Abstract
In this study the membrane orientation of a tryptophan-flanked model peptide, WALP23, was determined by using peptides that were labeled at different positions along the sequence with the environmentally sensitive fluorescent label BADAN. The fluorescence properties, reflecting the local polarity, were used to determine the tilt and rotation angles of the peptide based on an ideal α-helix model. For WALP23 inserted in dioleoylphosphatidylcholine (DOPC), an estimated tilt angle of the helix with respect to the bilayer normal of 24° ± 5° was obtained. When the peptides were inserted into bilayers with different acyl chain lengths or containing different concentrations of cholesterol, small changes in tilt angle were observed as response to hydrophobic mismatch, whereas the rotation angle appeared to be independent of lipid composition. In all cases, the tilt angles were significantly larger than those previously determined from 2H NMR experiments, supporting recent suggestions that the relatively long timescale of 2H NMR measurements may result in an underestimation of tilt angles due to partial motional averaging. It is concluded that although the fluorescence technique has a rather low resolution and limited accuracy, it can be used to resolve the discrepancies observed between previous 2H NMR experiments and molecular-dynamics simulations.
Introduction
Membrane proteins fulfill many essential functions for the survival of a cell. These functions include signaling, transport of molecules across the membrane, and transduction of energy, which by definition all require at least a temporal change in the conformation of the membrane protein. It has been suggested that the function and therefore most probably the conformation of certain membrane proteins depend on membrane properties such as bilayer thickness, lipid packing, or the presence of microdomains (1–3). In many cases, the underlying mechanism of the interaction between membrane proteins and the lipid environment is still far from clear, partly due to the difficulties involved in studying these complex hydrophobic systems.
To avoid some of these problems, simple model systems composed of either natural or artificial peptides in synthetic lipid bilayers have been utilized to obtain deeper insights into the basic principles of peptide-lipid interactions. One example of a frequently used natural model peptide is the M13 coat protein (4). It was shown that the transmembrane part of this protein responds to a decreasing bilayer thickness by increasing its tilt angle (5). Similar results have been reported for other small natural membrane peptides/proteins, such as the transmembrane segment of Vpu (6), cell-signaling peptides (7), and alamethicin (8).
In addition to natural peptides, synthetic model peptides with well-defined structures have been used for experimental and modeling studies to elucidate the basic principles of peptide-lipid interactions (9–13). These peptides are advantageous because they allow systematic variation of peptide parameters, such as the hydrophobic length or hydrophobicity, and easy incorporation of labels via peptide synthesis. An example is the family of WALP peptides, which consist of a hydrophobic stretch of alternating leucines and alanines flanked by a pair of tryptophans at the N- and C-termini. These and other transmembrane model peptides are now widely used in systematic approaches to investigate the consequences of hydrophobic mismatch, such as helix tilt.
It appears that for WALP peptides, measurement of tilt angles is not straightforward. In a study of WALP23 peptides, a recently developed approach using 2H NMR spectroscopy on deuterated alanines revealed a very small but systematic increase in tilt angle with decreasing bilayer thickness (14,15). However, recent molecular-dynamics (MD) studies predicted much larger tilt angles for WALP23 (16) and related model peptides (17,18). This discrepancy may arise from the fact that only limited motion was included in the models used for analysis of the 2H NMR data, which may not have been sufficient to account for averaging effects, resulting in an underestimation of the tilt angle (16,17,19). Alternatively, the MD simulations may need improvement, such as by the use of longer timescales. Clearly, it is important to resolve this issue because accurate determination of the tilt angle is essential for understanding the basic principles of peptide-lipid interactions.
One approach to obtain accurate tilt angles would be to include different types of NMR labels combined with different dynamic models. Polarization inversion spin exchange at magic angle (PISEMA) methods (20,21) are a step in this direction. Alternatively, one could use methods with shorter timescales, which would reduce signal averaging due to peptide motions. Here, we chose the latter approach by using steady-state fluorescence spectroscopy. For this purpose, a set of WALP23 peptides with single cysteine replacements at different positions in the peptide sequence were labeled with the fluorescent label BADAN, which reports the polarity of the local environment (22). Analysis of the fluorescence results for WALP23 peptides in bilayers yielded similar rotation angles but much larger tilt angles than determined from 2H NMR experiments, which indeed suggests an underestimation of the tilt angle due to motional averaging. Furthermore, WALP23 was found to respond to changes in the thickness of the bilayers by only partly adapting its tilt angle. This is in agreement with the observation that tilting is not the only response of WALP peptides to mismatch; other responses, such as stretching or disordering of the lipids and an increased tendency to self-associate, can occur simultaneously (9).
Materials and Methods
Materials
Cholesterol, 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (14:1PC), 1,2-dipalmitoleoyl-sn-glycero-3-phosphocholine (16:1PC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (18:1PC), 1,2-dieicosenoyl-sn-glycero-3-phosphocholine (20:1PC), and 1,2-dierucoyl-sn-glycero-3-phosphocholine (22:1PC) were purchased as lyophilized powders from Avanti Polar Lipids (Alabaster, AL) and used without further purification. All other chemicals used were of analytical grade, and the water used was deionized and purified with a Milli-Q Gradient water purification system from Millipore (Billerica, MA).
The peptides WALP23-C0, WALP23-A11C, WALP23-L12C, WALP23-A13C, and WALP23-C24 were synthesized using Fmoc/tBu solid-phase peptide synthesis as described elsewhere for related KALP peptides (23). All other peptides were synthesized using manual solid-phase synthesis protocols developed by SynPep (Dublin, CA). The peptide sequences are given in Table 1.
Table 1.
