Abstract
Amplified ribosomal DNA restriction analysis (ARDRA) and restriction fragment length polymorphism were originally used for strain typing and for screening clone libraries to identify phylogenetic clusters within a microbial community. Here we used ARDRA as a model to examine the capacity of restriction-based techniques for clone identification, and the possibility of deriving phylogenetic information from ARDRA-based dendrograms. ARDRA was performed in silico on 48,759 sequences from the Ribosomal Database Project, and it was found that the fragmentation profiles were not necessarily unique for each sequence in the database, resulting in different species sharing fragmentation profiles. Although ARDRA-based clusters separated clones into different genera, these phylogenetic clusters did not overlap with trees constructed according to sequence alignment, calling into question the intra-genus ARDRA-based phylogeny. It is thus suggested that the prediction power of ARDRA clusters in identifying clone phylogeny be regarded with caution.
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-009-9380-1) contains supplementary material, which is available to authorized users.
Keywords: ARDRA, In-silico fragmentation, 16S subunit ribosomal DNA, Ribosomal database project
Amplified ribosomal DNA restriction analysis (ARDRA) is a commonly used tool to study microbial diversity that relies on DNA polymorphism (Deng et al. 2008). Clones containing 16S rDNA gene fragments, obtained by applying either universal or genus-specific primer sets, are amplified and digested by restriction endonucleases (REs), followed by separation of the resulting fragments on high-density agarose or acrylamide gels. The emerging profiles are then used either to cluster the community into genotypic groups or for strain typing (Tiedje et al. 1999).
Attempts to use ARDRA to identify species within particular genera have only been partially successful. Mycoplasma species isolated from cats (Criado-Fornelio et al. 2003) and humans (Stakenborg et al. 2005) were identified using the ARDRA technique, though their identity was not confirmed by sequencing. ARDRA fingerprinting could not distinguish between genomic species of Acinetobacter, even at a low cut-off level (Koeleman et al. 1998), while identification of Bdellovibrio and Bdellovibrio-like microorganisms using ARDRA grouping mostly reflected diversity and phylogenetic affiliation when compared to sequencing of the 16S rDNA gene (Davidov and Jurkevitch 2004). Rhizobia species isolated from nodules of Vicia were identified using 16S ARDRA, restriction fragment length polymorphism (RFLP) of 16S–23S internally transcribed spacer (ITS), and sequencing of the 16S rDNA. The derived phylogenetic relationships mostly supported the relationships estimated by the ARDRA and ITS-RFLP, albeit some discrepancies were detected (Lei et al. 2008). Lactobacillus strains isolated from grape must and wine (Rodas et al. 2003), dairy products (Giraffa et al. 1998), and faecal samples (Ventura et al. 2000) were identified by ARDRA fingerprinting. However, the tested isolates were verified by partial sequencing and by comparing the ARDRA patterns to predicted profiles of known strains of lactobacilli. Attempts to apply ARDRA profiles using a panel of six enzymes in order to discriminate Ralstonia and Pandoraea strains isolated from the respiratory tract of cystic fibrosis patients showed that all the Ralstonia, but not the Pandoraea strains tested could be differentiated (Segonds et al. 2003). Brevibacillus species isolated from clinical, dairy and industrial environments were distinguished using ARDRA (the amplicons were digested with five REs), but comparison of the emerging profiles to those obtained with several phenotypic methods and sequence analysis revealed inconsistencies (Logan et al. 2002). However, a careful choice of REs enabled the use of the ARDRA technique to discriminate among Lactobacillus, Streptococcus and Bifidobacterium at the genus, but not species, level (Collado and Hernandez 2007).
Heyndrickx et al. (1996) studied the application of ARDRA in the clarification of the phylogeny and taxonomy of the genus Bacillus. They found several inter-specific phylogenetic relationships, as well as inter-group phylogenetic relationships, to be in accordance with 16S rDNA sequence analysis; thus, the ARDRA technique, based on the combination of five selected REs, was deemed reliable and valuable for phylogenetic and taxonomic studies of large sets of strains. However, some apparent phylogenetic relationships indicated by ARDRA were not supported by the sequence analysis results. It was postulated that this stemmed from the small phylogenetic distance between these rDNA groups (Vaneechoutte and Heyndrickx 2001).
