Abstract
Optical traps or “tweezers” use high-power, near-infrared laser beams to manipulate and apply forces to biological systems, ranging from individual molecules to cells. Although previous studies have established that optical tweezers induce photodamage in live cells, the effects of trap irradiation have yet to be examined in vitro, at the single-molecule level. In this study, we investigate trap-induced damage in a simple system consisting of DNA molecules tethered between optically trapped polystyrene microspheres. We show that exposure to the trapping light affects the lifetime of the tethers, the efficiency with which they can be formed, and their structure. Moreover, we establish that these irreversible effects are caused by oxidative damage from singlet oxygen. This reactive state of molecular oxygen is generated locally by the optical traps in the presence of a sensitizer, which we identify as the trapped polystyrene microspheres. Trap-induced oxidative damage can be reduced greatly by working under anaerobic conditions, using additives that quench singlet oxygen, or trapping microspheres lacking the sensitizers necessary for singlet state photoexcitation. Our findings are relevant to a broad range of trap-based single-molecule experiments—the most common biological application of optical tweezers—and may guide the development of more robust experimental protocols.
Introduction
Single molecule techniques have emerged as a powerful tool in molecular biology, biochemistry, and biophysics. In particular, optical traps or “tweezers” use optical forces generated by focused laser light to manipulate microscopic objects (1)—typically polystyrene or latex microspheres—and to detect the minuscule biological forces exerted on them by individual molecules. This technique has been instrumental in addressing fundamental biological problems. For example, optical tweezers have been used to understand the mechanical properties of nucleic acid structures and proteins, sensitively probe protein-nucleic acid interactions, and decipher the mechanisms of many cytoskeletal and nucleic acid molecular motors (2–4).
Generation of the large optical forces necessary to efficiently trap microscopic objects and to counteract the forces exerted by biological systems (typically in the 1–100 pN range) requires both a high photon flux and tight focus of light to a diffraction limited spot (5). The high light intensity at the optical trap (>1 MW/cm2) thus poses a risk for optical damage to the biological systems of interest. An early finding in the development of this technique was that near-infrared (NIR) wavelengths (800–1100 nm) were more biocompatible compared to those in the visible spectrum, due to decreased absorption by cellular molecules and proteins in the NIR spectrum (6). NIR light is now used exclusively in biological applications of optical tweezers, with 1064 nm the most common wavelength in the field due in large part to the availability of high-power YAG lasers at this wavelength (7).
Despite these findings and widespread use of NIR optical traps, it is well-documented that NIR wavelengths can still cause photodamage in irradiated Escherichia coli, HeLa, and CHO cells (7–9) as deduced from reduced motility and cloning efficiency. Although the exact mechanism for this process has not been well established, evidence suggests excitation of molecular oxygen into reactive oxidative species (ROS) by the NIR light via sensitizer molecules in the cell (7–9). Studies of trap-induced optical damage have so far been limited to cells, yet the most common biological applications of optical traps involve single molecules studied in vitro. Thus, the potential for photodamage in the most widespread optical tweezers assays has not yet been adequately examined.
In this study, we investigate the effects of NIR (1064-nm) optical tweezers on a simple molecular system consisting of individual DNA molecules tethered between two trapped, functionalized microspheres, an arrangement common to many DNA- and RNA-based optical trap experiments (10). With this simple assay, we establish that the effects of the trapping light are detrimental, irreversible, and pervasive, affecting both the longevity of tethers and the efficiency with which they can be formed, and in certain cases the structure of the DNA molecules themselves. We further identify the components of this simple system most prone to damage. Moreover, we show that the source of damage is the highly reactive singlet state of molecular oxygen generated by the trapping light, and reveal the identity of the molecular sensitizers necessary for its photoexcitation.
Materials and Methods
See the Supporting Material for Materials and Methods.
Results
Tether longevity
To investigate the effect of NIR traps in vitro, we developed a simple assay that captures essential features common to a large class of optical tweezers experiments. Optical trap measurements of nucleic acids and the proteins with which they interact usually involve tethering a molecule between two attachment points: either an optically trapped microsphere and the surface of a fluidic chamber or micropipette, or two optically trapped microspheres (4,11,12). Typically, this is achieved by modifying the ends of the DNA molecule with different chemical moieties that can make specific linkages with the functionalized microsphere or surface. Biotin, which interacts tightly with streptavidin, and a small molecule like the hapten digoxigenin, which binds to its antibody, are common approaches to forming specific linkages (13).
In this study, we measured the properties of single DNA tethers with a 1064-nm dual trap optical tweezers (14). Double-stranded DNA (dsDNA) molecules of 3.4-kilobase (kb) contour length were synthesized with a single 5′-biotin and 5′-digoxigenin modification at each end (see Materials and Methods in the Supporting Material). Inside a custom flow cell, these bifunctional molecules were tethered (see Materials and Methods in the Supporting Material) to 0.79-μm streptavidin (SA) and 0.86-μm anti-digoxigenin (AD) antibody-coated microspheres each held in an optical trap, as shown schematically in Fig. 1 A. Measurements were carried out in an experimental buffer of 50 mM Tris with 150 mM NaCl. In our first experiment, we measured the longevity of these tethers under a constant range of tensions (14 ± 3 pN; mean ± SD) as a function of the intensity of 1064-nm light in both traps. In Fig. 1 B, the average tether lifetime is seen to be strongly affected by increasing trap power (measured at the sample plane in the trap holding the AD microsphere; see Materials and Methods in the Supporting Material), decreasing as a power law with an exponent of −1.66 ± 0.12 (χ2 = 13.7). Tether breakage was probably irreversible, based on the limited success in reforming tethers once broken. At all tested trap powers, lifetimes were exponentially distributed, in agreement with previous reports (13), and were independent of the tether history—whether the molecule was held at different tensions or exposed to different light intensities before lifetime measurement (data not shown)—indicative of a process determined by a single rate-limiting step. Lifetimes also exhibited a weak dependence on tension, as reported previously (13), which was identical across the range of trap powers investigated (lifetimes were well-fit to an exponential exp(− FΔx/kBT), with Δx = 0.30 ± 0.1 nm; data not shown).
Figure 1.
