Abstract
Background
Genetic markers for thyroid cancers identified by microarray analysis have offered limited predictive accuracy so far because of the few classes of thyroid lesions usually taken into account. To improve diagnostic relevance, we have simultaneously analyzed microarray data from six public datasets covering a total of 347 thyroid tissue samples representing 12 histological classes of follicular lesions and normal thyroid tissue. Our own dataset, containing about half the thyroid tissue samples, included all categories of thyroid lesions.
Methodology/Principal Findings
Classifier predictions were strongly affected by similarities between classes and by the number of classes in the training sets. In each dataset, sample prediction was improved by separating the samples into three groups according to class similarities. The cross-validation of differential genes revealed four clusters with functional enrichments. The analysis of six of these genes (APOD, APOE, CLGN, CRABP1, SDHA and TIMP1) in 49 new samples showed consistent gene and protein profiles with the class similarities observed. Focusing on four subclasses of follicular tumor, we explored the diagnostic potential of 12 selected markers (CASP10, CDH16, CLGN, CRABP1, HMGB2, ALPL2, ADAMTS2, CABIN1, ALDH1A3, USP13, NR2F2, KRTHB5) by real-time quantitative RT-PCR on 32 other new samples. The gene expression profiles of follicular tumors were examined with reference to the mutational status of the Pax8-PPARγ, TSHR, GNAS and NRAS genes.
Conclusion/Significance
We show that diagnostic tools defined on the basis of microarray data are more relevant when a large number of samples and tissue classes are used. Taking into account the relationships between the thyroid tumor pathologies, together with the main biological functions and pathways involved, improved the diagnostic accuracy of the samples. Our approach was particularly relevant for the classification of microfollicular adenomas.
Introduction
Over the past few years, the use of microarray technologies has contributed to the identification of new markers for the diagnosis and prognosis of human tumors. Cancer research usually involves the study of a single class of tumor and the corresponding normal tissue. Increasing the number of binary studies does not necessarily improve the relevance of the molecular signature. Thus, a meta-analysis comparing 40 types of cancer in various tissues relative to their normal counterpart allowed the identification of a common signature essential to carcinogenesis but may fail to distinguish between different classes of tumor affecting a given organ, which may therefore have specific prognoses [1]. Moreover, the same signature may also appear in a variety of other cellular contexts such as inflammatory processes.
Thyroid nodules are extremely common in the adult population, but less than 20% of the nodules are malignant [2]. Papillary thyroid carcinoma (PTC), diagnosed on the basis of characteristic nuclear features, is the most frequent malignant thyroid tumor. According to the 2004 WHO report, the diagnosis of minimal invasive follicular thyroid carcinoma (FTC) is problematic because of its morphological and molecular similarities to benign follicular thyroid adenoma (FTA) [3]. Moreover, atypical or oncocytic features render the differential diagnosis of follicular tumors difficult on histologic examination and call for new molecular or biological markers [4], [5]. To date, microarray analyses of thyroid tumors have essentially compared two classes of tissue [6]–[11]. These studies have either searched for specific markers by comparing a particular class of thyroid tumor to the corresponding normal tissue, or looked for markers of malignancy by examining the most frequent benign and malignant classes of thyroid tumor (usually the FTA and PTC classes). The predictive accuracy of the markers identified is therefore rather limited with regard to tumors belonging to other classes. For example, the CITED1 gene, which was claimed to be a significant marker distinguishing PTC from normal tissue [9] turned out to be less specific when data from FTC samples were included [8]. Furthermore, this gene did not even figure among the 42 best PTC marker genes detected by a meta-analysis that included benign tumors [12]. Meta-analyses may increase not only the number of classes required to define more relevant markers but also increase the imbalance in the representation of some classes. In a recent analysis, cross-validated marker genes from 21 studies differentiated benign from malignant thyroid tissues [13]. However, the majority of these were pertinent to the diagnosis of PTC since these thyroid cancers accounted for more than 40% of the samples studied. This highlights the bias due to the recruitment of thyroid samples for microarray studies and the consequent failure in identifying classifiers for clinical applications despite the large number of analyses exploited.
Few studies have simultaneously compared more than four types of thyroid tissue [5], [14]–[17]. In one of these [15], we were able to refine the diagnosis of tumors of uncertain malignancy by the simultaneous analysis of eight types of thyroid tissue. These promising results encouraged further research on relevant markers of thyroid tumors, taking into account more classes and subclasses of interest, as some authors have recommended [18]. In the present study, we have explored gene-expression signatures from the majority of the differentiated follicular pathologies of thyroid tissue. We have used published data or data available from the Gene Expression Omnibus (GEO) of the U. S. National Center for Biotechnology Information, as well as specifically generated data, to simultaneously analyze several datasets containing information on various pathologies, including datasets with many histological subclasses. Our analysis, covering the major subtypes of thyroid tissue, may be expected to enhance microarray data concerning thyroid tumors and lead to the definition of more reliable tissue-specific markers for thyroid lesions.
Results
We have simultaneously analyzed 347 thyroid tissue samples from six datasets (Table 1). The Fontaine dataset, generated by our own laboratory [15], comprised 166 thyroid tissue samples, i.e. about half the total number of samples.