A study assessing the applicability of ARDRA to the identification of operational taxonomic units based on their ARDRA profiles was carried out more than a decade ago (Moyer et al. 1996), in which a detailed analysis of the types of REs that provide the best differentiating power was performed. In addition, Moyer et al. (1996) compared phylogenetic trees based on 16S rDNA sequences and on ARDRA profiles, and reached the conclusion that using ten REs will yield 76–100% success in obtaining accurate phylogenetic affiliations. However, that study used the very narrow range of sequences available at the time, while today the databases have increased many-fold and the questions have once again arisen: Are ARDRA profiles sufficient for clone identification? Are phylogenetic relationships described by ARDRA sufficiently representative of the “true” relationships determined by the 16S rDNA sequences?
In the current study, we re-evaluated the predictive power of ARDRA, by assessing two ways in which ARDRA can be used to foresee the identity or phylogeny of clones: (a) environmental clone identification via profile matching to theoretically computed fragmentation profiles and (b) clustering of ARDRA fragmentation profiles in comparison to parallel sequence-based clustering.
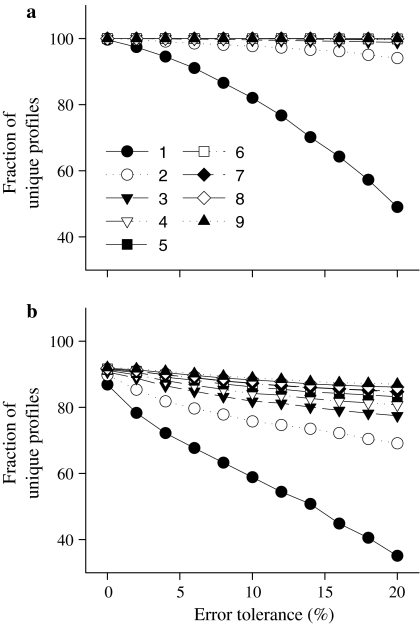
A total of 48,759 sequences from the Ribosomal Database Project (RDP) (Maidak et al. 2001) were taken for in-silico ARDRA (see “Supplementary material” for detailed methods). Profiles were found to be unique to genera for two or more REs (Fig. 1a), even at a high error level of 20% in the sizing of the restriction fragments (as might occur in agarose gels). However, at least three enzymes were necessary to differentiate species (Fig. 1b). Even then, while most patterns referred to unique sequences, exceptions were observed, e.g. species of Salmonella (one species), Citrobacter (3), Klebsiella (3) and Enterobacter (7) shared patterns, as did species of Kocuria (1), Micrococcus (3), Arthrobacter (4) and Streptomyces (3). When all ten REs were used, only four patterns were shared by more than one genus: Citrobacter (4) and Klebsiella (1); Legionella (3) and Fluoribacter (1); Saccharothrix (1) and Actinosynnema (1); Raoultella (1) and Citrobacter (1). These results agree with those obtained by Moyer et al. (1996); thus, we found their conclusions to be applicable, even when a large number of sequences is considered (48,759 vs. 106 sequences), for several combinations of the tested REs (data not shown). It is important to point out that the conclusions reached above come from a global perspective; it is possible that for specific genera, different sets of three REs would be better for inter-genera differentiation. When a specific genus is of interest, a specific set of REs may produce a higher resolution even with less than three REs. The scripts written by the authors can be used to this end and can be obtained on request.
Fig. 1.
Proportion of profiles unique to a single genus (a) and to a single species (b), defined as the number of profiles specific to a single genus or species, respectively, divided by the total number of profiles. Both are dependent on the error tolerance (the x-axis) and on the number of restriction enzymes used. The number indicates the number of restriction enzymes used
Dendrograms calculated from the theoretical fragmentation profiles (see “Supplementary material”) of random collections of species, both intra and inter-genus, were found to have little relationship with the phylogenetic clustering based on the corresponding 16S rDNA sequence. Trees based on 16S rDNA sequences and on ARDRA fragmentation were compared, and the average distance between the ARDRA-based and sequence-based trees was 77 ± 2.6, out of a maximum distance of 94 (see “Supplementary material”). In a parallel study, 20 groups of 85 sequences each were used for comparison of ARDRA to 16S rDNA sequence-based phylogenies (see details in “Supplementary material”). The average similarity between the sequence- and ARDRA-based clusters was 2.9%, while the maximum similarity was 7.3%.