Peptide sequences and masses of peptides after BADAN labeling from MALDI-TOF characterization
| Pos. | Peptide | Sequence | Theor. mass | Observed mass from MALDI-TOF |
|---|---|---|---|---|
| 0 | WALP23-C0 | Acetyl-C∗GWWLALALALALALALALALWWA-amide | 2831.6 | 2855.4, M+Na |
| 2 | WALP23-W2C | Acetyl-GC∗WLALALALALALALALALWWA-amide | 2645.5 | 2669.4, M+Na |
| 4 | WALP23-L4C | Acetyl-GWWC∗ALALALALALALALALWWA-amide | 2718.5 | 2741.7, M+Na |
| 6 | WALP23-L6C | Acetyl-GWWLAC∗ALALALALALALALWWA-amide | 2718.5 | 2742.3, M+Na |
| 8 | WALP23-L8C | Acetyl-GWWLALAC∗ALALALALALALWWA-amide | 2718.5 | 2742.3, M+Na |
| 10 | WALP23-L10C | Acetyl-GWWLALALAC∗ALALALALALWWA-amide | 2718.5 | 2742.0, M+Na |
| 11 | WALP23-A11C† | Acetyl-GWWLALALALC∗LALALALALWWA-amide | 2764.6 | 2786.7, M+Na |
| 12 | WALP23-L12C† | Acetyl-GWWLALALALAC∗ALALALALWWA-amide | 2722.5 | 2746.0, M+Na |
| 13 | WALP23-A13C† | Acetyl-GWWLALALALALC∗LALALALWWA-amide | 2764.6 | 2788.3, M+Na |
| 14 | WALP23-L14C | Acetyl-GWWLALALALALAC∗ALALALWWA-amide | 2718.5 | 2742.3, M+Na |
| 16 | WALP23-L16C | Acetyl-GWWLALALALALALAC∗ALALWWA-amide | 2718.5 | 2741.7, M+Na |
| 18 | WALP23-L18C | Acetyl-GWWLALALALALALALAC∗ALWWA-amide | 2718.5 | 2742.7, M+Na |
| 20 | WALP23-L20C | Acetyl-GWWLALALALALALALALAC∗WWA-amide | 2718.5 | 2742.4, M+Na |
| 22 | WALP23-W22C | Acetyl-GWWLALALALALALALALALWC∗A-amide | 2645.5 | 2768.7, M+Na |
| 24 | WALP23-C24 | Acetyl-GWWLALALALALALALALALWWGC∗-amide | 2817.6 | 2841.6, M+Na |
Cysteine labeled with BADAN.
Peptide includes a d4-alanine.
Methods
Labeling of peptides with BADAN
First, ∼1 mg of each peptide was weighed into an Eppendorf tube and dissolved in 200 μL trifluoroethanol (TFE). Subsequently, 10 μL H2O were added and the peptide solution was deoxygenized by bubbling with N2 gas for several minutes. Still under N2 atmosphere, 2 μL of triethylamine and 1.5 equivalents of BADAN (6-bromoacetyl-2-dimethylaminonaphthalene; Molecular Probes, Invitrogen, Carlsbad, CA), dissolved in methanol and purged with N2, were added. After the reaction mixture was stirred in the dark for 3 days at 4°C, the peptides were precipitated in 10 mL of cold methyl tert-butyl ether/n-hexane (1:1; −20°C) to remove unbound BADAN label. The precipitate was collected by centrifugation, the supernatant containing the unreacted BADAN label was decanted, and the precipitate was washed once again with methyl tert-butyl ether/n-hexane (1:1). Removal of unbound BADAN label was confirmed by thin-layer chromatography using chloroform/methanol/water (65:25:4) as the running solvent. No free BADAN label was visible under ultraviolet light.
The purity of the peptides was analyzed by analytical high-performance liquid chromatography (HPLC) with a C4 reverse phase column (Reprosil 300 C4 5 μm, 250 × 4.6 mm) using a linear solvent gradient from 10% to 100% methanol containing 0.1% TFA (trifluoroacetic acid) over 30 min. Before injection, the peptides were dissolved in TFE. If the purity was <90%, the labeled peptides were purified using a solvent gradient from 60% to 100% methanol containing 0.1% TFA over 40 min. For the purified peptides, the HPLC solvents were evaporated and residual TFA was removed by repeated cycles of dissolving the peptides in TFE followed by evaporation under high vacuum. The identity of all BADAN-labeled WALP23 peptides was verified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry using α-cyano-4-hydroxycinnamic acid as the matrix. The observed peptide masses are listed in Table 1.
Sample preparation
Stock solutions of ∼5 mM lipids in chloroform were prepared by weight. The phospholipid concentrations were determined using the Rouser phosphorus assay. Stock solutions of BADAN-labeled WALP23 peptides were prepared with a concentration of ∼25 μM in TFE. The peptide concentration was determined by absorption spectroscopy using an extinction coefficient of 22400 M−1cm−1 at 280 nm for WALP23. The main absorption band of the BADAN label attached to the WALP peptides has a maximum at 387 nm, but also low absorbance at 280 nm. We corrected for this by subtracting a pure BADAN absorption spectrum of equal intensity. From the peptide absorption spectra, a typical labeling efficiency of 80–90% was estimated.
After the solutions were mixed with appropriate amounts of peptide and phospholipids/cholesterol, the organic solvents were evaporated under a stream of N2 gas and were further removed under vacuum overnight (∼1 × 10−2 mbar). All fluorescence experiments were performed with a peptide/lipid molar ratio of 1/500 and a final peptide concentration of ∼1 μM. The samples were hydrated with buffer (25 mM HEPES, 100 mM NaCl, pH 7.0) and vortexed. Large unilamellar vesicles were produced by extrusion through inorganic membrane filters with 200 nm pore size (Anotop 10; Whatman International, Maidstone, England).
Fluorescence measurements
After preparation, the samples (1.2 mL) were directly transferred to a 10 mm quartz cuvette and measured at room temperature on a Fluorolog 3.22 fluorimeter (Jobin Yvon-Spex, Edison, NJ), using excitation light at 380 nm and a band pass of 2 nm in both excitation and detection light paths. The emission spectra were corrected for wavelength-dependent deviations in the detection system by means of the instrument-specific correction file. All spectra were also corrected for background signals using a blank sample containing unlabeled WALP23 peptides with the same peptide/lipid ratio. Inner filter effects were negligible because all samples measured had an absorbance of <0.025.
Analysis of the fluorescence spectra
Fluorescent labels containing a DAN moiety like BADAN exhibit a dual fluorescence behavior with two different excited states: the locally excited (LE) or Franck Condon state, and the intramolecular charge transfer (ICT) state, which evolves from the short-lived LE state (24). The complex line shape of BADAN emission spectra can be explained by the presence of different emitting ICT states: a non-hydrogen-bonded ICT state and hydrogen-bonded ICT (HICT) states in an immobilized environment (HICTi) and a mobile environment (HICTm), all possessing different energy levels and therefore emitting at different wavelengths (see Koehorst et al. (25) for an energy level scheme). The fraction of each emission state on the total emission depends on the local environment of the label.