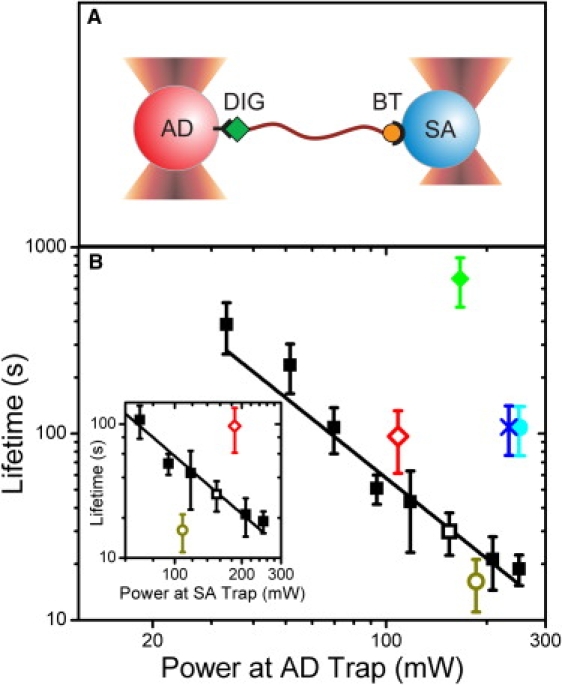
Dependence of tether lifetime on trap power. (A) Schematic representation of a dsDNA tether with 5′-digoxigenin (labeled DIG) and 5′-biotin (BT) modifications held between an anti-digoxigenin (AD, red) and a streptavidin (SA, blue) microsphere. (B) Tether lifetime versus laser power measured at the AD microsphere. Average lifetimes of tethers formed between 0.86-μm AD and 0.79-μm SA microspheres in traps of identical power (black squares, N = 18–55), and asymmetric power, with the AD microsphere in the low-power trap (open red diamond, N = 23) and high-power trap (open dark yellow circle, N = 22). Open symbols represent tethers under identical total trap power. Average lifetime of tethers formed between 2.1-μm AD and SA microspheres (cyan circle, N = 29), between a 2.1-μm AD microsphere and a 0.79-μm SA microsphere (blue X, N = 16) and between two 0.79-μm SA microspheres through dual biotin-streptavidin linkages (green diamond, N = 8). Inset: Above data plotted as a function of laser power measured at the SA microsphere. All tethers were held at tensions of 10–20 pN (14 ± 3 pN; mean ± SD, N = 268). Error bars = SE. Power-law fit to tether lifetime versus trap power measured at the AD microsphere yields the equation t = A × PB with A = (3.9 ± 2.0) × 105 s/mW and B = −1.66 ± 0.12, (χ2 = 13.7).
To determine which components of the tethers were most prone to breakage, we carried out two tests of the attachment moieties. In the first, we applied asymmetrical light intensities to the dual traps (120 mW in one versus 230 mW in the other) and measured the tether lifetimes in the two possible geometries: SA microspheres in the strong trap and AD microspheres in the weak trap, or vice versa. As shown in Fig. 1 B (red open diamond and dark yellow open circle), the tether lifetimes correlated strongly with the light intensity in the trap holding the AD microsphere, corresponding to the abscissa in that plot. In contrast, lifetimes correlated only weakly with the intensity in the trap holding the SA microsphere (Fig. 1 B, inset) or the total intensity (data not shown). In the second test, DNA molecules with 5′-biotin modifications at both ends were tethered to two SA microspheres in traps of equal power (see Materials and Methods in the Supporting Material). In comparison to bifunctional molecules exposed to the same trapping light intensity, the lifetimes of dual-biotin tethers were enhanced by a factor of >20 (Fig. 1 B, green diamond). These two results establish that the digoxigenin-anti-digoxigenin linkage is prone to breakage, and responsible for the tether lifetime. This is consistent with the many-fold slower dissociation rate of streptavidin-biotin in comparison to that of digoxigenin-anti-digoxigenin observed in bulk studies (15,16).
The strong power law dependence of tether lifetime with trapping light intensity is suggestive of photodamage. Although the 1064-nm light of the traps is absorbed by the aqueous buffer in the experimental flow cell, leading to heating, the temperature increase with trap power is small (∼1.0–1.45°C per 100 mW (8,17)) and unlikely to elicit the dramatic effect on tether lifetime observed. Fits to the data in Fig. 1 B with the Arrhenius equation, as expected for a temperature-dependent effect, are poor compared to a power law (χ2 = 1005; data not shown). Moreover, as shown previously (18), heating by optical traps is not localized to the laser focus, but extends spatially in a weak, logarithmic decay. It follows that the temperature of the region surrounding the dual traps in these experiments (that are separated by at most ∼2 μm) is approximately uniform, and determined by the total light intensity in the two traps. However, our measurements indicate that trap-mediated tether breakage is a highly local effect as shown by the fact that tethers under identical total trap power exhibit such disparate lifetimes depending on whether the SA or AD microsphere is exposed to more light (Fig. 1 B, open symbols). This point is further corroborated by the observation that the lifetimes of tethers attached to two large, 2.1-μm microspheres are sixfold longer than those of tethers on smaller microspheres at identical trap powers (110 ± 31 s compared to 20 ± 3.5 s; mean ± SE; Fig. 1 B, cyan circle). In addition, tethers held between a small, 0.79-μm SA microsphere and a large, 2.1-μm AD microsphere lasted as long as those attached to two large microspheres (110 ± 32 s; mean ± SE; Fig. 1 B, blue X), corroborating the view that tether lifetime is determined by the digoxigenin-AD linkage. Thus, although we cannot rule out that temperature may play a minor role, our data are more consistent with local optical damage as the primary cause of tether breakage, a claim further confirmed by additional studies detailed below.
Tethering efficiency
For our next experiment we characterized the ability of trapped microspheres to form DNA tethers as a function of exposure to trapping light. To determine if one linkage was more sensitive than the other, we tested two configurations: one in which the SA microsphere was coated with DNA and the AD microsphere was bare, and vice versa. An attempt was made to form a tether for each microsphere in the two configurations independently by bringing its complementary microsphere in contact using the optical traps (see Materials and Methods in the Supporting Material). For each type of microsphere we determined the tethering efficiency—defined as the fraction of trials that formed tethers—at low laser power (100 mW; power measured at each trap unless otherwise noted) both before and after 10 min of exposure to high-intensity (350 mW) light. To isolate the effect of NIR irradiation to one microsphere, we used a new, unexposed complementary microsphere for each time point.