Table 1. Datasets, classes and samples.
| Datasets | Fontaine et al. 2008 | Giordano et al.2006 | Weber et al.2005 | Jarzab et al. 2005 | He et al. 2005 | Reyes et al. 2006 | |
| Microarray platform | Single channel cDNA microarrays | Affymetrix GeneChip HG U133A | Affymetrix GeneChip HG U133A | Affymetrix GeneChip HG U133A | Affymetrix GeneChip HG U133 Plus 2.0 | Affymetrix GeneChip HG U133 Plus 2.0 | |
| (Probes) | (9,216) | (22,283) | (22,283) | (22,283) | (54,681) | (54,681) | |
| Tissue samples | Symbol | 166 | 93 | 24 | 32 | 18 | 14 |
| Wild Type tissue | WT | 24 | 4 | - | 16 | 9 | 7 |
| Autoimmune Thyroiditis | AT | 5 | - | - | - | - | - |
| Grave's Disease | GD | 5 | - | - | - | - | - |
| Follicular Thyroid Adenoma | FTA | 26 | 10 | 12 | - | - | - |
| Microfollicular Thyroid Adenoma | FTAb | 17 | - | - | - | - | - |
| Multi Nodular Goitre | MNG | 24 | - | - | - | - | - |
| Follicular Thyroid Carcinoma | FTC | 3 | - | 6 | - | - | - |
| FTC with PAX8/PPARγ fusion | FTC+ | - | 7 | - | - | - | - |
| FTC without PAX8/PPARγ fusion | FTC- | - | 6 | - | - | - | - |
| Oncocytic Thyroid Adenoma | OTA | 30 | 7 | - | - | - | - |
| Oncocytic Thyroid Carcinoma | OTC | 4 | 8 | 6 | - | - | - |
| Oncocytic Tumour of Uncertain | OTUM | 5 | - | - | - | - | - |
| Malignancy | |||||||
| Papillary Thyroid Carcinoma | PTC | 13 | 51 | - | 16 | 9 | 7 |
| Tumour of Uncertain Malignancy | TUM | 10 | - | - | - | - | - |
Molecular classification of thyroid pathologies
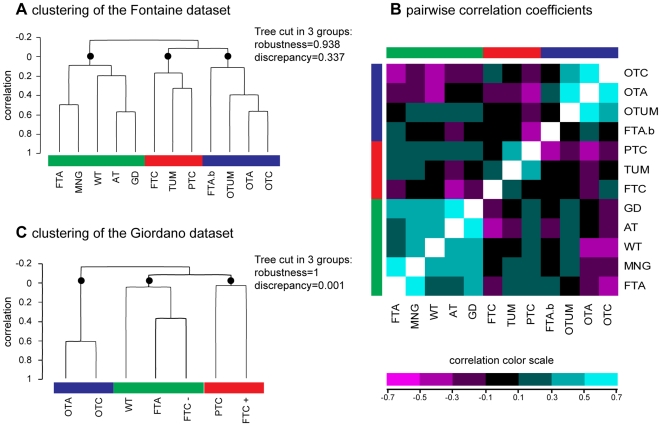
To analyze the global classification of the follicular thyroid lesions in the Fontaine dataset, we defined the centroid of each class of tissue by a composite signature of mean gene-expression levels. The hierarchical clustering of the class centroids (Figure 1A) revealed three groups. The first group comprised benign thyroid lesions and normal tissues (green boxes for FTA, MNG, WT, AT, and GD); the second contained malignant lesions (red boxes for FTC, TUM and PTC); and the third included oncocytic tumors and microfollicular adenomas (blue boxes for FTAb, OTUM, OTA and OTC). These groups, supported by a high robustness index of 0.938 and a discrepancy index of 0.337, are respectively referred to as the ‘benign’, ‘malignant’ and ‘oncocytic’ groups. The three groups, cross-validated in the Giordano dataset (Figure 1C), presented almost perfect robustness ( = 1) and discrepancy indices ( = 0.001). This dataset showed a highly correlated oncocytic group (OTA, OTC, correlation >0.6) and a group of malignant tumors (PTC, FTC+). The third group, predominantly composed of benign tumors and normal tissues (WT, FTA), also included the FTC- group.
Figure 1. Molecular classification of thyroid tissues.
The centroid signatures of the thyroid tissues were compared to each other in the Fontaine dataset generated by our laboratory [15], and compared to the Giordano dataset [5]. Centroids were defined as the mean gene-expression signature of each tissue class over all the genes. Panel A shows the hierarchical clustering of the tissues in the Fontaine dataset. The dendrogram can be cut into three robust branches (black circles) defining three groups of classes. The groups contain benign lesions and normal tissue (green boxes), malignant tumors (red boxes), or oncocytic tumors and microfollicular adenomas (blue boxes). Panel B shows pairwise correlation coefficients between the tissue classes from the Fontaine dataset. The heatmap represents a symmetrical matrix of color-coded correlation coefficients. Tissue classes are ordered as in the hierarchical clustering and marked with the same three colors (green, red and blue). Panel C shows the hierarchical clustering of the tissues in the Giordano dataset. The dendrogram can be cut into three robust branches (black circles) defining three groups of classes. The groups contain heterogeneous lesions and normal tissue (green boxes), malignant tumors (red boxes), or oncocytic tumors (blue boxes). The two datasets show three groups containing similar tissue classes. WT: wild type tissue; PTC: papillary thyroid carcinoma; FTA: macrofollicular thyroid adenoma; FTAb: microfollicular thyroid adenoma; GMN, multinodular goiter; FTC: follicular thyroid carcinoma (FTC+: with Pax8/PPARγ translocation; FTC-: without Pax8/PPARγ translocation); OTA: oncocytic thyroid adenoma; OTC: oncocytic thyroid carcinoma; TUM: tumor of uncertain malignancy; OTUM: oncocytic tumor of uncertain malignancy; GD: Grave's disease; and AT: autoimmune thyroïditis.