For a quantitative analysis, the fluorescence spectra were decomposed into the different spectral components described above using three Gaussians, each characterized by a width, intensity, and peak position. The fraction of each spectral component was then calculated from the product of the width and intensity, relative to the total of all spectral components. The fraction of total hydrogen-bonded labels fHB was found by adding the spectral fractions of mobile and immobile hydrogen-bonded BADAN labels. Although the peak position of the HICTm component highly depends on the local polarity at the label position, the HICTi and ICT components were found to be relatively independent in the case of BADAN (25). This allows one to simplify the spectral decomposition by fixing the peak position of the ICT state and the HICTi state at 23,400 cm−1 and 22,140 cm−1, respectively. These numbers were determined from emission spectra with a large fraction of the respective spectral component. From the emission maxima as a function of label position, the tilt and rotation angles were determined by fitting the values to those calculated for a tilted α-helix, as detailed below and in Section S1 of the Supporting Material.
Results
Spectral shape depends on label position
A series of WALP23 peptides with cysteine replacements in different positions along the α-helix were labeled with the environmentally sensitive fluorophore BADAN. First, the labeled peptides were inserted in 18:1PC and fluorescence spectra were recorded (Fig. 1). The spectra for WALP23 peptides labeled at positions close to the peptide termini are clearly different from spectra from peptides labeled at positions in the hydrophobic stretch, and show abrupt changes in polarity between positions 4 and 6, and between positions 18 and 20. The differences in spectral shape are due to the sensitivity of BADAN to the polarity of the local environment. Across a lipid bilayer, the polarity of the environment changes from very polar for the water phase and lipid headgroups to apolar for the lipid acyl chain region with a steep gradient across the interfacial region (26). The emission maximum exhibits a large red shift of 80 nm for labels positioned close to the peptide termini, indicating a more polar environment for labels at the N- or C-terminus of the peptide. These findings confirm the transmembrane insertion of the WALP23 peptide in 18:1PC bilayers.
Figure 1.
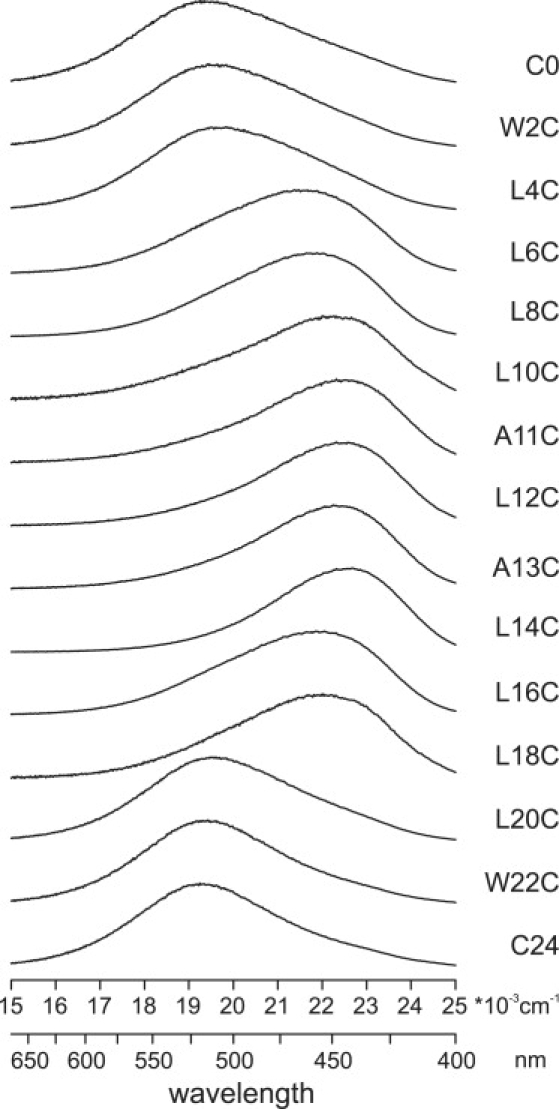
Emission spectra for a series of BADAN-labeled WALP23 peptides incorporated into 18:1PC using a peptide/lipid ratio of 1:500. The final concentration of peptide was ∼1 μM. All samples were measured at room temperature.
Quantification of the spectral effects
Next, the effects were quantified by spectral decomposition of the emission spectra, which yielded different types of information as described previously (25). In Fig. 2, the decomposition is illustrated for a label positioned in the bilayer interior (position 14, upper spectrum) and a label located at the lipid/water interface (position 0, N-terminus, lower spectrum). A comparison of the spectra reveals striking differences in the spectral components. For position 14, all components are almost equally present (Fig. 2 A), whereas for a label at the N-terminus a large fraction of mobile hydrogen-bonded labels is observed (Fig. 2 B). Furthermore, the emission maximum of the mobile hydrogen-bonded labels shifts (from 20,690 cm−1 at position 14 to 19,470 cm−1 at the N-terminus), which reflects changes in the polarity of the local label environment (25). The fraction of immobilized labels emitting with a maximum of 22,140 cm−1 is larger for position 14, which indicates that labels located in the bilayer interior are more restricted in their motion. Finally, the spectral fraction of non-hydrogen-bonded labels emitting with a maximum of 23,400 cm−1 is much larger for the label positioned in the interior of the bilayer, as expected because there are less water molecules in the interior of the bilayer compared to the membrane/water interface.
Figure 2.
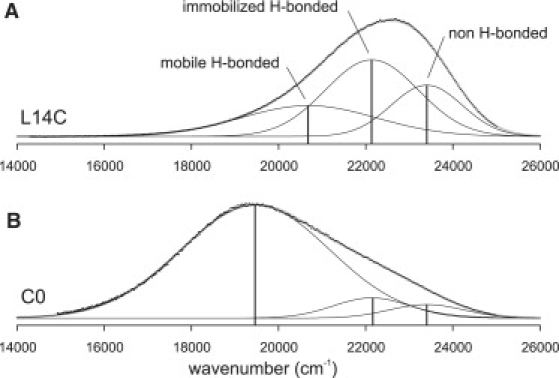
Spectral decomposition for emission spectra (dots) for a BADAN label in position L14C (A) and at the N-terminus (B). For both fits (thick gray line), the peak positions of the spectral components were allowed to vary for the HICTm state (between 18,000 and 22,000 cm−1) and were fixed for the HICTi and ICT states at 22,140 cm−1 and 23,400 cm−1, respectively (thin lines).