Although the initial tethering efficiencies were high for all microsphere types (Fig. 2: SA with DNA: blue bar 1, bare AD: blue bar 4, bare SA: red bar 1, and AD with DNA: red bar 4), they decreased significantly on microsphere exposure to 350-mW light in certain cases. DNA-coated SA microspheres, for instance, exhibited much lower tethering efficiencies compared to bare AD microspheres after irradiation (Fig. 2, compare blue bars 2 and 5). In control experiments with 10 min irradiation with low light intensities (100 mW; Fig. 2, blue bars 3 and 6), the efficiencies were indistinguishable from their initial values. Interestingly, irradiating the SA or AD microsphere for a longer time in proportion to the light intensity (∼40 min for 100 mW light) eventually reduced the tethering efficiency to 0 (data not shown), suggesting that the rate of decay is determined by the total dosage of photons. When bare SA microspheres were irradiated, the decrease in tethering efficiency was much more dramatic (Fig. 2, red bar 2); no tethers were formed in a set of 16 microspheres. Here, moreover, controls at low power (Fig. 2, red bar 3) also exhibited a reduced efficiency. Experiments on DNA-coated AD microspheres displayed a similar pattern (Fig. 2, red bar 5 and 6), though less severe. In these experiments, tether lifetimes in instances when tethers were formed (27.3 ± 3.7 s; mean ± SE) were comparable, within SE, to those of unexposed microsphere pairs (26.3 ± 4.5 s; mean ± SE), suggesting that this fraction of molecules was not affected by trap irradiation.
Figure 2.
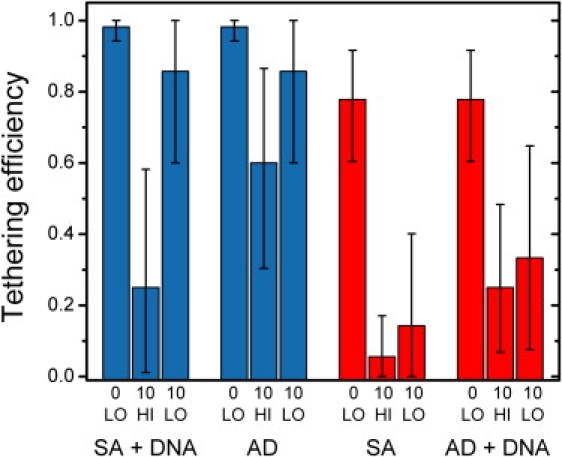
Dependence of tether forming efficiencies on trap irradiation. Tether formation was attempted in microsphere pairs in two configurations: 0.79-μm DNA-coated SA with 0.86-μm bare AD microspheres (blue bars; ∼20 mol/microsphere), or DNA-coated AD with bare SA microspheres (red bars; ∼30 mol/microsphere). Tethering efficiencies were measured for DNA-coated SA (blue bars 1–3), AD (blue bars 4–6), SA (red bars 1–3), and DNA-coated AD (red bars 4–6) microspheres under the following conditions: after initial trapping at low, 100 mW laser power (denoted 0 LO), after 10 min of exposure to high, 350 mW power (10 HI), or after 10 min of exposure to low, 100 mW power (10 LO). Each trial involved a tethering attempt with a new, unexposed complementary microsphere. Tethering efficiencies were calculated using the Laplace best estimator (50) (S + 1)/(N + 2), where S is the number of successes and N is the number of trials. This estimator is considered better than the maximum-likelihood S/N, when N is small. Error bars = 95% confidence intervals from the adjusted Wald method (51).
In the two cases where bare SA (Fig. 2, red bars 1–3) and AD (Fig. 2, blue bars 4–6) microspheres were exposed, the data clearly indicate that the SA microspheres were prone to rapid, irreversible photodamage even at modest laser powers, whereas the AD microspheres were relatively insensitive to irradiation by NIR light. The results for DNA-coated SA (Fig. 2, blue bars 1–3) and AD (Fig. 2, red bars 4–6) microspheres are more difficult to interpret. Based on our experiments on tether lifetime implicating tether breakage at the microsphere-DNA linkage, one possible interpretation for the data is that the DNA detached from the microspheres. Alternatively, there may have been irreversible damage to the DNA or the microsphere attachment moieties, resulting in lowered tethering efficiency.
To test these possible interpretations, we developed an assay to monitor the amount of DNA on trapped microspheres in real time. Microspheres undergoing random Brownian motion in the harmonic potential of an optical trap normally exhibit a characteristic Lorentzian noise power spectrum (19). We discovered that microspheres coated with DNA displayed excess noise at low frequencies (<100 Hz) that increased with the amount of DNA (Fig. 3 A). Presumably, this excess noise is caused by hydrodynamic interactions of the DNA molecules with the surrounding solvent. As shown in Fig. 3 B, this excess noise can be calibrated against the amount of DNA coating the microspheres (see Materials and Methods in the Supporting Material); thus, by periodically monitoring the noise characteristics of DNA-coated microspheres, we determined the amount of DNA on the microspheres as a function of time.
Figure 3.
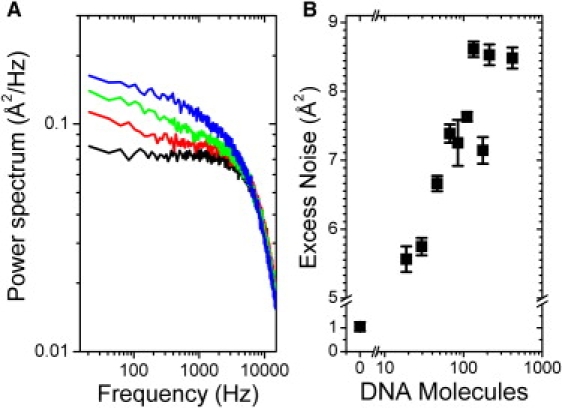
Low frequency noise as a function of DNA on microspheres. (A) Power spectra for a SA microsphere coated with 0 (black), ∼30 (red), ∼80 (green), and ∼400 molecules (blue) of DNA exposed to 300 mW laser power. (B) Excess integrated noise between 0 and 100 Hz as a function of the number DNA molecules on the microsphere. Each data point represents the average from nine power spectra from three separate microspheres. Error bars = SE.