We computed the prediction accuracy for the benign, malignant and oncocytic groups of thyroid tissue classes as defined above. The signature of each group was trained versus all the other samples, and its accuracy was compared with the predictive accuracy of individual lesions (Table 2). In both datasets, the positive accuracy for each group was greater than the best accuracy reported for an individual lesion within the group. The high proportion of PTC (79%) in the malignant group from the Giordano dataset may explain the good accuracy for this group despite the rate of discrepancy shown in the Figure 1C. The separation of the samples into three groups on the basis of class similarities led to improved sample prediction in the two datasets.
Table 2. Prediction of samples by considering three groups of tissue classes. Values can be compared to the range of accuracies for individual lesions that composed each group.
| Fontaine dataset | Giordano dataset | |||
| Group | Individual lesions | Group | Individual lesions | |
| Benign group | 0,79 | 0.11–0.71 | 0,76 | 0.31–0.47 |
| Malignant group | 0,69 | 0.24–0.67 | 0,93 | 0.93–0.94 |
| Oncocytic group | 0,75 | 0.02–0.64 | 0,86 | 0.52–0.62 |
The matrix of 66 pairwise correlation coefficients between the classes (Figure 1B) allowed a detailed observation of the similarities and dissimilarities in the Fontaine dataset. All the classes in the benign/normal group were positively correlated with each other. Some pairs of classes were highly correlated (correlation coefficient >0.4), e.g. FTA with MNG, and AT with GD. All the classes in the malignant group were positively correlated with each other. The TUM class had a stronger correlation with PTC than with FTC (respectively 0.354 and 0.274). This similarity between TUM and PTC has previously been demonstrated within this dataset and with independent samples [15]. In the oncocytic group, the classes of oncocytic tumors were positively correlated with each other, except for the FTAb class. Finally, we were able to identify 220 non-redundant genes for the molecular classification of each of the 11 subcategories of follicular thyroid pathologies explored (Table 3).
Table 3. Classifier genes from the Fontaine dataset (n = 220 genes).
| Symbol | Clone ID | Class | Cross validation support (%) | Parametric p-value | t-value | Fold-change |
| ABCB9 | 36371 | AT | 100 | 1,00E-07 | 6,81 | 3,28 |
| SEP-02 | 729542 | AT | 100 | 2,67E-01 | −1,11 | 0,75 |
| SEP-05 | 448163 | AT | 100 | 1,00E-07 | 7,03 | 4,96 |
| AAAS | 740617 | AT | 99 | 3,00E-07 | 5,36 | 3,34 |
| ABCA3 | 52740 | AT | 97 | 4,99E-01 | 0,68 | 1,13 |
| ABCA8 | 284828 | AT | 97 | 1,86E-01 | −1,33 | 0,74 |
| ABCB1 | 1837488 | AT | 97 | 2,26E-01 | −1,22 | 0,72 |
| ABCB8 | 238705 | AT | 97 | 3,51E-01 | −0,94 | 0,84 |
| ABCC3 | 208097 | AT | 96 | 1,54E-01 | 1,43 | 1,64 |
| ABCF2 | 1086914 | AT | 96 | 1,00E-07 | 6,91 | 3,37 |
| BATF | 1929371 | AT | 83 | 1,00E-07 | 10,52 | 6,03 |
| C18orf51 | 742649 | AT | 62 | 1,00E-07 | 9,87 | 7,47 |
| CARD11 | 665651 | AT | 46 | 1,00E-07 | 10,85 | 4,72 |
| CBFA2T2 | 43629 | AT | 39 | 1,00E-07 | 10,18 | 10,03 |
