cases were kinked or curved. In addition, there was frequently a large kink in the femur, even though the transgene was not expressed in this tissue. The wings tended to be smaller, more delicate and slightly curved. Finally, we noticed a slight bend in the middle tibia corresponding to the boundary of the Dll expression domain with the segments distal to this point being larger in general to those proximal to the boundary.
Figure 3. Overexpression of Rheb in distal leg segments results in a strongly penetrant malformed leg phenotype.
(A) Model of the PI3K/Tor signaling pathway. Details of the pathway are found in the text. (B) Control animals, in this case heterozygous for UAS-PI3K92E.CAAX, but without a GAL4 driver, show normal leg morphology in live adults. Similarly, Dll-GAL4/+ and UAS-Rheb/+ adults have no leg malformations (not shown). (C) UAS-PI3K92E.CAAX/+; Dll-GAL4/+ animals express activated PI3K in distal leg segments (distal to the arrow) that results in a growth advantage giving the adults a “Popeye” leg phenotype. The most common morphogenesis defect is a slightly short, fat first tarsal segment (arrowhead). (D and E) Dll-GAL4/+; UAS-Rheb/+ adults show a highly penetrant malformed leg phenotype independent of the “Popeye” growth phenotype. Mild phenotypes include moderate to extreme short fat distal tibia and tarsal segments (arrowhead in E), whereas more extreme phenotypes include severely misshapen and twisted segments (arrowhead in D). The edges of the wings are often curved while the overall size of the wing is reduced. These defects are often accompanied by severe bends in the femur (white asterisk in E), even though the transgene is not expressed in this segment.
Rheb encodes a small GTPase known to activate Tor, which in turn regulates cellular growth via increased protein synthesis and inhibition of autophagy (reviewed in [39]). These proteins act in the highly conserved PI3K/Tor signaling pathway (Figure 3A). PI3 kinase (PI3K) is recruited to the plasma membrane in response to activation of the insulin receptor. At the membrane PI3K catalyzes the conversion of phophatidylinositol 4, 5-bisphosphate into phosphatidylinositol 3, 4, 5-triphosphate (PIP3). Increased levels of PIP3 at the membrane lead to the recruitment of the serine/threonine kinase Akt to the membrane. Akt then negatively regulates the GTPase activating proteins TSC1 and TSC2, which negatively regulate Rheb. Thus recruiting PI3K to the membrane ultimately results in the activation of Tor via Rheb. Since overexpressing Rheb may be sufficient to activate Tor and thereby increase growth, we wondered whether these phenotypes were simply due to an increase in cell size for the cells in the Dll domain. To address this we expressed P{UAS-PI3K92E.CAAX} with P{GawB}Dllmd23. PI3K92E.CAAX encodes the PI3K92E coding sequence with a farnesylation signal to target the recombinant protein to the membrane and thereby produces a constitutively active kinase. Flies expressing PI3K92E.CAAX in the Dll domain all showed the enlarged distal tibia and tarsal segments (a phenotype we referred to as “Popeye” legs), but very rarely showed any other malformations. The most extreme malformations we typically observed were the shorter fatter tarsal segments depicted in Figure 3C. Thus the severe malformed leg phenotypes associated with overexpression of Rheb are novel and independent of those produced by activating the Tor signaling pathway.