Fig. 4, A and B, show that DNA indeed dissociates with exposure to high trap light intensities (350 mW). Not surprisingly, given that the digoxigenin-AD linkage was more prone to breakage in the tether longevity measurements, DNA molecules detached from the AD microspheres more rapidly than from SA microspheres (Fig. 4 B, red circles and blue squares, respectively,). These data suggest that dissociation of DNA from AD microspheres may explain the decrease in tethering efficiency in DNA-coated AD microspheres (Fig. 2, red bars 4–6) because they occur on similar timescales. However, DNA detaches from SA microspheres much too slowly to account for the rapid decay in efficiency in DNA-coated SA microspheres (Fig. 2, blue bars 1–3); it takes ∼30 min for half the molecules to detach from SA microspheres compared to 10 min to completely abolish tethering efficiency. This result indicates that the DNA itself—most likely the digoxigenin linkage moiety—is being photodamaged with irradiation. These measurements taken together thus highlight which components of the two-microsphere DNA-tether system are most affected by the optical traps. Damage to digoxigenin likely accounts for several observed behaviors: reduced tether lifetime (Fig. 1 B), decreased tethering efficiency in exposed DNA-coated SA microspheres (Fig. 2, blue bar 2), and dissociation of DNA from AD microspheres (Fig. 4, red circles). Streptavidin is also prone to damage, as attested by the dramatic and rapid decrease in tethering efficiency with irradiation (Fig. 2, red bars 2 and 3). However, in contrast to digoxigenin, photodamage to SA does not lead to fast dissociation of DNA (Fig. 4, blue squares), suggesting that SA may be protected if bound to a complementary biotin.
Figure 4.
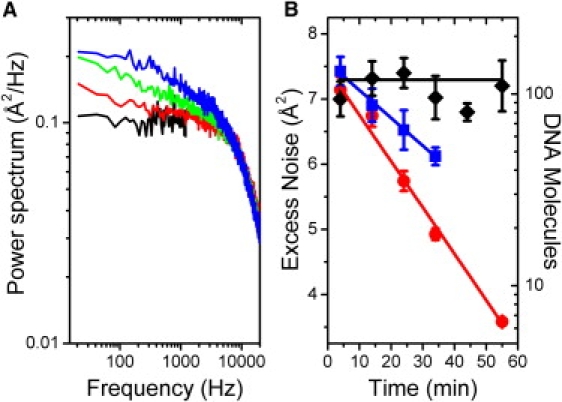
DNA dissociation from microspheres. (A) Power spectra of a heavily DNA-coated AD microsphere (∼230 mol/ microsphere) at t = 4 min (blue), 14 min (green), and 34 min (red) and an AD microsphere with no DNA (black). (B) Excess integrated noise between 0 and 100 Hz as a function of time for 0.79-μm SA (blue squares), 0.86-μm AD (red circles), and 0.97-μm SA silica (black diamonds) DNA-coated microspheres exposed to 350 mW of laser power. Error bars = SE from five power spectra. Red, blue, and black lines are trend lines to guide the eye.
Cause of photodamage
The observed decrease in tether longevity and efficiency with exposure to NIR trapping light points to damage of the attachment moieties in the DNA tethers. In studies of trap-induced optical damage in cells, the underlying mechanism is believed to entail generation of ROS by the NIR trapping light (7–9). To determine if a similar mechanism is involved, we repeated the above experiments under anaerobic conditions, using two different oxygen scavenging systems: the protocatechuic acid-protocatechate 3,4-dioxygenase (PCA/PCD) system and the glucose oxidase-catalase (GODCAT) coupled enzyme system (20,21) (see Materials and Methods in the Supporting Material). In the absence of oxygen, the detrimental effects of the NIR trapping light are dramatically reduced, indicating an analogous mechanism at play in this simplified, in vitro assay. Dissociation of the digoxigenin-anti-digoxigenin linkage is reduced, leading to longer tether lifetimes (350 ± 110 s with PCA/PCD compared to 21 ± 10 s without; mean ± SE) and improved tethering efficiency in experiments where DNA coats the AD microsphere, and irreversible damage to the SA microspheres is all but eliminated (the tethering efficiency for SA and DNA-coated AD microspheres remained high after over 1 h of exposure at 350 mW; data not shown). Table 1 summarizes the relative benefits of the PCA/PCD and GODCAT oxygen scavenging systems, as measured by the improvement in tether longevity at a high trap power; the concentrations of enzymes and substrates used in both systems reflect standardized conditions from the literature (20,21). Interestingly, the PCA/PCD system seems to elicit a larger improvement in tether longevity compared to GODCAT, consistent with its reported higher efficiency of oxygen depletion (21).
Table 1.
Increase in tether lifetime with chemical additives
| Method (N) | Concentration | Relative lifetime |
|---|---|---|
| PCA/PCD (28)∗ | 100 mM PCA; 10 nM PCD | 17.0 ± 9.5 |
| GODCAT (21)∗ | 100 nM glucose oxidase; 1.5 mM catalase; 56 mM glucose | 7.6 ± 4.3 |
| Ascorbic acid (41)† | 12.5 mM | 5.2 ± 1.9 |
| Sodium azide (31)† | 100 mM | 3.8 ± 1.0 |
| Lipoic acid (18)† | 3.1 mM | 2.1 ± 1.3 |
| Tris-Cl (25)‡ | 200 mM | 1.1 ± 0.4 |
| Mannitol (35)‡ | 200 mM | 1.0 ± 0.4 |
Tether lifetimes with additive were measured relative to those in standard TS buffer at the same trap power (range = 150–300 mW). Errors = SE. GODCAT, glucose oxidase/catalase; PCA, protocatechuic acid; PCD, protocatechate 3,4-dioxygenase.
Increase in tether lifetime on addition of oxygen scavengers.
Increase in tether lifetime on addition of singlet oxygen quenchers.
Increase in tether lifetime on addition of hydroxyl radical quenchers.
The above results suggest that, as observed in vivo, trap-mediated damage occurs through the excitation of molecular oxygen into ROS, leading to oxidative damage of the DNA linkages. Several kinds of ROS can in principle be generated from molecular oxygen: superoxide anion, hydrogen peroxide, hydroxyl radicals, and singlet oxygen (22) to name a few examples. Studies carried out in vivo indicate that hydroxyl radicals and singlet oxygen are two ROS generated by laser irradiation (9). To determine which ROS is the dominant source of damage in our assays, we carried out two tests. In the first, we measured the improvement in tether longevity at high trap power on addition of known singlet oxygen quenchers—the antioxidants ascorbic acid, lipoic acid, and sodium azide (23–25)—and hydroxyl radical quenchers Tris and mannitol (26) (see Materials and Methods in the Supporting Material). Table 1 summarizes the results. Although all three singlet oxygen quenchers increased tether lifetimes, with ascorbic acid (12.5 mM) eliciting the largest improvement comparable to that of GODCAT (an ∼6-fold improvement), the hydroxyl radical quenchers had little effect, implicating singlet oxygen as the ROS generated by the optical traps.