| CD38 | 1352408 | AT | 29 | 1,00E-07 | 10,18 | 7,22 |
| CD79A | 1056782 | AT | 25 | 1,00E-07 | 11,14 | 8,58 |
| CD79B | 155717 | AT | 25 | 1,00E-07 | 11,91 | 10,51 |
| CXCL6 | 2108870 | AT | 14 | 1,00E-07 | 10,13 | 4,29 |
| EPPB9 | 668454 | AT | 7 | 1,00E-07 | 9,67 | 9,03 |
| DKFZp667B0210 | 1844689 | AT | 3 | 1,00E-07 | 9,30 | 4,83 |
| PLXNB2 | 1420676 | FTA | 65 | 6,00E-07 | 5,21 | 1,70 |
| TBC1D17 | 739955 | FTA | 62 | 4,00E-07 | 5,27 | 1,56 |
| ZNF76 | 745003 | FTA | 59 | 8,00E-07 | 5,15 | 1,51 |
| KIAA1196 | 738938 | FTA | 52 | 1,40E-06 | 5,01 | 1,52 |
| ASGR1 | 25883 | FTA | 48 | 1,25E-05 | 4,51 | 1,75 |
| BCL2L11 | 300194 | FTA | 42 | 6,00E-06 | 4,68 | 1,42 |
| DAP | 725371 | FTA | 41 | 6,20E-06 | 4,67 | 1,51 |
| CABIN1 | 1844968 | FTA | 40 | 1,12E-05 | 4,53 | 1,59 |
| FLJ14981 | 24532 | FTA | 31 | 1,84E-04 | 3,83 | 1,50 |
| RAD51L1 | 295412 | FTA | 30 | 4,23E-05 | 4,21 | 1,66 |
| TSPAN1 | 376003 | FTA | 26 | 3,52E-05 | 4,25 | 1,57 |
| IMAGE:745465 | 745465 | FTA | 25 | 2,58E-02 | 2,25 | 1,24 |
| PLCG1 | 1174287 | FTA | 25 | 8,36E-05 | −4,04 | 0,67 |
| COL18A1 | 359202 | FTA | 24 | 3,20E-05 | 4,28 | 1,60 |
| RPS6KA2 | 22711 | FTA | 23 | 3,77E-05 | 4,24 | 1,57 |
| CaMKIINalpha | 173820 | FTA | 22 | 9,52E-04 | 3,37 | 1,35 |
| KLF1 | 208991 | FTA | 16 | 3,01E-03 | 3,01 | 1,49 |
| BAT3 | 24392 | FTA | 13 | 8,11E-04 | 3,41 | 1,42 |
| PDK4 | 487379 | FTA | 11 | 1,00E-02 | −2,61 | 0,70 |
| IMAGE:24065 | 24065 | FTA | 6 | 4,52E-04 | −3,58 | 0,75 |
| KRTHB5 | 364569 | FTAb | 86 | 1,00E-07 | 6,02 | 1,94 |
| METRN | 487475 | FTAb | 86 | 1,00E-07 | 5,68 | 2,15 |
| DCI | 667892 | FTAb | 74 | 6,00E-07 | 5,19 | 1,63 |
| IFI30 | 740931 | FTAb | 68 | 4,90E-06 | −4,73 | 0,41 |
| ALCAM | 172828 | FTAb | 64 | 1,21E-05 | −4,51 | 0,68 |
| ADAMTS2 | 364844 | FTAb | 55 | 1,61E-04 | 3,86 | 1,41 |
| ETF1 | 146976 | FTAb | 54 | 1,12E-04 | 3,96 | 1,54 |
| TP53I11 | 667514 | FTAb | 44 | 1,22E-04 | 3,94 | 1,94 |
| IMAGE:487086 | 487086 | FTAb | 42 | 1,52E-04 | −3,88 | 0,67 |
| IMAGE:666315 | 666315 | FTAb | 35 | 5,63E-04 | 3,52 | 1,52 |
| GRB10 | 564994 | FTAb | 33 | 1,80E-04 | 3,83 | 1,42 |
| ISG20 | 740604 | FTAb | 33 | 3,51E-05 | −4,26 | 0,60 |
| SPARC | 267358 | FTAb | 33 | 1,09E-04 | 3,97 | 1,47 |
| HAS3 | 667533 | FTAb | 28 | 1,07E-04 | 3,97 | 1,52 |
| USP13 | 666007 | FTAb | 20 | 9,96E-05 | 3,99 | 1,56 |
| KRT4 | 1173570 | FTAb | 13 | 2,87E-04 | 3,71 | 1,65 |
| LHFP | 591534 | FTAb | 9 | 2,24E-03 | −3,11 | 0,74 |
| PCDHB15 | 730593 | FTAb | 9 | 1,06E-03 | −3,33 | 0,74 |
| TPM2 | 740620 | FTAb | 9 | 9,35E-03 | 2,63 | 1,45 |
| DKFZp434M202 | 743118 | FTAb | 2 | 5,77E-03 | −2,80 | 0,76 |
| C22orf9 | 667250 | FTC | 89 | 2,00E-07 | −5,43 | 0,34 |
| SUCLG2 | 687551 | FTC | 83 | 7,00E-07 | −5,18 | 0,27 |
| LAPTM4A | 726684 | FTC | 77 | 4,00E-07 | −5,26 | 0,24 |
| KIAA1576 | 743426 | FTC | 76 | 1,00E-07 | 5,70 | 3,33 |
| NR2F2 | 72744 | FTC | 65 | 2,60E-06 | 4,87 | 3,11 |
| CHST3 | 665327 | FTC | 62 | 2,90E-06 | −4,85 | 0,32 |
| PAI-RBP1 | 236055 | FTC | 55 | 5,30E-06 | −4,71 | 0,29 |
| IMAGE:729896 | 729896 | FTC | 54 | 3,00E-07 | 5,34 | 3,22 |
| ZBTB16 | 2467442 | FTC | 52 | 4,72E-05 | 4,18 | 4,68 |
| ZNF364 | 471834 | FTC | 52 | 5,80E-06 | −4,69 | 0,39 |