Assessing potential RhoGEFs involved in leg morphogenesis
RhoGEF2 is the only potential Rho-specific GEF that definitively showed dose-sensitive interactions with Rho1 during leg morphogenesis, although two additional genomic intervals containing potential RhoGEF genes also interacted with Rho1. Flybase [40] lists 26 genes as having potential RhoGEF activity based upon electronic annotation. Of these, 22 have Dbl homology (DH) domains, and 15 have both DH and PH domains. Since most well-described Rho-specific GEFs possess both a DH and a PH domain [41], we focused our attention on these 15 genes. One of these genes is identified only as a cDNA (GH16492), and thus is not annotated to a particular chromosomal location. We also included pebble (pbl), a known RhoGEF involved in cytokenesis that has a DH domain and a BRCA1 carboxyl-terminal domain [42]. Of these 15 genes, 7 of them are predicted to be deleted in at least one of the Exelixis deficiencies used in the primary screen. Six of these deficiencies showed no SSNC with Rho1E(br)246, whereas Df(2R)Exel6065 showed strong SSNC with Rho1 and is deficient for RhoGEF2 (Table 4). To test the remaining 8 potential RhoGEF genes, we obtained deficiencies predicted to uncover these genes from the Bloomington Drosophila stock center. Two of the deficiencies were molecularly defined, whereas the remaining five were relatively large, randomly generated deficiencies (4 X-ray induced and one generated by imprecise P element excision) with the potential RhoGEF genes well within the predicted endpoints of the deficiencies (Df(3L)pbl-X1 uncovers both pbl and CG33275). Three of these seven deficiencies produced at least 10% malformed legs when doubly heterozygous with Rho1E(br)246 (Table 4). Df(3R)ED5092 showed a SSNC of 12% with Rho1E(br)246 and uncovered the potential RhoGEF Cdep. Cdep encodes a protein with a FERM domain in addition to DH and PH domains. We failed to detect an interaction with a piggybac allele of Cdep (Table 4), although this allele is likely not amorphic.We also observed a very strong SSNC between Rho1E(br)246 and both Df(2R)nap11 and Df(2R)ED3636 (Table 4), raising the possibility that CG30440 and/or CG30115 may act as GEFs for Rho1 during leg morphogenesis. Unfortunately there are no specific mutations for either gene, and we have therefore not been able to test these genes directly. We have also thus far not been able to refine these deficiency intervals, although we have tested several specific mutations within the deficiencies and have not identified candidate Rho1-interacting genes (data not shown). Interestingly, Df(2R)nap11 is predicted to uncover genes through 42F including the ecdysone receptor gene EcR. Since we observed a strong genetic interaction between the ecdysone-inducible transcription factor br and Rho1, we suspected that the interaction between Df(2R)nap11 and Rho1E(br)246 might be due to EcR. In contrast to this notion, however, we found no SSNC between Rho1E(br)246 and the amorphic allele EcRm554fs (data not shown). Finally, Df(2L)64j showed an overall interaction with Rho1E(br)246 of 8% and uncovers Son of Sevenless (Sos), a Ras GEF that also contains DH and PH domains. We therefore tested an amorphic allele of Sos, Sos34Ea-6 and found no interaction with Rho1E(br)246 (Table 4).
Table 4. SSNC tests to assess potential RhoGEF genes.
| Potential RhoGEF | Cytologya | Protein domainsb | Df or mutation tested | Cytologya | % malf. (n)c |
| CG8557 | 16A5-B1 | DH, PH | Df(1)BK10 | 15F2; 16C10 | 0 (91) |
| vav | 18B6-7 | DH, PH, SH2, SH3, C1, CH | Df(1)Exel9068 | 18B4; 18B6 | 0 (64) |
| Sos | 34D1 | DH, PH, RasGEF | Df(2L)64j | 34D1; 35C1 | 8 (255) |
| Sos34Ea-6 | 34D1 | 2 (133) | |||
| CG10188 | 37E4-5 | DH, PH | Df(2L)Exel8041 | 37D7; 37F2 | 0 (106) |
| rtGEF | 38C5 | DH, PH, SH3 | Df(2L)Exel6046 | 38C2; 38C7 | 5 (21) |
| CG30440 | 41F2 | DH, PH | Df(2R)nap11 | 41E3; 42A10 | 54 (126) |
| RhoGEF2 | 53E4-F1 | DH, PH, PDZ, C1 | Df(2R)Exel6065 | 53D14; 53F9 | 17 (163) |
| CG30115 | 55D3-4 | DH, PH | Df(2R)ED3636 | 55B8; 55E3 | 67 (27) |
| RhoGEF3 | 61B3-C1 | DH, PH, SH3 | Df(3L)Exel6084 | 61B2; 61C1 | 6 (242) |
| trio | 61E1-2 | DH, PH, spectrin | Df(3L)Exel6086 | 61C9; 61E1 | 0 (208) |
| sif | 64E1-5 | DH, PH, PDZ | Df(3L)Exel6106 | 64D6; 64E2 | 4 (116) |
| RhoGEF4 | 65F4 | DH, PH | Df(3L)BSC33 | 65E10;65F6 | 4 (138) |
| pbl | 66A18-19 | DH, BRCT | Df(3L)pbl-X1 | 65F3; 66B10 | 5 (147) |
| CG33275 | 66A6-8 | DH, PH | Df(3L)pbl-X1 | 65F3; 66B10 | 5 (147) |
| Cdep | 82E2-3 | DH, PH, FERM | Df(3R)ED5092 | 82A3; 82E8 | 12 (104) |
| Pbac{5HPw+}CdepB122 | 82E2-3 | 4 (110) |
Cytology is based upon flybase annotations as of January 2009 (reflects release 5 of the Drosophila genome). bDH, Dbl homology domain; PH, pleckstrin homology domain; SH2, Src homology 2 domain; SH3, Src homology 3 domain; C1, Protein kinase C conserved region 1 domain; CH, Calponin homology domain; RasGEF, Ras guanine nucleotide exchange factor domain; PDZ, domain found in PSD-95, Dlg and ZO 1/2; BRCT, BRCA1 C-terminal domain; FERM, Protein 4.1, Ezrin, Radixin, Moesin homology domain. c% malformed indicates the percentage of animals heterozygous for the indicated Rho1 allele and heterozygous for the indicated deficiency or specific mutation showing the malformed leg phenotype in at least one leg. n, total number of flies of the indicated genotype that were scored.