This conclusion is further confirmed by our second test, in which singlet oxygen was directly detected with 3-(10-(2-carboxy-ethyl)-anthracen-9-yl)-propionic acid (CEAPA). This anthracene derivative exhibits specific reactivity for singlet oxygen by forming a stable epoxide derivative via a Diels-Alder cycloaddition across its middle ring, and also acts as a singlet oxygen sensitizer (27). CEAPA dissolved in methanol was flowed into a custom sample chamber in the absence of microspheres, exposed to a high intensity of our trapping light (1.6 W; total power) for a period of 360 min, collected, and tested by electrospray ionization-mass spectrometry (see Materials and Methods in the Supporting Material). After exposure to the trapping light, electrospray ionization-mass spectrometry of CEAPA showed a peak at a mass-to-charge (m/z) value of 355.1, corresponding to a molecular mass of 354.1 g/mol, precisely one O2 molecule more than that of unexposed CEAPA, providing direct evidence for photoexcitation of singlet oxygen by the optical traps.
Although the energy required to excite ground state molecular oxygen into its singlet excited state (E = 0.98 eV; λ = 1270 nm) is consistent with the energy provided by the NIR trapping light (E = 1.17 eV; λ = 1064 nm), this transition is strictly forbidden by spin, symmetry, and Laporte selection rules (28). As a result, singlet oxygen can only be produced by energy transfer to molecular oxygen through a triplet sensitizer (29). This sensitizer molecule must be present to accept energy, store it in the form of vibrations, and transfer it to ground state oxygen, exciting the molecule to its singlet state. Molecules capable of storing energy in vibrational form are typically rich in π-bonded electrons and highly aromatic. Inside the cellular environment, molecules of this character are likely plentiful, facilitating the generation of singlet oxygen in cells exposed to NIR trapping light. In the case of our in vitro tether assays, however, the only likely sensitizer exhibiting significant π-bond character and aromaticity are the polystyrene microspheres. (In the experiments with CEAPA, the anthracene derivative itself acted as a sensitizer.) To test this hypothesis, we developed a hybrid fluorescence-optical tweezers assay, using the singlet-oxygen sensor green (SOSG) fluorescent probe, which emits light at 525 nm in the presence of singlet oxygen (30). Our apparatus could switch between brightfield images of trapped microspheres and fluorescence images of the specimen plane (excitation, 488 nm; emission, 525 nm; see Materials and Methods in the Supporting Material), allowing us to localize singlet oxygen generation at the optical traps and determine precisely the conditions for its generation.
The results are summarized in Fig. 5, A–E. In control experiments, optically trapped 0.79-μm polystyrene microspheres in the absence of SOSG produced the expected brightfield microsphere images but no fluorescence (Fig. 5 A). In the presence of SOSG, the optical trap itself (not trapping a microsphere) produced no fluorescence signal (Fig. 5 B). Only when a polystyrene microsphere was trapped in buffer containing SOSG did we detect significant fluorescence localized at the trapped microsphere position (Fig. 5, C and D), in support of our conjecture that the microspheres provide the sensitizers required for singlet oxygen generation. Interestingly, the intensity profiles of the SOSG fluorescence for two microspheres of different sizes (compare 0.79-μm microsphere in Fig. 5 C with 2.1-μm microsphere in Fig. 5 D) correlate well with the microsphere diameter and suggest that fluorescence is localized at the surface. Moreover, the total fluorescence intensity from the larger 2.1-μm microsphere, normalized by the microsphere surface area, is consistent with the higher tether lifetimes observed with larger microspheres (Fig. 1 B). The ratio of fluorescence intensity per unit area is ∼6 (small microsphere/large microsphere) and the ratio of the tether lifetimes (large microsphere/small microsphere) is ∼6. Finally, when trapping a comparably sized (0.78 μm) small microsphere made of silica—a material lacking the aromaticity of polystyrene—SOSG fluorescence was not observed (Fig. 5 E). This observation would suggest that oxidative damage is reduced in silica microspheres. Indeed, DNA-coated SA silica microspheres of a similar size (0.97 μm) exhibited dramatically reduced levels of photodamage. There was no detectable dissociation of DNA from the microspheres over 1 h of exposure to high laser power (350 mW; Fig. 4 B, black diamonds). These results show that the generation of singlet oxygen is mediated by the polystyrene microspheres, which act as a triplet sensitizer.
Figure 5.
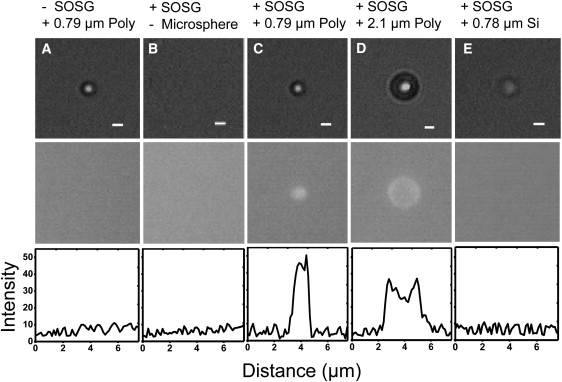
SOSG fluorescence in optically trapped microspheres. (Top) Brightfield images. (Center) Fluorescence images at 535 nm. (Bottom) Fluorescence image line scans. (A) 0.79-μm SA polystyrene microsphere without SOSG. (B) No microsphere with SOSG. (C) 0.79-μm SA polystyrene microsphere with SOSG. (D) 2.1-μm SA polystyrene microsphere with SOSG. (E) 0.78-μm silica microsphere with SOSG. Scale bar = 1 μm. A trap power of 390 mW was used in all the images.
Photodamage to nucleic acids
The results detailed above indicate that trap-mediated oxidative damage is local and targets the biotin-SA and digoxigenin-AD linkages in the tethered molecule. However, singlet oxygen is also known to oxidize certain nucleic acids (guanine, thymine, and uracil) irreversibly (31). Moreover, many studies have reported singlet oxygen-induced damage to single- and double-stranded DNA (32–37). To investigate the effect of trapping light on nucleic acids more directly, we carried out experiments on a 3.1-kb DNA molecule containing an 89-bp hairpin sequence (see Materials and Methods in the Supporting Material). This construct allowed us to expose its bases at will by unwinding the hairpin with tension, and probe changes to its secondary structure through its force-extension behavior.
Fig. 6, A and B, display typical hairpin force-extension curves taken in our standard tether assay buffer and in a buffer depleted of oxygen by the GODCAT coupled enzyme system, respectively. Other than the presence or absence of oxygen, the measurements were taken under identical conditions (140 mW, 2 pN/s pulling rate). Under aerobic conditions, where generation of singlet oxygen by the traps is possible, the force-extension curves exhibit irreversible hysteresis that grows with time (Fig. 6 A). Interestingly, whereas the unfolding curves of the hairpin (where tension is increased) are the same, the refolding curves (where tension is decreased) display a progressively lower refolding force, suggesting that the folded hairpin configurations are identical, but that energetic barriers to refolding become progressively larger over time with continued exposure. In contrast, all force-extension curves are reversible under anaerobic conditions, displaying no hysteresis for extended periods of time (Fig. 6 B), demonstrating a mechanism that also involves oxygen-dependent damage.