| GHITM | 25077 | FTC | 50 | 5,30E-06 | −4,71 | 0,31 |
| LRP8 | 415554 | FTC | 49 | 1,43E-04 | 3,90 | 2,74 |
| CEECAM1 | 666784 | FTC | 45 | 2,67E-04 | 3,73 | 2,22 |
| HIST1H4C | 682451 | FTC | 43 | 1,57E-05 | −4,45 | 0,25 |
| TIE1 | 743043 | FTC | 35 | 5,46E-05 | 4,14 | 2,52 |
| SH3RF2 | 744797 | FTC | 22 | 5,37E-03 | 2,82 | 2,18 |
| MGC5395 | 238840 | FTC | 15 | 5,07E-04 | 3,55 | 2,66 |
| EMX2 | 365121 | FTC | 14 | 6,95E-03 | −2,73 | 0,51 |
| LRRC28 | 26519 | FTC | 13 | 1,51E-03 | −3,23 | 0,40 |
| GFRA3 | 2716748 | FTC | 11 | 4,52E-03 | 2,88 | 1,93 |
| TTC19 | 136366 | GD | 72 | 1,57E-04 | 3,87 | 1,87 |
| ATP6V1G2 | 726424 | GD | 61 | 1,91E-04 | 3,82 | 2,22 |
| IMAGE:23817 | 23817 | GD | 61 | 3,65E-04 | −3,64 | 0,52 |
| IMAGE:178161 | 178161 | GD | 52 | 6,99E-04 | 3,46 | 2,53 |
| BDKRB2 | 665674 | GD | 50 | 6,16E-03 | 2,78 | 2,15 |
| IMAGE:744385 | 744385 | GD | 48 | 1,37E-02 | 2,49 | 1,70 |
| LOC150837 | 258698 | GD | 47 | 5,84E-04 | 3,51 | 1,81 |
| SSX1 | 262894 | GD | 47 | 5,97E-04 | 3,50 | 1,76 |
| DHX57 | 24623 | GD | 46 | 1,69E-03 | −3,19 | 0,59 |
| RAI14 | 731711 | GD | 46 | 8,19E-03 | 2,68 | 1,56 |
| IMAGE:180851 | 180851 | GD | 45 | 5,36E-03 | −2,82 | 0,61 |
| MGC5566 | 53119 | GD | 43 | 1,74E-03 | 3,18 | 1,85 |
| D21S2056E | 740140 | GD | 36 | 2,44E-03 | 3,08 | 1,72 |
| IMAGE:729489 | 729489 | GD | 31 | 1,28E-03 | 3,28 | 2,95 |
| KIAA1737 | 122063 | GD | 29 | 1,78E-03 | 3,18 | 1,67 |
| MLH3 | 250771 | GD | 21 | 8,59E-03 | 2,66 | 1,59 |
| IMAGE:53081 | 53081 | GD | 18 | 3,35E-03 | 2,98 | 1,77 |
| TERF1 | 135773 | GD | 16 | 2,81E-02 | 2,22 | 1,45 |
| DCXR | 724596 | GD | 15 | 3,39E-03 | −2,97 | 0,61 |
| IMAGE:136686 | 136686 | GD | 15 | 6,29E-02 | 1,87 | 1,48 |
| FLJ11127 | 668625 | MNG | 88 | 2,30E-06 | 4,90 | 1,54 |
| RODH | 471641 | MNG | 83 | 1,51E-05 | 4,46 | 2,57 |
| MGC17299 | 713422 | MNG | 59 | 6,54E-05 | 4,10 | 1,37 |
| NR2F2 | 72744 | MNG | 57 | 3,10E-04 | 3,69 | 1,40 |
| ZNF83 | 486356 | MNG | 50 | 3,72E-04 | 3,64 | 1,32 |
| IMAGE:744505 | 744505 | MNG | 43 | 2,51E-04 | 3,74 | 1,38 |
| IMAGE:745187 | 745187 | MNG | 42 | 1,15E-03 | −3,31 | 0,78 |
| CAV1 | 309645 | MNG | 41 | 3,38E-04 | 3,66 | 1,59 |
| CNTN6 | 257773 | MNG | 41 | 2,34E-04 | 3,76 | 1,38 |
| C10orf116 | 740941 | MNG | 40 | 2,82E-04 | 3,71 | 1,72 |
| CALM1 | 594510 | MNG | 39 | 1,21E-03 | −3,30 | 0,77 |
| IMAGE:726782 | 726782 | MNG | 39 | 3,97E-04 | 3,62 | 1,38 |
| IMAGE:250463 | 250463 | MNG | 28 | 6,76E-03 | 2,74 | 1,29 |
| IMAGE:731726 | 731726 | MNG | 28 | 3,90E-04 | 3,62 | 1,52 |
| TTC17 | 665668 | MNG | 28 | 5,24E-04 | 3,54 | 1,31 |
| PRKCA | 768246 | MNG | 25 | 9,12E-04 | −3,38 | 0,71 |
| NR4A1 | 1073288 | MNG | 19 | 1,88E-03 | −3,16 | 0,64 |
| PDLIM1 | 135689 | MNG | 14 | 9,76E-04 | 3,36 | 1,48 |
| IMAGE:731292 | 731292 | MNG | 11 | 2,65E-02 | −2,24 | 0,68 |
| SLC17A2 | 207920 | MNG | 9 | 2,70E-02 | −2,23 | 0,67 |
| MLL | 80688 | OTA | 76 | 1,00E-07 | −8,18 | 0,55 |
| ALDH1A3 | 486189 | OTA | 67 | 1,00E-07 | 6,38 | 1,79 |
| CLECSF12 | 258865 | OTA | 63 | 1,00E-07 | 7,66 | 3,51 |
| IMAGE:485104 | 485104 | OTA | 60 | 1,00E-07 | 7,32 | 2,25 |
| DCN | 666410 | OTA | 49 | 1,00E-07 | −7,19 | 0,50 |