Discussion
A genetic screen for modifiers of Rho signaling during leg morphogenesis
Using a deficiency-based genetic modifier screen that enabled us to test ∼50% of the genome, we identified 12 chromosomal regions that contain Rho1-interacting genes necessary for leg imaginal disc morphogenesis during metamorphosis in Drosophila. Through extensive secondary screening we were able to identify the likely Rho1-interacting gene for six of these intervals. Three of these genes, RhoGEF2, broad and stubbloid, had previously been shown to interact with Rho1 during leg development, validating the screen. We additionally identified Cdc42, Rheb and Sc2 as new Rho1-interacting genes necessary for leg imaginal disc morphogenesis, and observed a possible interaction between Rho1 and the Target of rapamycin (Tor) in this process.
Two aspects of the screen greatly contributed to its success. First, we identified an allele of Rho1, Rho1E(br)246, that has a very low background of leg malformations when heterozygous and can be strongly enhanced by second-site mutations in interacting genes. We determined that this is an amorphic allele of Rho1, which makes this a truly dose-sensitive interaction screen. The second key to the success of this screen was the use of the Exelixis deficiency collection. The near isogenic background of the stocks in this collection produced essentially no leg malformations, which resulted in a relatively high signal to noise ratio. When we remove the 18 intervals that were deemed to have passed the primary screen, the average percentage of malformed legs in the SSNC class for the remaining lines was 1.8% (as compared to the 1.7% malformations we observed in the cross between Rho1E(br)246 and w1118; Table 1). Thus, in conjunction with Rho1E(br)246, the identification of an interacting deficiency with even a modest 6-8 fold increase over background was significant as demonstrated by the fact that 67% of the intervals that were identified in the primary screen were confirmed through the secondary screen. We considered the screen to be successful based upon recovering all possible previously identified Rho1-interacting genes involved in leg morphogenesis (RhoGEF2; [23], br; [25], and sbd; [26]). We would have also expected to identify deficiencies uncovering zip (in 60E12 on 2R; [24]), and Rho kinase (in 14F2 on X; [26]), but the Exelixis collection did not contain deficiencies for these intervals. As a means to further characterize potential Rho1-interacting genes we tested each interacting deficiency, as well as specific mutations in candidate genes, for SSNC with alleles of zip and RhoGEF2 (Table 3). Nine of the twelve Rho1-interacting deficiencies showed SSNC with at least one of these mutations, as did specific mutations in br, sbd, and Cdc42, with Rheb and Sc2 showing an interaction that was just below our stated threshold (RhoGEF2 had already been shown to interact with zip and was not tested). Taken together, these results indicate that the screen was very successful in identifying genes that play important roles in Rho1 signaling during imaginal disc morphogenesis, and that the cloning and characterization of the remaining Rho1-interacting genes will likely provide important novel insights into this critical signaling pathway. We will next describe the newly identified Rho1-interacting genes and comment on their role in leg morphogenesis.