Figure 6.
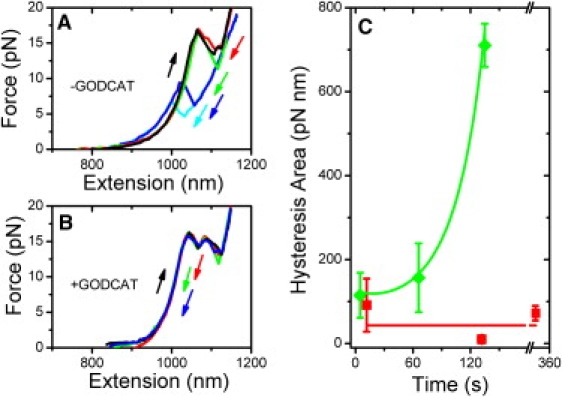
Oxidative damage to DNA hairpin. (A) Hairpin force-extension behavior under aerobic conditions (without GODCAT): stretching curves (black), relaxation curves after holding the hairpin folded at t = 0 s (red), 75 s (green), 200 s (blue), and 220 s (cyan). (B) Hairpin force-extension behavior under anaerobic conditions (with GODCAT): stretching curves (black), and relaxation curves at t = 0 s (red), 150 s (green), and 330 s (blue). All force-extension curves obtained at a pulling rate of 2 pN/s. (C) Increase in hysteresis area as a function of time for a DNA hairpin stretched under anaerobic condition (red squares), and aerobic conditions (green diamonds). Error bars = SE from 49 force-extension curves of two tethers. Red line and green curve are trend lines to guide the eye.
Plotting the hysteresis area—the difference between the unfolding and refolding force-extension areas—as a function of time summarizes these results. In Fig. 6 C, the hysteresis area increases (Fig. 6 C, green diamonds) with time under aerobic conditions, but remains low under anaerobic conditions (Fig. 6 C, red squares). These results indicate that bases are prone to irreversible oxidative damage when exposed to the surrounding oxygen-rich buffer, an effect we attribute to the production of singlet oxygen by the NIR laser. Though our experiment does not test for damage to duplex DNA, it is possible that bases in a duplex DNA conformation may also be prone to oxidative damage.
Discussion
In this article, we study the effect of NIR (1064-nm) optical traps in a simplified in vitro assay consisting of a single DNA molecule tethered between two trapped microspheres, an arrangement that captures many generic features of trap-based single molecule assays. We show that optical traps generate singlet oxygen via sensitizers in the polystyrene microspheres, and show that the oxidative damage is wide ranging, affecting the chemical moieties that link the DNA tether to the microspheres and the DNA bases themselves. Our measurements pinpoint two likely areas of damage to the microsphere-DNA linkages: digoxigenin and streptavidin. Oxidative damage of digoxigenin is likely responsible for the observed decrease in tether lifetimes (and tethering efficiency) with irradiation. Damage to streptavidin, on the other hand, dramatically reduces the efficiency but surprisingly does not reflect the slow rate of DNA dissociation observed. This result suggests that although streptavidin may be prone to damage, its binding sites are protected if bound to a complementary biotin.
Several results indicate that trap-mediated photodamage is a local effect: the dependence of tether lifetime on which microsphere is exposed to high light intensities, the increased tether lifetime when using larger microspheres, and the localization of SOSG fluorescence to the microspheres. Given the diffusion constant of molecular O2 in water (2 × 10−5 cm2/s (38)) and the lifetime of singlet oxygen in water (2 μs (39)), we estimate that this ROS should be localized to ∼100 nm surrounding the trapped microspheres, consistent with this picture. This likely explains why oxidative damage appears manifested most at the microsphere-DNA linkages of our dsDNA tethers. However, the DNA hairpin construct also exhibits signs of photodamage despite not being localized to the microsphere surfaces (the hairpin is separated from both microspheres by two 1.5-kb, or 510-nm, dsDNA spacer “handles”). It is possible that in cases of high sensitivity to oxidation such as with exposed bases, damage need not be limited to the region surrounding the microsphere surfaces.
Our SOSG fluorescence images and controls with silica microspheres show that generation of singlet oxygen is mediated by the polystyrene microspheres. Although they seem to act as sensitizers, polystyrene is not known to absorb in the NIR (40). One possibility is that impurities in the microspheres play a role. Another possibility is that the chemistry for linking the attachment moieties to the microspheres helps play a role in the sensitization of the singlet oxygen excitation process. However, due to the lack of aromaticity of these chemical entities, it is unlikely that they act as the sensitizer for singlet oxygen. The current experimental approach is insensitive to the effects of these attachment moieties in the role of singlet oxygen production, and further measurements will be necessary to investigate these effects on tether stability and singlet oxygen production. Alternatively, sensitization may occur through a two-photon process; it has been shown that polystyrene microspheres do absorb at visible wavelengths (40). The observed exponent of −1.66 ± 0.12 in the power-law dependence of tether lifetime on trapping light intensity could indicate that photodamage involves one- and two-photon absorption processes. It is also conceivable that a two-photon mechanism corresponds to excitation of molecular oxygen to its second excited triplet state (E = 6.9 eV; λ = 757.1 nm) that is known to decay rapidly to the longer-lived reactive singlet state (28). It is interesting to note that in studies of trap photodamage in E. coli cells (7), a smaller exponent (albeit >1) was measured. The fact that the sensitizers involved in singlet state generation in cells are undoubtedly different than in our in vitro assay may account for the discrepancies in the two measurements, however.
Our findings are relevant to a large class of in vitro optical trap experiments that involve tethering nucleic acids. Short tether lifetimes severely limit the duration and throughput of experiments. More importantly, trap-mediated photodamage requires that an excess of DNA be placed on microspheres to form tethers with a reasonable efficiency. As we have shown in the microsphere power spectra, however, excess DNA coating the microspheres also leads to increased noise. Recent advances in optical tweezers design have led to new high-resolution instruments capable of detecting conformational motion on the scale of one basepair of DNA (14,41,42). Excess noise from DNA-coated microspheres may thus be an important consideration in measurements requiring high resolution. Finally, although we have limited the scope of our study to DNA tethers, ROS may have significant effects on the activity and structure of other biomolecules such as RNA, lipids, and, in particular, proteins (43). In many experiments probing protein-nucleic acid interactions or molecular motors translocating along nucleic acids, the proteins are either linked directly or are in close proximity to the trapped microspheres, and thus subject to the same conditions that lead to photodamage of our DNA tethers. Proteins can undergo conformational changes, experience changes in refolding rates, exhibit reduced activity, and form cross-linked aggregates when exposed to singlet oxygen (44,45). Histidine, tyrosine, methionine, and cysteine are particularly vulnerable to oxidation by singlet oxygen (45).