| PROZ | 430471 | OTA | 47 | 1,00E-07 | 7,05 | 2,76 |
| CHRDL2 | 485872 | OTA | 46 | 1,00E-07 | 6,72 | 1,58 |
| B3GNTL1 | 53341 | OTA | 39 | 9,00E-07 | 5,10 | 2,04 |
| SNRP70 | 729971 | OTA | 39 | 1,00E-07 | −7,00 | 0,52 |
| HSPC159 | 365045 | OTA | 38 | 1,00E-07 | 6,86 | 1,70 |
| C4A | 724366 | OTA | 32 | 1,00E-07 | −6,30 | 0,39 |
| SLC6A8 | 725877 | OTA | 31 | 1,00E-07 | −7,00 | 0,23 |
| OBSCN | 730926 | OTA | 29 | 1,00E-07 | 6,95 | 1,55 |
| IMAGE:731616 | 731616 | OTA | 27 | 1,00E-07 | 6,88 | 2,42 |
| DC12 | 724895 | OTA | 24 | 1,00E-07 | 6,67 | 1,71 |
| UBA52 | 530069 | OTA | 16 | 4,40E-06 | −4,75 | 0,29 |
| KCNQ2 | 179534 | OTA | 14 | 1,00E-07 | 6,43 | 1,77 |
| HCNGP | 365934 | OTA | 11 | 1,92E-05 | 4,40 | 1,40 |
| C9orf72 | 726849 | OTA | 10 | 4,40E-06 | −4,75 | 0,69 |
| GNAI1 | 753215 | OTA | 6 | 8,90E-06 | 4,59 | 1,48 |
| SDHA | 40304 | OTC | 95 | 1,00E-07 | 6,29 | 3,16 |
| DNALI1 | 782688 | OTC | 72 | 1,00E-07 | −5,53 | 0,24 |
| IMAGE:382423 | 382423 | OTC | 50 | 1,50E-06 | 5,00 | 3,52 |
| ATP11C | 667991 | OTC | 47 | 2,30E-05 | 4,36 | 2,17 |
| C1QBP | 173371 | OTC | 46 | 2,70E-06 | 4,86 | 2,47 |
| DCN | 666410 | OTC | 43 | 6,80E-06 | −4,65 | 0,29 |
| MT1F | 78353 | OTC | 41 | 3,70E-06 | −4,79 | 0,16 |
| KIAA0543 | 175080 | OTC | 38 | 9,15E-05 | 4,01 | 2,03 |
| FBXO46 | 471664 | OTC | 37 | 1,67E-05 | 4,44 | 2,85 |
| CD33 | 1917430 | OTC | 35 | 1,06E-05 | 4,55 | 3,71 |
| TH | 813654 | OTC | 32 | 8,32E-04 | 3,41 | 1,95 |
| IMAGE:742061 | 742061 | OTC | 31 | 1,99E-05 | 4,39 | 2,73 |
| IDH3B | 723755 | OTC | 27 | 1,90E-05 | 4,41 | 2,27 |
| IMAGE:359454 | 359454 | OTC | 23 | 1,31E-05 | −4,50 | 0,27 |
| SST | 39593 | OTC | 23 | 1,34E-03 | 3,26 | 1,81 |
| DHRS6 | 364412 | OTC | 20 | 4,03E-04 | −3,61 | 0,48 |
| IMAGE:136933 | 136933 | OTC | 19 | 6,25E-03 | 2,77 | 2,19 |
| TPM2 | 740620 | OTC | 18 | 1,59E-05 | −4,45 | 0,30 |
| TM7SF3 | 666928 | OTC | 14 | 3,58E-04 | 3,65 | 2,00 |
| PREB | 740347 | OTC | 5 | 1,29E-02 | −2,51 | 0,33 |
| LRRIQ2 | 487152 | OTUM | 59 | 1,29E-04 | 3,92 | 2,01 |
| LRRC28 | 26519 | OTUM | 53 | 2,04E-04 | −3,80 | 0,43 |
| NDUFC1 | 796513 | OTUM | 53 | 2,65E-03 | −3,05 | 0,61 |
| NUMBL | 1855110 | OTUM | 53 | 2,94E-04 | −3,70 | 0,56 |
| IMAGE:667527 | 667527 | OTUM | 48 | 2,31E-04 | 3,77 | 2,45 |
| MAK | 382002 | OTUM | 43 | 3,71E-04 | 3,64 | 2,14 |
| PCGF4 | 740457 | OTUM | 43 | 3,06E-04 | 3,69 | 2,61 |
| IFITM3 | 713623 | OTUM | 42 | 1,74E-02 | 2,40 | 1,63 |
| CLGN | 1049033 | OTUM | 40 | 1,52E-03 | 3,23 | 1,86 |
| DEFB1 | 665086 | OTUM | 40 | 9,17E-04 | 3,38 | 2,79 |
| IMAGE:669098 | 669098 | OTUM | 35 | 9,47E-04 | 3,37 | 1,85 |
| GNAS | 382791 | OTUM | 31 | 6,58E-04 | 3,47 | 2,75 |
| PTTG1 | 742935 | OTUM | 29 | 7,53E-03 | −2,71 | 0,52 |
| USP14 | 376462 | OTUM | 28 | 9,21E-03 | −2,64 | 0,65 |
| SNF7DC2 | 730401 | OTUM | 25 | 1,36E-02 | 2,49 | 1,51 |
| C21orf84 | 730814 | OTUM | 19 | 1,24E-03 | 3,29 | 2,08 |
| TCTE3 | 136862 | OTUM | 19 | 2,48E-02 | −2,27 | 0,69 |
| TH | 813654 | OTUM | 19 | 3,81E-02 | −2,09 | 0,69 |
| BAK1 | 235938 | OTUM | 16 | 1,12E-03 | 3,32 | 1,80 |
| PAX6 | 230882 | OTUM | 12 | 1,28E-02 | −2,52 | 0,66 |
| CDH3 | 359051 | PTC | 99 | 1,00E-07 | 9,16 | 3,95 |