Cdc42
Cdc421 showed very strong interactions with all three of the Rho1 alleles that we tested, as well as with RhoGEF211-3b and zipE(br). Although Cdc421 is reported to be an antimorphic allele [14], it does not have a dominant effect on leg morphogenesis (data not shown). In addition, the weaker hypomorphic allele Cdc423 also showed SSNC with all three alleles of Rho1. Together these results support the identification of Cdc42 as the Rho1-interacting gene responsible for the interacting interval identified in 18D-F on the X chromosome.
Cdc42 and Rho1 are closely related small GTPases that likely regulate unique, but complementary processes required for leg morphogenesis. In general, Cdc42 regulates the cytoskeleton by controlling actin polymerization either through the formin mDia2 or by regulating actin branching through WASP and the ARP2/3 complex [43]. Rho can also regulate actin polymerization directly through mDia, but is also likely affecting leg morphogenesis through its regulation of myosin activity, a pathway dependent on Rho kinase. It is therefore possible that during leg morphogenesis Cdc42 regulates the actin cytoskeleton while Rho regulates myosin to affect cell shape changes and cell rearrangements. Another intriguing possibility is that Cdc42 and Rho1 are each regulating distinct aspects of adherens junction plasticity during the cell rearrangements that are occurring during the early stages of leg morphogenesis. Cell rearrangements necessitate the redistribution of junctional material as the border between two adjacent cells either expands or shrinks. It was recently shown that Cdc42 is required for the endocytosis of DE-cadherin and other adherens junction proteins in the early pupal notum, and that it carries out this function in conjunction with Par6 and atypical Protein Kinase C, through its downstream effectors WASP and ARP2/3 [44], [45]. Although the pupal notum is a fairly homeostatic tissue at this time, this function of Cdc42 is likely also critical during the more dynamic stages of tissue morphogenesis occurring 12 hours prior. Rho1 has also been shown to regulate the adherens junction. Specifically, Rho1 localizes to the adherens junction through its interaction with α-catenin and p120 catenin [33], and has been shown to regulate the distribution of DE-cadherin at adherens junctions during embryonic morphogenesis [46], likely through a mechanism dependent upon its downstream effector diaphanous [47].
Rheb
During the primary screen we identified a potential Rho1-interacting interval in 83A-B that we were not able to verify with an overlapping deficiency, although we did limit the potential interval to 17 protein-coding genes through non-interacting deficiencies. In the course of testing 8 of these potential Rho1-interacting genes, we identified a hypomorphic allele of Rheb that showed SSNC with Rho1E(br)233 and was close to reaching the threshold for SSNC with Rho1E(br)246 (Table 2). Although we failed to observe an interaction between three additional P-element insertion alleles of Rheb and Rho1E(br)246, we suggest that Rheb does interact with Rho1 during imaginal disc morphogenesis for the following reasons. First, RhebAV4 behaved similarly in pattern and level of interaction to Df(3R)Exel6144 with all three Rho1 alleles tested (Table 2). Second, zipE(br)/+;RhebAV4/+ and RhoGEF211-3b/+;RhebAV4/+ produced 19% and 9% malformed legs, respectively, which although just below the threshold we established for interaction, raises the possibility that Rheb interacts with the Rho signaling pathway (Table 3). Finally, overexpression of Rheb in distal leg segments resulted in a nearly completely penetrant malformed leg phenotype (Figure 3D, E).
An intriguing possibility is that there is an interaction between the Rho1 and Target of rapamycin (Tor) signaling pathways during imaginal disc morphogenesis. We observed an interaction between Df(2L)Exel7055 and all three tested alleles of Rho1. We subsequently limited the Rho1-interacting interval to 13 potential genes including Tor (Table 2). Although we did not detect an obvious interaction with Tor, we consistently observed a trend of ∼5% SSNC between TorΔP and all three Rho1 alleles (Table 2). In addition, we observed nearly the same level of interaction between another weak P element allele of Tor and Rho1E(br)246. Given the low background in this study, these results stood out even if they did not reach our threshold. In higher eukaryotes, a single TOR protein participates in two functionally distinct protein complexes: TOR complex 1 (TORC1) and TOR complex 2 (TORC2) (reviewed in [48]). TORC1 is a central regulator of cellular growth that integrates a number of growth promoting stimuli including growth factors, low energy and nutrients to regulate ribosome biogenesis and protein synthesis. TORC2 may also regulate cell growth, but importantly also appears to regulate cell morphology by mediating actin cytoskeletal modifications in response to growth factors. Recent evidence indicates that it carries out this function through the activation of Rac and Rho [49]. It is conceivable that TORC2 acts upstream of the Rho1 signaling pathway to regulate the actin cytoskeleton during the cell shape changes and cell rearrangements that drive leg morphogenesis.