For further analysis, we used the peak position Pm of the mobile hydrogen-bonded fraction of labels. In principle, the total fraction of hydrogen-bonded labels fHB (see Section S2 of the Supporting Material for details) can also be used to retrieve the tilt angle. Here, we focused on Pm because it is more sensitive to changes across the membrane.
Determination of the peptide orientation
The polarity profile as sensed by the BADAN labels roughly follows a sigmoidal-shaped curve (see data points in Fig. 3 A), as expected for the polarity profile of a bilayer (26). Thus, the emission spectrum of a BADAN label attached to the WALP23 peptide strongly reflects the location, i.e., the depth of insertion into the bilayer. If the peptide is tilted with respect to the membrane normal, the location of any label in the bilayer will depend on the extent of the tilt and the rotation of the helix. Tilt introduces a periodicity on the depth of insertion into the bilayer, and the phase of the periodicity is determined by the rotation of the helix. Closer inspection of the Pm data set for WALP23 peptides in 18:1PC indeed suggests such a periodicity (Fig. 3 A, black line) superimposed on a sigmoidal curve (gray line in Fig. 3 A, representing the expected profile for a nontilted helix). From this, quantitative information can be retrieved about the orientation of the WALP23 peptide in a manner similar to that previously used to determine the tilt angle for M13 coat protein (5).
Figure 3.
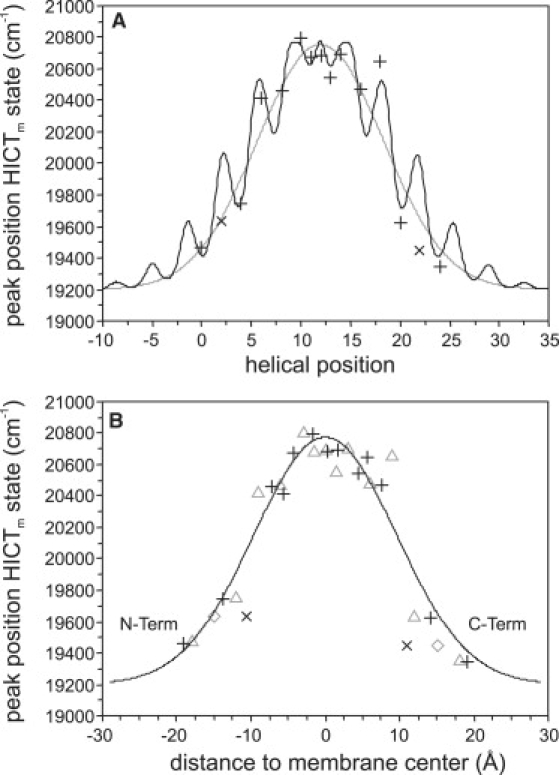
Fits to the peak positions of the HICTm state of BADAN-labeled WALP23 peptides in 18:1PC depending on the helical position of the label (A) and on the distance of the label to the center of the membrane (B). The best solution (τ = 23.6°, ρ = 107°) is indicated with a black line in A, and with black symbols in B (×: position of Trp; +: other positions). For comparison, the result with an imposed tilt angle τ = 0° is depicted in A with a gray line, and in B with gray symbols (open diamond, position of Trp; open triangle, other positions).
To obtain the tilt and rotation angles, the WALP23 peptide is modeled as an ideal α-helix. We also assume that the peptide tilt and rotation angle are not biased by the presence of the label. The validity of these assumptions will be discussed later. In addition, an estimation of the distance between the label and the helix axis is needed to calculate the depth of insertion of the label into the bilayer. The label is connected to the peptide via a flexible linker chain, leading to a distance distribution. As a first approximation, we estimate the average orthogonal distance of the label to the helix axis to 7.5 Å. The fit procedures are further detailed and discussed in Sections S1 and S3 of the Supporting Material. In brief, choosing a larger distance will result in a smaller tilt angle, and vice versa.
Using the model described above, the insertion depths of labels attached to different positions can be calculated for any given combination of tilt angle τ and rotation angle ρ (for details see Section S1 of the Supporting Material). These depths of insertion can be translated to wavenumber peak positions using a polarity profile for the bilayer. Here, we used a linear relationship between emission peak positions and polarity (see Section S4 of the Supporting Material). In previous studies, the sigmoidal function was used to model the polarity profile of the bilayer (5,27). Here, we used a Gaussian to describe the polarity profile of the bilayer because it requires one fit parameter less, and hence allows a more reliable comparison. However, very similar results were obtained when the results were fitted with a sigmoidal function (data not shown). The calculated peak positions were fitted to the peak position data sets using least-square minimization to retrieve the best solution for tilt and rotation angles.
Fig. 3 A (black line) shows the best fit (tilt angle τ = 23.6° and a rotation angle ρ = 107°) to the Pm data set of WALP23 peptides incorporated into bilayers of 18:1PC as function of the helical position, and Fig. 3 B shows the fit that depends on the insertion depths of the labels. To obtain this fit, the data points for positions 2 and 22, where interfacial tryptophans were replaced by cysteines labeled with BADAN, were omitted (Fig. 3, ×), because these points showed large deviations in comparison with all other data points. We believe that this is a direct result of substituting tryptophans, because these may be a dominant factor in determining the global orientation of the peptide (28). The contour plot of the sum of squared deviations (SSD) for fits with different combinations of tilt and rotation angles shown in Fig. 4 illustrates the overall quality of the fit.
Figure 4.