| DPP4 | 343987 | PTC | 98 | 1,00E-07 | 8,72 | 4,22 |
| CLDN1 | 594279 | PTC | 86 | 1,00E-07 | 7,88 | 8,98 |
| SEP-02 | 729542 | PTC | 83 | 2,60E-01 | 1,13 | 1,20 |
| ABCC3 | 208097 | PTC | 74 | 1,00E-07 | 6,30 | 3,48 |
| ECM1 | 301122 | PTC | 67 | 1,00E-07 | 8,24 | 6,00 |
| TSC | 745490 | PTC | 60 | 1,00E-07 | 7,70 | 4,65 |
| QPCT | 711918 | PTC | 57 | 1,00E-07 | 7,27 | 3,36 |
| ADAMTS9 | 376153 | PTC | 48 | 6,40E-06 | 4,66 | 1,95 |
| IMAGE:687667 | 687667 | PTC | 48 | 1,00E-07 | 7,10 | 3,74 |
| SLPI | 378813 | PTC | 46 | 1,00E-07 | 7,03 | 2,98 |
| MATN1 | 1624260 | PTC | 43 | 1,00E-07 | −6,74 | 0,42 |
| CITED1 | 265558 | PTC | 40 | 1,00E-07 | 6,84 | 4,88 |
| CCND1 | 324079 | PTC | 38 | 1,00E-07 | 6,39 | 2,55 |
| IMAGE:136976 | 136976 | PTC | 23 | 1,00E-07 | 6,15 | 2,11 |
| PET112L | 743125 | PTC | 20 | 1,00E-07 | 6,27 | 1,96 |
| DAF | 627107 | PTC | 16 | 1,00E-07 | 5,68 | 1,89 |
| GSS | 140405 | PTC | 7 | 1,00E-07 | 5,73 | 2,05 |
| CASP3 | 823680 | PTC | 5 | 6,00E-07 | 5,20 | 2,08 |
| CD9 | 727251 | PTC | 1 | 1,45E-05 | 4,47 | 1,57 |
| IMAGE:687667 | 687667 | TUM | 94 | 1,00E-07 | 6,33 | 3,90 |
| CREB3L2 | 136399 | TUM | 63 | 4,90E-06 | −4,73 | 0,48 |
| EIF1AY | 380394 | TUM | 55 | 4,02E-05 | −4,22 | 0,57 |
| DJ462O23.2 | 738970 | TUM | 51 | 1,64E-05 | 4,44 | 1,63 |
| CITED1 | 265558 | TUM | 49 | 1,21E-05 | 4,51 | 3,53 |
| KLF1 | 208991 | TUM | 46 | 3,72E-05 | −4,24 | 0,43 |
| SNCB | 50202 | TUM | 45 | 4,68E-05 | 4,18 | 1,99 |
| FLJ10748 | 726830 | TUM | 44 | 4,24E-05 | 4,21 | 1,71 |
| ANK2 | 2139315 | TUM | 43 | 1,31E-03 | 3,27 | 1,68 |
| IGFBP3 | 269873 | TUM | 39 | 9,68E-05 | 4,00 | 2,06 |
| WDR1 | 714196 | TUM | 37 | 2,06E-04 | −3,80 | 0,65 |
| IFNAR2 | 123950 | TUM | 36 | 2,56E-04 | −3,74 | 0,61 |
| SCNN1A | 741305 | TUM | 36 | 2,37E-05 | −4,35 | 0,41 |
| TSC | 745490 | TUM | 27 | 1,95E-04 | 3,81 | 2,61 |
| NBL1 | 503874 | TUM | 24 | 2,47E-04 | −3,75 | 0,35 |
| ETF1 | 146976 | TUM | 18 | 6,36E-04 | 3,48 | 1,63 |
| ICOSLG | 2074228 | TUM | 18 | 1,12E-03 | 3,32 | 1,55 |
| NCB5OR | 743367 | TUM | 15 | 2,03E-03 | −3,14 | 0,56 |
| ZNF415 | 23939 | TUM | 14 | 4,94E-03 | −2,85 | 0,63 |
| KLK2 | 1102600 | TUM | 9 | 1,39E-03 | −3,25 | 0,66 |
AT: autoimmune thyroiditis; FTA: macrofollicular thyroid adenoma; FTAb: microfollicular thyroid adenoma; FTC: follicular thyroid carcinoma; GD, Graves' disease; MNG: multinodular goiter; OTA: oncocytic thyroid adenoma; OTC: oncocytic thyroid carcinoma; PTC: papillary thyroid carcinoma; TUM: tumor of uncertain malignancy; and OTUM:, oncocytic tumor of uncertain malignancy.
Class prediction from binary and from complete training
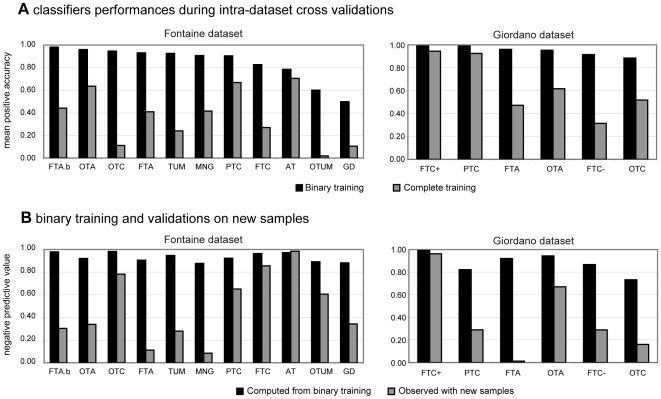
We compared classifiers defined from many classes, i.e. the entire dataset (complete training), and two classes, i.e. one pathological class versus normal thyroid tissue (binary training). The predictive accuracy for the samples of a class, i.e. the positive accuracy, was always higher after binary training than after complete training in the two main datasets (Figure 2A). With regard to complete training, only two classes in each dataset had similar performances: PTC and AT in the Fontaine dataset, FTC+ and PTC in the Giordano dataset.