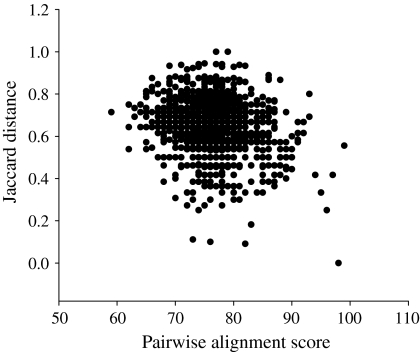
Since clustering is based on pairwise distance or similarity between sequences, two distance parameters will result in similar trees if the two parameters correlate to a certain degree. When random groups of 50 species were used, a slight negative correlation was found between the pairwise alignment scores and the calculated Jaccard distances (for details see “Supplementary material”), with an average Pearson correlation coefficient of −0.23 ± 0.078 (Fig. 2). The negative direction of the correlation was expected, since pairwise alignment scores measure similarity and the Jaccard distance measures dissimilarity; however, the low value of the Pearson correlation coefficient indicated a weak correlation. Consequently, the phylogenetic information that could be attributed to the ARDRA clustering, based on the Jaccard distance between profiles, was limited.
Fig. 2.
Correlation between the pairwise sequence analysis scores and the calculated Jaccard distances for a random group of 50 species
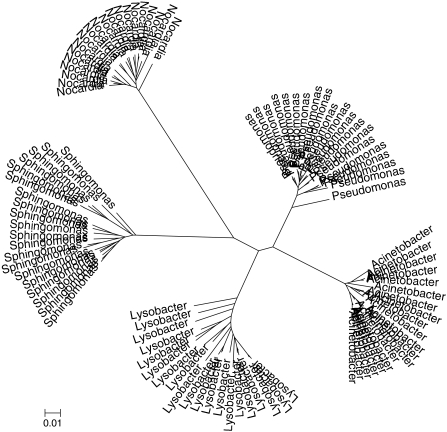
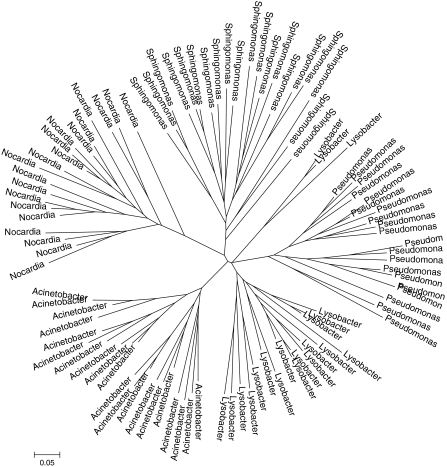
Phylogenetic trees were constructed for 20 representative OTUs from each of five genera with increasing inter-genera distances (Fig. S1), based on their 16S rDNA sequence (Fig. 3) and on the ARDRA fragmentation profile (Fig. 4). The sequence-based tree (Fig. 3) produced a clear division between the genera, with inter-genera distances reflecting the expected differences based on the taxonomic identity of the genera. However, while the ARDRA tree (Fig. 4) did differentiate between the OTUs (with one exception), it did not maintain the taxonomic structure of the genera (Fig. S1).
Fig. 3.
Sequence-based phylogenetic tree of 20 representatives from each of the genera listed in Fig. S1
Fig. 4.
ARDRA-based phylogenetic tree of 20 representatives from each of the genera listed in Fig. S1
To conclude, ARDRA can be a suitable tool for genus differentiation of environmental clones based on in-silico fragmentation. Moreover, in-silico profiles may be used for species identification provided caution is taken in the type and number of REs selected. Differentiation of strains requires more stringent measures, which are so time-consuming that the applicability of ARDRA to that end can be called into question. In addition, ARDRA-based dendrograms may not mirror 16S rDNA sequence-based phylogenetic trees.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ARDRA
Amplified ribosomal DNA restriction analysis
- RE
Restriction endonuclease
Contributor Information
Menachem Y. Sklarz, Phone: +972-8-6563513, FAX: +972-8-6596831, Email: Sklarz@bgu.ac.il
Roey Angel, Phone: +49-17661278588, Email: angel@mpi-marburg.mpg.de.
Osnat Gillor, Phone: +972-8-6596986, FAX: +972-8-6596909, Email: gilloro@bgu.ac.il.