An alternative possibility is that Rheb interacts with Rho1 during leg morphogenesis in a pathway that is independent of its role in regulating Tor. Our overexpression studies support this notion. Overexpression of an activated PI3 kinase in distal leg segments results in a highly penetrant growth advantage in those tissues, producing larger distal tibias and tarsal segments with very little overt malformations (Figure 3C). In contrast, overexpressing a wild type Rheb transgene gave a consistently strong malformed leg phenotype in addition to the growth advantage (Figure 3D, E). We observed nearly identical results using two independent Rheb transgenic lines. In addition, recent evidence suggests that although Rheb has a well-described role as a key upstream activator of TORC1, it may not activate TORC2, and in fact may have an indirect inhibitory effect on TORC2 [50]. Furthermore, clonal analysis of loss of function Rheb alleles in Drosophila imaginal discs revealed unusual stretched and odd shaped clones that caused the authors of the study to suggest that Rheb has other functions in addition to growth control [51]. It will be interesting to determine if the Rheb GTPase has effectors other than Tor and how these potential effortors may intersect with the Rho1 signaling pathway in imaginal disc morphogenesis.
Sc2
We identified a single hypomorphic allele of Sc2, Sc21, which showed SSNC with all three Rho1 alleles, and also showed a low level interaction with zipE(br). Four additional alleles of Sc2 did not show an interaction with Rho1E(br)246, raising the possibility that this potential interaction could be due to a second-site mutation on the Sc21chromosome. The strongest evidence that this is not the case is that Sc2 is one of only four genes deleted by two overlapping Rho1-interacting deficiencies, and was the only one of these genes to show an interaction with Rho1. Specifically, SSNC tests indicated no interaction between Rho1E(br)246 and ida or mge (Table S2). The final gene in this interval, Eip63F-1, is not essential (Vaskova 2000), and is thus less likely to be the Rho1-interacting gene. Sc2 encodes a 302 amino acid protein with two conserved domains, an amino-terminal ubiquitin domain (IPR000626) and a carboxyl-terminal 3-oxo-5-alpha-steroid 4-dehydrogenase domain (IPR001104). This latter domain is highly conserved and catalyzes the terminal step in the microsomal fatty acyl elongation cycle for long chain and very long chain fatty acids (palmitic acid, C16, and larger) [52]. How the synthesis of very long chain fatty acids influences Rho1 signaling is unclear, especially since these enzymes are not required for the synthesis of isoprenoids, and thus not involved in the lipid modification of Rho proteins directly. Rather, it might relate to the overall lipid composition of the plasma membrane. Lipid rafts are specialized membrane microdomains enriched in glycosphingolipids and cholesterol. Numerous membrane proteins involved in signal transduction events, including proteins that regulating Rho signaling, are enriched in these membrane domains [53]. Perhaps the regulated expression of long chain fatty acids via Sc2 modulates lipid raft size and composition, and thereby regulates signal transduction events through Rho1. Additional work will be required to address this hypothesis.
RhoGEF2 is a key regulator of Rho signaling during leg morphogenesis
One question we were interested in addressing as part of this study is which RhoGEF genes were likely functioning during imaginal disc morphogenesis. Our results clearly demonstrate that RhoGEF2 interacts genetically with Rho1 in this process (Table 2), supporting previous work from Halsell et al. [23] and Bayer et al. [26]. We found no evidence to definitively support any other potential RhoGEF as interacting with Rho1 during leg development, although we did identify two deficiencies that uncover putative RhoGEF genes that strongly enhance the malformed leg phenotype of Rho1E(br)246 (Table 4). Since there are no loss of function alleles for these genes, we have not been able to address them directly, and thus future studies are needed to test potential roles for CG30440 and CG30115 in leg morphogenesis. It should be noted that although we have no evidence to support a role for other RhoGEF genes, it is not possible to exclude them as functioning in leg morphogenesis if, for example, they are playing a redundant role or are expressed at levels that render them less dose sensitive.