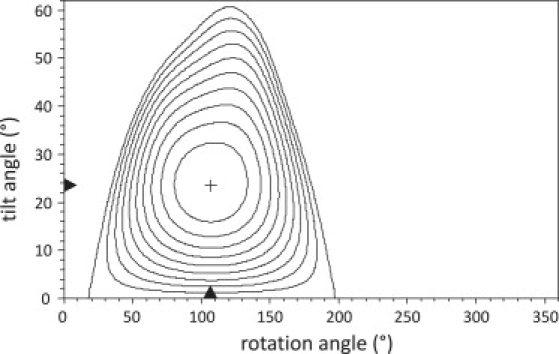
Contour plot of the SSD resulting from fits to the peak position of the HICTm state of BADAN-labeled WALP23 peptides in 18:1PC. The global minimum is marked by the plus sign and denotes the best solution. The isolines represent linearly spaced intervals based on the SSD of the best solution and the SSD of the solution with an imposed tilt angle τ = 0°, i.e., the helix oriented parallel to the membrane normal. The combinations of tilt and rotation angles not enclosed by the isolines give SSDs larger than the SSD obtained for τ = 0°.
Effects of hydrophobic mismatch
To investigate the effects of hydrophobic mismatch, fluorescence spectra were recorded for BADAN-labeled WALP23 peptides inserted into bilayers of unsaturated phospholipids with varying thickness. The results of the spectral decomposition (i.e., Pm) are depicted in Fig. 5 A. A lower value of Pm corresponds to a more polar environment, and vice versa. For the thinner bilayers, the local polarity sensed by the BADAN labels is higher in the bilayer interior, which can be explained by the lower water content in thicker hydrocarbon layers. However, for the thickest bilayer, 22:1PC, we observed a relatively high polarity for some labels positioned in the middle of the α-helix. This may be due to water molecules trapped in the center of the bilayer as a consequence of peptide-induced bilayer distortions under conditions of large negative mismatch.
Figure 5.
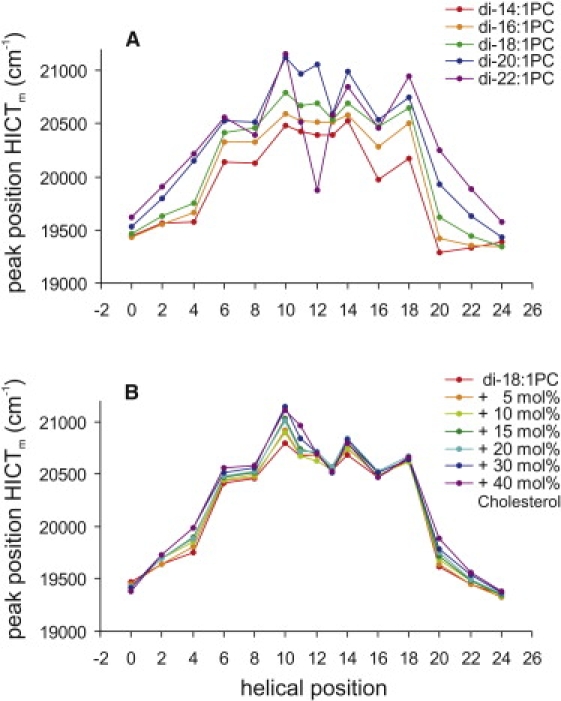
Wavenumber peak position Pm of the mobile hydrogen-bonded fraction of labels obtained from spectral decomposition of emission spectra of BADAN-labeled WALP23 peptides incorporated into bilayers of unsaturated phospholipids with different thicknesses (A) and bilayers of 18:1PC containing different cholesterol concentrations (B). The lines are drawn to guide the eyes, since only some positions in the WALP23 peptide were labeled with BADAN.
The polarity profiles for the different bilayers were fitted to determine tilt and rotation angles for WALP23 peptides incorporated into bilayers of different thickness. The results are listed in Table 2. No reliable fits could be obtained for the thinnest (14:1PC) and thickest (22:1PC) bilayers, possibly because of distortions in the local lipid environment or the peptide structure under these extreme mismatch conditions. Table 2 shows that the tilt angles slightly decrease with increasing bilayer thickness, whereas the rotation angle remains about the same, suggesting a preferred orientation of the peptide that is independent of the tilt.
Table 2.
Tilt and rotation angles estimated for WALP23 peptides in bilayers of different thicknesses
| Bilayer | τ (°) | ρ(°) | dC∗ (Å) |
|---|---|---|---|
| 16:1PC | 24.8 | 96 | 26.5 |
| 18:1PC | 23.6 | 107 | 29.6 |
| 20:1PC | 19.8 | 108 | 33.1 |
Thickness of the bilayer hydrophobic core derived from Kucerka et al. (29).
The adaptations of tilt angle are much less than expected for a complete adaptation to mismatch based on geometrical considerations. Indeed, if the peptide had a tilt angle of 23.6° in 18:1PC and would completely adapt to the thickness of the thinner bilayer in 16:1PC by further tilting, one would expect a tilt angle of 34.9° in the latter bilayer. Similarly, if the tilt angle of 24.8° that we observed in 16:1PC were a result of adaptation to mismatch, one would expect zero tilt in 18:1PC.
The general error of the tilt angles is quite large, since the precise distance between the label and the helix axis is not known (see Section S4 in the Supporting Material for details). Nevertheless, we believe the small changes in tilt angle to be significant because the “relative values” of the tilt angles are estimated to be accurate within <1° when the values for bilayers with different thicknesses are compared.
Effect of cholesterol
The addition of cholesterol is known to increase the bilayer thickness because of its ordering effect on the surrounding lipid acyl chains. This effect is most pronounced for saturated lipids, but unsaturated lipids also are slightly ordered and therefore thickened (29). BADAN-labeled peptides were incorporated into 18:1PC bilayers containing cholesterol concentrations of up to 40 mol %. It was found that the polarity sensed by the BADAN labels decreased slightly with increasing cholesterol concentration (Fig. 5 B), suggesting that the more ordered bilayer contains less water molecules in the hydrocarbon region. The changes in the local polarity as sensed by the BADAN labels were much smaller for changes in cholesterol concentrations than for changes in bilayer thickness.
The polarity data sets for the different cholesterol concentrations were fitted according to the procedure above, and reliable fits were obtained for all cholesterol concentrations used. As summarized in Table 3, the tilt angles show a slight decrease with increasing cholesterol concentration, whereas the rotation angle remains almost unchanged. The results suggest that the effect of cholesterol can be attributed to its effect on bilayer thickness, and that the presence of cholesterol by itself does not affect the behavior of the peptide in the bilayer.
Table 3.