As we have shown, oxidative damage can be largely mitigated by working under anaerobic conditions with the appropriate oxygen scavenging systems. In certain cases, it may not be permissive to operate under oxygen free conditions (for example in applications where trapping is attempted in vivo (46,47)), and we have shown that certain antioxidant additives can reduce damage almost as efficiently. The microsphere composition can also have a large effect on oxidative damage. A promising direction for the future will be the examination of alternative microsphere compositions and the development of less sensitive attachment moieties—for example covalent linkages using amide, carboxyl, and sulfhydryl chemistry (13)—that completely abolish trap-mediated oxidative damage in optical tweezers assays. Finally, the choice of trapping wavelength may have a strong effect on the degree of damage, as observed in studies of photodamage in vivo (7). Although we limited our studies to a single wavelength, 1064 nm, due to its common usage in the field, optical traps at other IR wavelengths are also used. Singlet oxygen production likely decreases away from the peak in the oxygen absorption band (1270 nm) (48). However, two-photon excitation of the short-lived triplet excited state by short IR wavelengths (590–880 nm) is also possible, depending on the sensitizer (49). Future work will be necessary to determine if the observed effects are as severe at other wavelengths.
Supporting Material
Materials and methods and references are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01314-9.
Supporting Material
Acknowledgments
The authors thank Dr. Matthias Selke, Dr. James Tsay, Daniel Robbins, Jeffrey Moffitt, and all the members of the Ha and Chemla labs for helpful discussions and for their generous advice.
This work was supported by the National Science Foundation (grant 082265, Physics Frontier Center: Center for the Physics of Living Cells). Y.R.C. was supported by Burroughs-Wellcome Fund-Career Awards at the Scientific Interface. M.P.L. acknowledges a University of Illinois Graduate College Fellowship.
References
- 1.Ashkin A., Dziedzic J.M., Bjorkholm J.E., Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante C., Bryant Z., Smith S.B. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 3.Mehta A.D., Rief M., Spudich J.A., Smith D.A., Simmons R.M. Single-molecule biomechanics with optical methods. Science. 1999;283:1689–1695. doi: 10.1126/science.283.5408.1689. [DOI] [PubMed] [Google Scholar]
- 4.Moffitt J.R., Chemla Y.R., Smith S.B., Bustamante C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 5.Neuman K.C., Block S.M. Optical trapping. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashkin A., Dziedzic J.M., Yamane T. Optical trapping and manipulation of single cells using infrared laser beams. Nature. 1987;330:769–771. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- 7.Neuman K.C., Chadd E.H., Liou G.F., Bergman K., Block S.M. Characterization of photodamage to Escherichia coli in optical traps. Biophys. J. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Sonek G.J., Berns M.W., Tromberg B.J. Physiological monitoring of optically trapped cells: assessing the effects of confinement by 1064-nm laser tweezers using microfluorometry. Biophys. J. 1996;71:2158–2167. doi: 10.1016/S0006-3495(96)79417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohanty S.K., Sharma M., Gupta P.K. Generation of ROS in cells on exposure to CW and pulsed near-infrared laser tweezers. Photochem. Photobiol. Sci. 2006;5:134–139. doi: 10.1039/b506061c. [DOI] [PubMed] [Google Scholar]
- 10.Williams M.C., Rouzina I. Force spectroscopy of single DNA and RNA molecules. Curr. Opin. Struct. Biol. 2002;12:330–336. doi: 10.1016/s0959-440x(02)00340-8. [DOI] [PubMed] [Google Scholar]
- 11.Bustamante C., Macosko J.C., Wuite G.J. Grabbing the cat by the tail: manipulating molecules one by one. Nat. Rev. Mol. Cell Biol. 2000;1:130–136. doi: 10.1038/35040072. [DOI] [PubMed] [Google Scholar]
- 12.Greenleaf W.J., Woodside M.T., Block S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller D.N., Gemmen G.J., Rickgauer J.P., Dupont A., Millin R. A general method for manipulating DNA sequences from any organism with optical tweezers. Nucleic Acids Res. 2006;34:e15. doi: 10.1093/nar/gnj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffitt J.R., Chemla Y.R., Izhaky D., Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G., Dubrawsky I., Mendez P., Georgiou G., Iverson B.L. In vitro scanning saturation mutagenesis of all the specificity determining residues in an antibody binding site. Protein Eng. 1999;12:349–356. doi: 10.1093/protein/12.4.349. [DOI] [PubMed] [Google Scholar]
- 16.Piran U., Riordan W.J. Dissociation rate-constant of the biotin-streptavidin complex. J. Immunol. Methods. 1990;133:141–143. doi: 10.1016/0022-1759(90)90328-s. [DOI] [PubMed] [Google Scholar]
- 17.Gross S.P. Application of optical traps in vivo. Methods Enzymol. 2003;361:162–174. doi: 10.1016/s0076-6879(03)61010-4. [DOI] [PubMed] [Google Scholar]
- 18.Mao H., Arias-Gonzalez J.R., Smith S.B., Tinoco I., Bustamante C. Temperature control methods in a laser tweezers system. Biophys. J. 2005;89:1308–1316. doi: 10.1529/biophysj.104.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visscher K., Gross S.P., Block S. Construction of multiple-beam optical traps with nanometer-resolution position sensing. IEEE J. Sel. Top. Quantum Electron. 1996;2:1066–1076. [Google Scholar]
- 20.Selvin P.R., Lougheed T., Tonks Hoffman M., Park H., Balci H. In Vitro and In Vivo FIONA and Other Acronyms for Watching Molecular Motors Walk. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2008. [Google Scholar]