Since it is clear that RhoGEF2 is functioning with Rho1 during imaginal disc morphogenesis, an important next question is how is RhoGEF2 activated in this process. Previous studies revealed that the subcellular localization of RhoGEF2 shifts to the apical region of cells destined to undergo apical constriction [54], [55]. Since RhoGEF2 has both PH and PDZ domains, its localization may be mediated by the PIP3 level of the plasma membrane or by the expression of a specific protein containing a PDZ binding site. Due to the potential involvement of the Tor signaling pathway in leg morphogenesis, we considered whether PI3 kinase was actively altering the PIP3 content of the plasma membrane in these cells. As an indirect means to assess the PIP3 content of the plasma membrane, we examined prepupal leg imaginal discs from flies expressing a fusion protein containing a PH domain linked to green fluorescent protein (tGPH; [56]). We found no obvious localization of the fusion protein to the plasma membrane in these cells indicating low levels of PIP3 in leg disc cells at these times, although as a control we could detect membrane localization of tGPH in larval salivary glands and fat bodies as expected (data not shown). Therefore these results suggest that RhoGEF2 is localized (and likely activated) through the binding of a specific protein. Perhaps the cloning of Rho1-interacting genes from the unidentified intervals may shed light on this issue in the future.
Leg morphogenesis requires a complex interplay of individual cell and collective tissue events, many of which are likely to be regulated by Rho signaling. The identification of three novel Rho1-interacting genes and six intervals containing novel Rho1-interacting genes, many of which interact genetically with other components of the Rho signaling pathway, provides a great opportunity to define the role of Rho signaling in these distinct morphogenetic events. Since this screen was predicated upon observing defects in leg morphogenesis that persisted until the final adult form of the appendage, it was not possible to determine which of these processes was most affected by the interaction between mutations in Rho1 and specific deficiencies or mutations. Thus our future challenge is to more accurately define the cellular events occurring during leg morphogenesis such that once we clone the relevant Rho1-interacting genes, we can use loss of function clonal analysis and induced RNA interference approaches to determine how these genes function during morphogenesis. In addition, we anticipate that more genetic screens will be performed to identify the Rho1-interacting genes playing critical roles at other stages of development such as during gastrulation, segment groove formation and head involution during embryogenesis. In this way we can define the myriad different Rho signaling modules that function during distinct morphogenetic processes during development, and may one day better understand how Rho signaling can be subverted in pathological states including tumor progression and metastasis.
Supporting Information
SSNC results of the primary screen between Rho1E(br)246 and Exelixis deficiencies
(0.47 MB DOC)
Secondary screen for all Exelixis deficiencies that showed 10% malformed legs when heterozygous with Rho1E(br)246/+
(0.30 MB DOC)
Acknowledgments
We thank Laurie von Kalm, Susan Halsell, Roger Karess, Greg Beitel and the Bloomington Drosophila Stock Center for fly stocks used in this study. We thank the Developmental Studies Hybridoma Bank for the antibodies used in this study. We are greatly indebted to Laurie von Kalm for stimulating conversations and sharing unpublished results. We thank Brian Ackley, Erik Lundquist and Robert Cohen for insightful discussions about the study, and anonymous reviewers for improving the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by NIH Grant Number P20 RR15563 and NIH Grant Number R01HD047570 from the National Center for Research Resources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, et al. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–4903. doi: 10.1242/dev.127.22.4891. [DOI] [PubMed] [Google Scholar]
- 4.Wei L, Roberts W, Wang L, Yamada M, Zhang S, et al. Rho kinases play an obligatory role in vertebrate embryonic organogenesis. Development. 2001;128:2953–2962. doi: 10.1242/dev.128.15.2953. [DOI] [PubMed] [Google Scholar]