Tilt and rotation angles estimated for WALP23 peptides in 18:1PC bilayers containing different concentrations of cholesterol
| Cholesterol (mol %) | τ (°) | ρ(°) | dC∗ (Å) |
|---|---|---|---|
| 0 | 23.6 | 107 | 29.6 |
| 5 | 23.3 | 110 | 29.9 |
| 10 | 23.1 | 109 | 30.2 |
| 15 | 22.3 | 111 | 30.7 |
| 20 | 23.0 | 108 | 31.3 |
| 30 | 20.7 | 111 | 32.7 |
| 40 | 18.4 | 108 | 34.6 |
Thickness of the bilayer hydrophobic core derived from Kucerka et al. (29).
Discussion
The main objective of this study was to investigate the tilt and rotation angles of the transmembrane model peptide WALP23 in a lipid bilayer. A series of WALP peptides were labeled with the environmentally sensitive fluorescent label BADAN in different positions and incorporated into bilayers of different thicknesses and cholesterol contents. Spectral decomposition of the emission spectra yielded information regarding the polarity and hydrogen-bonding capacities in the local vicinity of positions along the α-helix. These data profiles were used to retrieve the orientation of the transmembrane helix.
Analysis using the ideal α-helix model
The model used to fit the data profiles is based on a canonical α-helix. Previous circular dichroism and Fourier transform infrared spectroscopy experiments showed that WALP peptides adopt a regular α-helical conformation in various types of bilayers (23,30). In addition, MD simulation studies indicated that WALP23 in a matching bilayer of 14:0PC adopts a highly regular α-helical structure for the hydrophobic core region, and that unordered structures only occur for the residues at the N- and C-termini (16). In agreement with this, fits excluding the data points from the potentially unordered residues at the N- and C-termini of the helix yielded small SSD values. Furthermore, leaving out any data point within the hydrophobic region yielded very similar values of tilt and rotation angles (not shown), supporting the notion that the BADAN label does not interfere with the regularity of the α-helix.
The quality of the fits to the polarity profiles was best for WALP23 peptides incorporated into bilayers of 18:1PC, representing the situation of hydrophobic matching. For WALP23 peptides in bilayers of 16:1PC and 20:1PC, where the lipids may have been slightly adapted, the fit was still good as judged by the SSD. However, in bilayers of 14:1PC and 22:1PC, the quality of the fits was poor, even yielding local minima. This observation can be explained by the possible responses of the peptide and lipids to these extreme mismatch situations (9). In the case of 14:1PC, the hydrophobic region of the bilayer is much too thin for a WALP23 peptide, which may lead to a distortion of the peptide backbone, as suggested for WALP23 peptide in 12:0PC (15,31). Similarly, for WALP23 peptides in much too thick bilayers of 22:1PC, the α-helix may become partly unwound. Such distortions would prohibit the use of the ideal α-helix model for data analysis. The analysis could be further complicated by local distortions of the bilayer as a consequence of mismatch.
Finally, it is possible that only a limited amount of peptides can be incorporated into the bilayer in extreme mismatch situations (11,30). This would also hamper data analysis, since it is not possible to separate spectral fractions originating from such a nonincorporated population from those of the transmembrane population. However, at the low peptide/lipid ratios used in this study, we do not expect the latter effect to be significant.
Advantages and disadvantages of using fluorescent labels
Our approach based on fluorescence spectroscopy complements the well-established NMR methods to investigate the orientation of membrane-associated peptides, and presents several advantages as well as disadvantages. The most important advantage here is that fluorescence spectroscopy operates on much shorter timescales than NMR spectroscopy, preventing any influence due to (global) peptide motions. In our analysis, the instantaneous distributions of tilt and rotation angles are accounted for by fitting the spectral components with Gaussians, yielding a value for the mean orientation. Another advantage is that fluorescence spectroscopy is a sensitive method that requires only low concentrations of fluorescent molecules. This allows one to perform experiments at low peptide/lipid ratios and under physiologically relevant hydration conditions.
However, there are also disadvantages to our approach. A major disadvantage of the technique is the need for labels that might interfere with the orientation or conformation of the peptide. Furthermore, due to the assumptions that need to be made regarding the distribution of the labels and the distance between the label and the peptide helix axis, this technique has a rather low resolution and only limited accuracy. Nevertheless, this fluorescence approach is appropriate for resolving the discrepancies between previous 2H NMR experiments and MD simulations regarding the tilt angle of the WALP peptide.
In this study we used BADAN as environmentally sensitive label. Like the more frequently used AEDANS probe, it belongs to the family of fluorescent DAN moieties (see Fig. 6 for structures). However, whereas AEDANS has a single emission state, BADAN has relatively complex fluorescence properties, which provide additional information but also complicate interpretation of the results. The advantages of the BADAN label are that it is uncharged and has a shorter linker chain than AEDANS, and thus can be expected to introduce less bias for the orientation of the label in the hydrophobic environment of the membrane. For studies on natural peptides containing charged residues that strongly influence the local polarity, the longer linker chain of AEDANS can be advantageous because it allows one to probe the more distant membrane environment. Recently, the membrane topology of the M13 coat protein was successfully investigated using AEDANS (5).
Figure 6.
Structures of BADAN and 1,5-IAEDANS coupled to a cysteine.
WALP23 has a very regular amino acid sequence of alternating leucines and alanines in the hydrophobic stretch, and does not contain any charged residues. Thus, the influence of the amino acid sequence can be expected to be negligible, enabling use of the BADAN label. For all labeled positions, we assumed a homogeneous spatial distribution of label orientations due to the flexible linker chain, although we cannot exclude the possibility that the lipid environment biased its average orientation. As with any other label, we also cannot exclude the possibility that the label biased the orientation of the peptide. However, for WALP23, we expect that any influence of the label on the orientation of the peptide is small, as long as the label is positioned within the hydrophobic stretch flanked by the tryptophans. These tryptophans have been shown to strongly anchor to the membrane/water interface (32), most likely determining the preferred tilt and rotation angles, and dominating any effect of the label.
Membrane orientation of WALP peptides
In this study we found tilt angles of 24° ± 5° for WALP23 peptides in 18:1PC bilayers, which is much larger than the angles found in previous 2H NMR studies (14,15). In those studies, analysis of a set of quadrupolar splittings of WALP23 peptides, including a single deuterium-labeled alanine, with the geometric analysis of labeled alanines method yielded relatively small tilt angles of 4.8° and 5.2° for WALP23 peptides incorporated into matching bilayers of 18:1PC and 14:0PC, respectively (14). In 12:0PC, representing a positive mismatch situation, only a small increase to 8.1° was observed (14). Similar small tilt angles were observed using a different solid-state NMR method (PISEMA) on related peptides (20).