- 21.Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sies H. Oxidative stress: oxidants and antibiotics. Exp. Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 23.Machlin L.J., Bendich A. Free-radical tissue-damage—protective role of antioxidant nutrients. FASEB J. 1987;1:441–445. [PubMed] [Google Scholar]
- 24.Packer L., Witt E.H., Tritschler H.J. α-Lipoic acid as a biological antioxidant. Free Radic. Biol. Med. 1995;19:227–250. doi: 10.1016/0891-5849(95)00017-r. [DOI] [PubMed] [Google Scholar]
- 25.Devasagayam T.P., Sundquist A.R., Di Mascio P., Kaiser S., Sies H. Activity of thiols as singlet molecular oxygen quenchers. J. Photochem. Photobiol. B. 1991;9:105–116. doi: 10.1016/1011-1344(91)80008-6. [DOI] [PubMed] [Google Scholar]
- 26.Frati E., Khatib A.M., Front P., Panasyuk A., Aprile F. Degradation of hyaluronic acid by photosensitized riboflavin in vitro. Modulation of the effect by transition metals, radical quenchers, and metal chelators. Free Radic. Biol. Med. 1997;22:1139–1144. doi: 10.1016/s0891-5849(96)00525-4. [DOI] [PubMed] [Google Scholar]
- 27.Trabanco A.A., Montalban A.G., Rumbles G., Barrett A.G.M., Hoffman B.M. A seco-porphyrazine: Superb sensitizer for singlet oxygen generation and endoperoxide synthesis. Synlett. 2000:1010–1012. [Google Scholar]
- 28.Herzberg G. 2nd ed. VonNostrand; New York: 1950. Molecular Spectra and Molecular Structure I: Spectra of Diatomic Molecules. [Google Scholar]
- 29.Foote C.S., Denny R.W., Weaver M.S., Chang Y., Peters J. Quenching of singlet oxygen. Ann. N.Y. Acad. Sci. 1970;171:139–148. [Google Scholar]
- 30.Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U. Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, singlet oxygen sensor green (R) J. Exp. Bot. 2006;57:1725–1734. doi: 10.1093/jxb/erj181. [DOI] [PubMed] [Google Scholar]
- 31.Hallett F.R., Hallett B.P., Snipes W. Reactions between singlet oxygen and the constituents of nucleic acids. Importance of reactions in photodynamic processes. Biophys. J. 1970;10:305–315. doi: 10.1016/S0006-3495(70)86303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribeiro D.T., Madzak C., Sarasin A., Di Mascio P., Sies H. Singlet oxygen induced DNA damage and mutagenicity in a single-stranded SV40-based shuttle vector. Photochem. Photobiol. 1992;55:39–45. doi: 10.1111/j.1751-1097.1992.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 33.Ribeiro D.T., De Oliveira R.C., Di Mascio P., Menck C.F. Singlet oxygen induces predominantly G to T transversions on a single-stranded shuttle vector replicated in monkey cells. Free Radic. Res. 1994;21:75–83. doi: 10.3109/10715769409056559. [DOI] [PubMed] [Google Scholar]
- 34.Sies H., Menck C.F. Singlet oxygen induced DNA damage. Mutat. Res. 1992;275:367–375. doi: 10.1016/0921-8734(92)90039-r. [DOI] [PubMed] [Google Scholar]
- 35.Ravanat J.L., Di Mascio P., Martinez G.R., Medeiros M.H., Cadet J. Singlet oxygen induces oxidation of cellular DNA. J. Biol. Chem. 2000;275:40601–40604. doi: 10.1074/jbc.M006681200. [DOI] [PubMed] [Google Scholar]
- 36.Decuyper-Debergh D., Piette J., Van de Vorst A. Singlet oxygen-induced mutations in M13 lacZ phage DNA. EMBO J. 1987;6:3155–3161. doi: 10.1002/j.1460-2075.1987.tb02626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boegheim J.P., Lagerberg J.W., Dubbleman T.M., Van Steveninck J. Damaging action of photodynamic treatment in combination with hyperthermia on transmembrane transport in murine L929 fibroblasts. Biochim. Biophys. Acta. 1989;979:215–220. doi: 10.1016/0005-2736(89)90437-9. [DOI] [PubMed] [Google Scholar]
- 38.Bird R.B., Stewart W.E., Lightfoot E.N. John Wiley & Sons; New York: 1960. Transport Phenomena. [Google Scholar]
- 39.Merkel P.B., Kearns D.R. An experimental and theoretical study of electronic-to-vibrational energy transfer. Proc. Natl. Acad. Sci. USA. 1972;94:7244–7253. [Google Scholar]
- 40.Wittmershaus B., Baseler T., Beaumont G., Zhang Y. Excitation energy transfer from polystyrene to dye in 40-nm diameter microspheres. J. Lumin. 2002;96:107–118. [Google Scholar]
- 41.Abbondanzieri E.A., Greenleaf W.J., Shaevitz J.W., Landick R., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter A.R., Seol Y., Perkins T.T. Precision surface-coupled optical-trapping assay with one-basepair resolution. Biophys. J. 2009;96:2926–2934. doi: 10.1016/j.bpj.2008.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003;305:761–770. doi: 10.1016/s0006-291x(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 44.Prinsze C., Dubbelman T.M., Van Steveninck J. Protein damage, induced by small amounts of photodynamically generated singlet oxygen or hydroxyl radicals. Biochim. Biophys. Acta. 1990;1038:152–157. doi: 10.1016/0167-4838(90)90198-o. [DOI] [PubMed] [Google Scholar]
- 45.Davies M.J. Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 2004;3:17–25. doi: 10.1039/b307576c. [DOI] [PubMed] [Google Scholar]
- 46.Shubeita G.T., Tran S.L., Xu J., Vershinin M., Cermelli S. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chattopadhyay S., Moldovan R., Yeung C., Wu X.L. Swimming efficiency of bacterium Escherichia coli. Proc. Natl. Acad. Sci. USA. 2006;103:13712–13717. doi: 10.1073/pnas.0602043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krasnovsky A.A., Drozdova N.N., Ivanov A.V., Ambartsumian R.V. Activation of molecular oxygen by infrared laser radiation in pigment-free aerobic systems. Biochemistry (Mosc.) 2003;68:963–966. doi: 10.1023/a:1026052310563. [DOI] [PubMed] [Google Scholar]
- 49.Frederiksen P.K., Jorgensen M., Ogilby P.R. Two-photon photosensitized production of singlet oxygen. J. Am. Chem. Soc. 2001;123:1215–1221. doi: 10.1021/ja003468a. [DOI] [PubMed] [Google Scholar]
- 50.Lewis J.R., Sauro J. When 100% really isn't 100%: improving the accuracy of small-sample estimates of completion rates. JUS. 2006;1:136–150. [Google Scholar]
- 51.Agresti A., Coull B. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 1998;52:119–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