- 5.Causeret F, Hidalgo-Sanchez M, Fort P, Backer S, Popoff MR, et al. Distinct roles of Rac1/Cdc42 and Rho/Rock for axon outgrowth and nucleokinesis of precerebellar neurons toward netrin 1. Development. 2004;131:2841–2852. doi: 10.1242/dev.01162. [DOI] [PubMed] [Google Scholar]
- 6.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 7.Ng J, Nardine T, Harms M, Tzu J, Goldstein A, et al. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- 8.Garvalov BK, Flynn KC, Neukirchen D, Meyn L, Teusch N, et al. Cdc42 regulates cofilin during the establishment of neuronal polarity. J Neurosci. 2007;27:13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 10.Johndrow JE, Magie CR, Parkhurst SM. Rho GTPase function in flies: insights from a developmental and organismal perspective. Biochem Cell Biol. 2004;82:643–657. doi: 10.1139/o04-118. [DOI] [PubMed] [Google Scholar]
- 11.Crawford JM, Harden N, Leung T, Lim L, Kiehart DP. Cellularization in Drosophila melanogaster is disrupted by the inhibition of rho activity and the activation of Cdc42 function. Dev Biol. 1998;204:151–164. doi: 10.1006/dbio.1998.9061. [DOI] [PubMed] [Google Scholar]
- 12.Magie CR, Meyer MR, Gorsuch MS, Parkhurst SM. Mutations in the Rho1 small GTPase disrupt morphogenesis and segmentation during early Drosophila development. Development. 1999;126:5353–5364. doi: 10.1242/dev.126.23.5353. [DOI] [PubMed] [Google Scholar]
- 13.Magie CR, Parkhurst SM. Rho1 regulates signaling events required for proper Drosophila embryonic development. Dev Biol. 2005;278:144–154. doi: 10.1016/j.ydbio.2004.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genova JL, Jong S, Camp JT, Fehon RG. Functional analysis of Cdc42 in actin filament assembly, epithelial morphogenesis, and cell signaling during Drosophila development. Dev Biol. 2000;221:181–194. doi: 10.1006/dbio.2000.9671. [DOI] [PubMed] [Google Scholar]
- 15.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Cohen SM. Imaginal disc development. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. pp. 747–841. [Google Scholar]
- 17.Fristrom D, Fristrom JW. The metamorphic development of the adult epidermis. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 843–897. [Google Scholar]
- 18.Condic ML, Fristrom D, Fristrom JW. Apical cell shape changes during Drosophila imaginal leg disc elongation: a novel morphogenetic mechanism. Development. 1991;111:23–33. doi: 10.1242/dev.111.1.23. [DOI] [PubMed] [Google Scholar]
- 19.Fristrom D. The mechanism of evagination of imaginal discs of Drosophila melanogaster. III. Evidence for cell rearrangement. Dev Biol. 1976;54:163–171. doi: 10.1016/0012-1606(76)90296-7. [DOI] [PubMed] [Google Scholar]
- 20.Taylor J, Adler PN. Cell rearrangement and cell division during the tissue level morphogenesis of evaginating Drosophila imaginal discs. Dev Biol. 2008;313:739–751. doi: 10.1016/j.ydbio.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fristrom D, Fristrom JW. The mechanism of evagination of imaginal discs of Drosophila melanogaster. I. General considerations. Dev Biol. 1975;43:1–23. doi: 10.1016/0012-1606(75)90127-x. [DOI] [PubMed] [Google Scholar]
- 22.Chen GC, Gajowniczek P, Settleman J. Rho-LIM kinase signaling regulates ecdysone-induced gene expression and morphogenesis during Drosophila metamorphosis. Curr Biol. 2004;14:309–313. doi: 10.1016/j.cub.2004.01.056. [DOI] [PubMed] [Google Scholar]
- 23.Halsell SR, Chu BI, Kiehart DP. Genetic analysis demonstrates a direct link between rho signaling and nonmuscle myosin function during Drosophila morphogenesis. Genetics. 2000;155:1253–1265. doi: 10.1093/genetics/155.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halsell SR, Kiehart DP. Second-site noncomplementation identifies genomic regions required for Drosophila nonmuscle myosin function during morphogenesis. Genetics. 1998;148:1845–1863. doi: 10.1093/genetics/148.4.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward RE, Evans J, Thummel CS. Genetic modifier screens in Drosophila demonstrate a role for Rho1 signaling in ecdysone-triggered imaginal disc morphogenesis. Genetics. 2003;165:1397–1415. doi: 10.1093/genetics/165.3.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayer CA, Halsell SR, Fristrom JW, Kiehart DP, von Kalm L. Genetic interactions between the RhoA and Stubble-stubbloid loci suggest a role for a type II transmembrane serine protease in intracellular signaling during Drosophila imaginal disc morphogenesis. Genetics. 