M. Ines M. Soares, Phone: +972-8-6596834, FAX: +972-8-6596909, Email: soares@bgu.ac.il
References
- Collado MC, Hernandez M. Identification and differentiation of Lactobacillus, Streptococcus and Bifidobacterium species in fermented milk products with bifidobacteria. Microbiol Res. 2007;162:86–92. doi: 10.1016/j.micres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Criado-Fornelio A, Martinez-Marcos A, Buling-Saraña A, Barba-Carretero JC. Presence of Mycoplasma haemofelis, Mycoplasma haemominutum and piroplasmids in cats from southern Europe: a molecular study. Vet Microbiol. 2003;93:307–317. doi: 10.1016/S0378-1135(03)00044-0. [DOI] [PubMed] [Google Scholar]
- Davidov Y, Jurkevitch E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol. 2004;54:1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- Deng W, Xi D, Mao H, Wanapat M. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: a review. Mol Biol Rep. 2008;35:265–274. doi: 10.1007/s11033-007-9079-1. [DOI] [PubMed] [Google Scholar]
- Giraffa G, Vecchi PD, Rossetti L. Note: Identification of Lactobacillus delbrueckii subspecies bulgaricus and subspecies lactis dairy isolates by amplified rDNA restriction analysis. J Appl Microbiol. 1998;85:918–924. doi: 10.1046/j.1365-2672.1998.00606.x. [DOI] [PubMed] [Google Scholar]
- Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods. 1996;26:247–259. doi: 10.1016/0167-7012(96)00916-5. [DOI] [Google Scholar]
- Koeleman JGM, Stoof J, Biesmans DJ, Savelkoul PHM, Vandenbroucke-Grauls CMJE. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol. 1998;36:2522–2529. doi: 10.1128/jcm.36.9.2522-2529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Wang ET, Chen WF, Sui XH, Chen WX. Diverse bacteria isolated from root nodules of wild Vicia species grown in temperate region of China. Arch Microbiol. 2008;190:657–671. doi: 10.1007/s00203-008-0418-y. [DOI] [PubMed] [Google Scholar]
- Logan NA, Forsyth G, Lebbe L, et al. Polyphasic identification of Bacillus and Brevibacillus strains from clinical, dairy and industrial specimens and proposal of Brevibacillus invocatus sp. nov. Int J Syst Evol Microbiol. 2002;52:953–966. doi: 10.1099/ijs.0.02081-0. [DOI] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer CL, Tiedje JM, Dobbs FC, Karl DM. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodas AM, Ferrer S, Pardo I. 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Syst Appl Microbiol. 2003;26:412–422. doi: 10.1078/072320203322497446. [DOI] [PubMed] [Google Scholar]
- Segonds C, Paute S, Chabanon G. Use of amplified ribosomal DNA restriction analysis for identification of Ralstonia and Pandoraea species: interest in determination of the respiratory bacterial flora in patients with cystic fibrosis. J Clinic Microbiol. 2003;41:3415–3418. doi: 10.1128/JCM.41.7.3415-3418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakenborg T, Vicca J, Butaye P, Maes D, De Baere T, Verhelst R, Peeters J, de Kruif A, Haesebrouck F, Vaneechoutte M. Evaluation of amplified rDNA restriction analysis (ARDRA) for the identification of Mycoplasma species. BMC Infect Dis. 2005;5:46. doi: 10.1186/1471-2334-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje JM, Asuming-Brempong S, Nusslein K, Marsh TL, Flynn SJ. Opening the black box of soil microbial diversity. Appl Soil Ecol. 1999;13:109–122. doi: 10.1016/S0929-1393(99)00026-8. [DOI] [Google Scholar]
- Vaneechoutte M, Heyndrickx M. Application and analysis of ARDRA patterns in bacterial identification, taxonomy and phylogeny. In: Dijkshoorn L, Towner KJ, Struelens M, editors. New approaches for the generation and analysis of microbial typing data. The Netherlands: Elsevier; 2001. pp. 211–247. [Google Scholar]
- Ventura M, Casas IA, Morelli L, Callegari ML. Rapid amplified ribosomal DNA restriction analysis (ARDRA) identification of Lactobacillus spp. isolated from fecal and vaginal samples. Syst Appl Microbiol. 2000;23:504–509. doi: 10.1016/S0723-2020(00)80024-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.