Recently, it was suggested that the observation of relatively small tilt angles determined from 2H NMR experiments may be due to additional signal averaging, leading to an underestimation of the observed tilt angle (16,19,28). The larger tilt angles we obtained in this study using fluorescence methods are in line with the larger tilt angles (33.5°) suggested from MD simulation studies (16,17). If partial motional averaging were caused mainly by large amplitude fluctuations around the helix axis, one would expect that the rotation angles calculated from the 2H NMR experiments would not be significantly influenced by this motion. To compare the rotation angles obtained in fluorescence experiments with those acquired from previous 2H NMR results, we need to correct the rotation angle for method-inherent offsets. Due to the preferred orientation of the BADAN label along the Cα-Cβ bond, the fitted rotation angle differs from the rotation angle referenced to the Cα atom. We estimated the angle offset to 29° using a distance of 7.5 Å of the label to the helix axis and an angle of 43.3° between Cα orthogonally connected to the helix axis and the Cα-Cβ bond. The corrected rotation angle of 136° agrees well with the rotation angle of 146° obtained from 2H NMR experiments.
Our fluorescence experiments indicate that the response of the tilt angle of WALP23 to the changing bilayer thickness is insufficient to compensate for the effects of a hydrophobic mismatch. This points toward additional mismatch responses of these peptides and agrees with the observation that under conditions of positive hydrophobic mismatch, tryptophan-flanked peptides locally stretch the bilayer (32) and have an increased tendency to self-associate (33). This is in contrast to results from solid-state NMR studies on a peptide derived from Vpu, in which a complete compensation for hydrophobic mismatch by tilting was observed (6). Apparently, there is an energetic cost for tilting of the WALP peptides. This may be due to the presence of the tryptophan residues and is in line with the suggestion that interfacially localized tryptophans may “buffer” a transmembrane helix against changes in orientation due to changes in the bilayer thickness (34). Such an effect may also explain the absence of interfacial tryptophans in McsL. This channel protein undergoes large changes in tilt of α-helices upon opening, which is interfered with by the introduction of interfacial tryptophan residues (35).
To gain more information on the role of interfacial tryptophans in determining tilt and rotation angles, it would be interesting to study the KALP peptides, which are analogous to the WALP peptides but have a pair of lysines at both sides of the lipid/water interface instead of tryptophans. Indeed, these peptides do not seem to have strong anchoring interactions, as no complementary adaptation of the lipids is observed when a mismatch is introduced (32). Previous 2H NMR experiments suggested that WALP and KALP peptides both adopt small tilt angles (14). However, as discussed above, correct determination of tilt angles using 2H NMR requires information about motional averaging, and this may not be the same for both KALP peptides and WALP peptides. By using the approach presented above, one could in principle circumvent these problems. Unfortunately, however, this method most likely is not suitable for determining the tilt angles of KALP peptides, partly because the lack of strong interfacial anchoring may result in a larger influence of the label on the orientation of KALP than for WALP peptides. In addition, the charged lysine side chains in the KALP peptides may interfere with data analysis because they may have a large effect on the local polarity that is sensed by the BADAN label. Thus, further development of complementary methods, such as solid-state NMR techniques that take into account different types of possible motions, or MD approaches is necessary to fully understand the principles of how lipids affect the orientation of transmembrane helices.
Supporting Material
Four sections, five figures, a table, and references are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01305-8.
Supporting Material
Acknowledgments
This work was supported by a Marie Curie Early Stage Research Training Fellowship from the European Community's Sixth Framework Program (Biomem-MEST-CT 2004-007931 to A.H.), and a grant from the National Institutes of Health (1S10RR023065-01 to M.G.).
Footnotes
Andrea Holt's present address is The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Contributor Information
Andrea Holt, Email: a.holt@nki.nl.
J. Antoinette Killian, Email: j.a.killian@uu.nl.
References
- 1.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Booth P.J. Sane in the membrane: designing systems to modulate membrane proteins. Curr. Opin. Struct. Biol. 2005;15:435–440. doi: 10.1016/j.sbi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Nyholm T.K.M., Özdirekcan S., Killian J.A. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 4.Stopar D., Spruijt R.B., Hemminga M.A. Anchoring mechanisms of membrane-associated M13 major coat protein. Chem. Phys. Lipids. 2006;141:83–93. doi: 10.1016/j.chemphyslip.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Koehorst R.B.M., Spruijt R.B., Vergeldt F.J., Hemminga M.A. Lipid bilayer topology of the transmembrane α-helix of M13 major coat protein and bilayer polarity profile by site-directed fluorescence spectroscopy. Biophys. J. 2004;87:1445–1455. doi: 10.1529/biophysj.104.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S.H., Opella S.J. Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J. Mol. Biol. 2005;350:310–318. doi: 10.1016/j.jmb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Ramamoorthy A., Kandasamy S.K., Lee D.-K., Kidambi S., Larson R.G. Structure, topology, and tilt of cell-signaling peptides containing nuclear localization sequences in membrane bilayers determined by solid-state NMR and molecular dynamics simulation studies. Biochemistry. 2007;46:965–975. doi: 10.1021/bi061895g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh D., Jost M., Peggion C., Toniolo C. Lipid chain-length dependence for incorporation of alamethicin in membranes: electron paramagnetic resonance studies on TOAC-spin labeled analogs. Biophys. J. 2007;92:4002–4011. doi: 10.1529/biophysj.107.104026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killian J.A., Nyholm T.K.M. Peptides in lipid bilayers: the power of simple models. Curr. Opin. Struct. Biol. 2006;16:473–479. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Shahidullah K., London E. Effect of lipid composition on the topography of membrane-associated hydrophobic helices: stabilization of transmembrane topography by anionic lipids. J. Mol. Biol. 2008;379:704–718. doi: 10.1016/j.jmb.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mall S., Broadbridge R., Sharma R.P., Lee A.G., East J.M. Effects of aromatic residues at the ends of transmembrane α-helices on helix interactions with lipid bilayers. Biochemistry. 2000;39:2071–2078. doi: 10.1021/bi992205u. [DOI] [PubMed] [Google Scholar]