2003;165:1417–1432. doi: 10.1093/genetics/165.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–392. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 28.Gotwals PJ, Fristrom JW. Three neighboring genes interact with the Broad-Complex and the Stubble-stubbloid locus to affect imaginal disc morphogenesis in Drosophila. Genetics. 1991;127:747–759. doi: 10.1093/genetics/127.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le T, Liang Z, Patel H, Yu MH, Sivasubramaniam G, et al. A new family of Drosophila balancer chromosomes with a w- dfd-GMR yellow fluorescent protein marker. Genetics. 2006;174:2255–2257. doi: 10.1534/genetics.106.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karim FD, Thummel CS. Ecdysone coordinates the timing and amounts of E74A and E74B transcription in Drosophila. Genes Dev. 1991;5:1067–1079. doi: 10.1101/gad.5.6.1067. [DOI] [PubMed] [Google Scholar]
- 31.Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. In: Goldstein L, Fyrberg E, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. New York: Academic Press; 1994. pp. 565–573. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Magie CR, Pinto-Santini D, Parkhurst SM. Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development. 2002;129:3771–3782. doi: 10.1242/dev.129.16.3771. [DOI] [PubMed] [Google Scholar]
- 34.Patel PH, Thapar N, Guo L, Martinez M, Maris J, et al. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J Cell Sci. 2003;116:3601–3610. doi: 10.1242/jcs.00661. [DOI] [PubMed] [Google Scholar]
- 35.Wohlwill AD, Bonner JJ. Genetic analysis of chromosome region 63 of Drosophila melanogaster. Genetics. 1991;128:763–775. doi: 10.1093/genetics/128.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 38.Ward R, Reid P, Bashirullah A, D'Avino PP, Thummel C. GFP in living animals reveals dynamic developmental responses to ecdysone during Drosophila metamorphosis. Dev Biol. 2003;256:389–402. doi: 10.1016/s0012-1606(02)00100-8. [DOI] [PubMed] [Google Scholar]
- 39.Neufeld TP. Body building: regulation of shape and size by PI3K/TOR signaling during development. Mech Dev. 2003;120:1283–1296. doi: 10.1016/j.mod.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossman KL, Sondek J. Larger than Dbl: new structural insights into RhoA activation. Trends Biochem Sci. 2005;30:163–165. doi: 10.1016/j.tibs.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Prokopenko SN, Brumby A, O'Keefe L, Prior L, He Y, et al. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 44.Georgiou M, Marinari E, Burden J, Baum B. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr Biol. 2008;18:1631–1638. doi: 10.1016/j.cub.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 45.Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr Biol. 2008;18:1639–1648. doi: 10.1016/j.cub.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 46.Fox DT, Homem CC, Myster SH, Wang F, Bain EE, et al. Rho1 regulates Drosophila adherens junctions independently of p120ctn. Development. 2005;132:4819–4831. doi: 10.1242/dev.02056. [DOI] [PubMed] [Google Scholar]
- 47.Homem CC, Peifer M. Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development. 2008;135:1005–1018. doi: 10.1242/dev.016337. [DOI] [PubMed] [Google Scholar]
- 48.Jacinto E. What controls TOR? IUBMB Life. 2008;60:483–496. doi: 10.1002/iub.56. [DOI] [PubMed] [Google Scholar]
- 49.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 50.Yang Q, Inoki K, Kim E, Guan KL. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc Natl Acad Sci U S A. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 52.Moon YA, Horton JD. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J Biol Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- 53.Meiri KF. Lipid rafts and regulation of the cytoskeleton during T cell activation. Philos Trans R Soc Lond B Biol Sci. 2005;360:1663–1672. doi: 10.1098/rstb.2005.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 55.Mulinari S, Barmchi MP, Hacker U. DRhoGEF2 and diaphanous regulate contractile force during segmental groove morphogenesis in the Drosophila embryo. Mol Biol Cell. 2008;19:1883–1892. doi: 10.1091/mbc.E07-12-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SSNC results of the primary screen between Rho1E(br)246 and Exelixis deficiencies
(0.47 MB DOC)
Secondary screen for all Exelixis deficiencies that showed 10% malformed legs when heterozygous with Rho1E(br)246/+
(0.30 MB DOC)