- 12.Yano Y., Takemoto T., Kobayashi S., Yasui H., Sakurai H. Topological stability and self-association of a completely hydrophobic model transmembrane helix in lipid bilayers. Biochemistry. 2002;41:3073–3080. doi: 10.1021/bi011161y. [DOI] [PubMed] [Google Scholar]
- 13.Liu F., Lewis R.N.A.H., Hodges R.S., McElhaney R.N. Effect of variations in the structure of a polyleucine-based α-helical transmembrane peptide on its interaction with phosphatidylcholine bilayers. Biochemistry. 2002;41:9197–9207. doi: 10.1021/bi025661i. [DOI] [PubMed] [Google Scholar]
- 14.Özdirekcan S., Rijkers D.T.S., Liskamp R.M.J., Killian J.A. Influence of flanking residues on tilt and rotation angles of transmembrane peptides in lipid bilayers. a solid-state 2H NMR study. Biochemistry. 2005;44:1004–1012. doi: 10.1021/bi0481242. [DOI] [PubMed] [Google Scholar]
- 15.Strandberg E., Özdirekcan S., Rijkers D.T.S., van der Wel P.C.A., Koeppe R.E. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 2004;86:3709–3721. doi: 10.1529/biophysj.103.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Özdirekcan S., Etchebest C., Killian J.A., Fuchs P.F.J. On the orientation of a designed transmembrane peptide: toward the right tilt angle? J. Am. Chem. Soc. 2007;129:15174–15181. doi: 10.1021/ja073784q. [DOI] [PubMed] [Google Scholar]
- 17.Esteban-Martin S., Salgado J. Self-assembling of peptide/membrane complexes by atomistic molecular dynamics simulations. Biophys. J. 2007;92:903–912. doi: 10.1529/biophysj.106.093013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandasamy S.K., Larson R.G. Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers: a systematic investigation of hydrophobic mismatch. Biophys. J. 2006;90:2326–2343. doi: 10.1529/biophysj.105.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strandberg E., Esteban-Martin S., Salgado J., Ulrich A.S. Orientation and dynamics of peptides in membranes calculated from 2H-NMR data. Biophys. J. 2009;96:3223–3232. doi: 10.1016/j.bpj.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vostrikov V.V., Grant C.V., Daily A.E., Opella S.J., Koeppe R.E. Comparison of “polarization inversion with spin exchange at magic angle” and “geometric analysis of labeled alanines” methods for transmembrane helix alignment. J. Am. Chem. Soc. 2008;130:12584–12585. doi: 10.1021/ja803734k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteban-Martin S., Strandberg E., Fuertes G., Ulrich A.S., Salgado J. Influence of whole-body dynamics on 15N PISEMA NMR spectra of membrane proteins: a theoretical analysis. Biophys. J. 2009;96:3233–3241. doi: 10.1016/j.bpj.2008.12.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber G., Farris F.J. Synthesis and spectral properties of a hydrophobic fluorescent probe: 6-propionyl-2-(dimethylamino)naphthalene. Biochemistry. 1979;18:3075–3078. doi: 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- 23.de Planque M.R.R., Kruijtzer J.A.W., Liskamp R.M.J., Marsh D., Greathouse D.V. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane α-helical peptides. J. Biol. Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 24.Józefowicz M., Kozyra K.A., Heldt J.R., Heldt J. Effect of hydrogen bonding on the intramolecular charge transfer fluorescence of 6-dodecanoyl-2-dimethylaminonaphtalene. Chem. Phys. 2005;320:45–53. [Google Scholar]
- 25.Koehorst R.B.M., Spruijt R.B., Hemminga M.A. Site-directed fluorescence labeling of a membrane protein with BADAN: probing protein topology and local environment. Biophys. J. 2008;94:3945–3955. doi: 10.1529/biophysj.107.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White S.H., Wimley W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 27.Marsh D. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. USA. 2001;98:7777–7782. doi: 10.1073/pnas.131023798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteban-Martin S., Salgado J. The dynamic orientation of membrane-bound peptides: bridging simulations and experiments. Biophys. J. 2007;93:4278–4288. doi: 10.1529/biophysj.107.113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucerka N., Pencer J., Nieh M.-P., Katsaras J. Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles. Eur. Phys. J. E. 2007;23:247–254. doi: 10.1140/epje/i2007-10202-8. [DOI] [PubMed] [Google Scholar]
- 30.de Planque M.R.R., Goormaghtigh E., Greathouse D.V., Koeppe R.E., Kruijtzer J.A.W. Sensitivity of single membrane-spanning α-helical peptides to hydrophobic mismatch with a lipid bilayer: effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry. 2001;40:5000–5010. doi: 10.1021/bi000804r. [DOI] [PubMed] [Google Scholar]
- 31.Daily A.E., Greathouse D.V., van der Wel P.C.A., Koeppe R.E. Helical distortion in tryptophan- and lysine-anchored membrane-spanning α-helices as a function of hydrophobic mismatch: a solid-state deuterium NMR investigation using the geometric analysis of labeled alanines method. Biophys. J. 2008;94:480–491. doi: 10.1529/biophysj.106.097543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Planque M.R.R., Bonev B.B., Demmers J.A.A., Greathouse D.V., Koeppe R.E. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 33.Sparr E., Ash W.L., Nazarov P.V., Rijkers D.T.S., Hemminga M.A. Self-association of transmembrane α-helices in model membranes—importance of helix orientation and role of hydrophobic mismatch. J. Biol. Chem. 2005;280:39324–39331. doi: 10.1074/jbc.M502810200. [DOI] [PubMed] [Google Scholar]
- 34.Webb R.J., East J.M., Sharma R.P., Lee A.G. Hydrophobic mismatch and the incorporation of peptides into lipid bilayers: a possible mechanism for retention in the Golgi. Biochemistry. 1998;37:673–679. doi: 10.1021/bi972441+. [DOI] [PubMed] [Google Scholar]
- 35.Chiang C.-S., Shirinian L., Sukharev S. Capping transmembrane helices of MscL with aromatic residues changes channel response to membrane stretch. Biochemistry. 2005;44:12589–12597. doi: 10.1021/bi050750r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