Figure 2. Class prediction of the samples.
Panel A shows the mean positive accuracy of classifiers during intra-dataset cross-validations for the two main datasets, i.e. the Fontaine dataset and the Giordano dataset. Each classifier is composed of 20 genes selected by the Greedy Pairs algorithm and using Diagonal Linear Discriminant Analysis for training and prediction. Some classifiers were trained with samples from the selected class and the normal tissue, i.e. with binary training (black bars). Other classifiers were trained with all the samples of the dataset, i.e. with complete training (gray bars). Panel B shows the class prediction of new samples by binary-trained classifiers in the two datasets. All the new samples come from the same dataset and should have been predicted as negative. The negative predictive value which was deduced from binary training (black bars) is compared to observations on the new samples (gray bars). WT: wild type tissue; PTC: papillary thyroid carcinoma; FTA: macrofollicular thyroid adenoma; FTAb: microfollicular thyroid adenoma; MNG, multinodular goiter; FTC: follicular thyroid carcinoma (FTC+: with Pax8/PPARγ translocation; FTC-: without Pax8/PPARγ translocation); OTA: oncocytic thyroid adenoma; OTC: oncocytic thyroid carcinoma; TUM: tumor of uncertain malignancy; OTUM: oncocytic tumor of uncertain malignancy; GD, Graves' disease; and AT, autoimmune thyroïditis.
The classifiers defined by complete training in the Fontaine dataset were evaluated in five other thyroid datasets (Table 4). With respect to the positive accuracy computed from intra-dataset cross-validations, the PTC, FTC, FTA and OTC classifiers turned out to be more accurate than expected. The OTA classifier was less accurate than expected, with a value of 0.59 versus 0.64, but only one other dataset contained this class of thyroid tissue.
Table 4. Inter-dataset cross-validations. Positive accuracies of the classifiers defined by the Fontaine dataset in five other datasets.
| DATASET | PTC | FTC | FTA | OTC | OTA |
| Giordano et al. 2006 | 0.93 | 0.40 | 0.48 | 0.64 | 0.59 |
| Weber et al. 2005 | - | 0.77 | 0.96 | 0.72 | - |
| Jarzab et al. 2005 | 1.00 | - | - | - | - |
| Reyes et al. 2006 | 1.00 | - | - | - | - |
| He et al. 2005 | 1.00 | - | - | - | - |
FTA: macrofollicular thyroid adenoma; FTC: follicular thyroid carcinoma; OTA: oncocytic thyroid adenoma; OTC: oncocytic thyroid carcinoma; and PTC: papillary thyroid carcinoma.
Functional analysis of cross-validated differential genes
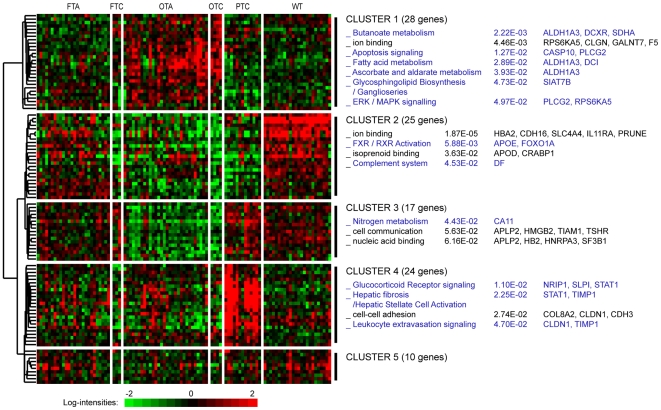
In the two largest datasets containing the main kinds of thyroid lesions [5], [15], we searched for cross-validated differential genes. The two lists of differential genes had an intersection of 104 genes that separated five gene clusters from the hierarchical clustering of classes shared by the two datasets (Figure 3 and Table 5). The functions and pathways identified were mainly related to oncocytic and papillary tumors (Clusters 1, 3 and 4). Gene ontologies were associated with oxido-reductive mechanisms (Cluster 1, 28 genes), nitrogen metabolism and cell communication (Cluster 3, 17 genes), glucocorticoid receptor signaling and leukocyte extravasation (Cluster 4, 24 genes). A cluster of genes showed a specific profile for tumors of malignant potential, for which the transcriptional factors involved in the primary anabolic and catabolic cell pathways were down-regulated (Cluster 2, 25 genes). However in Cluster 2, the heterogeneous profiles of some classes (FTA) were suspected of containing molecular subgroups whereas the profiles for WT were almost homogeneous. A cluster of 10 genes (Cluster 5) was not clearly specific to the tissue classes as their profiles differed between the two datasets. In the Fontaine dataset, 52% of the cross-validated genes belonged to at least two of the 11 classes of lesion (data not shown).
Figure 3. Hierarchical clustering of cross-validated differential genes.
The heatmap shows gene-expression levels in the Fontaine dataset for the tissue classes common to the Giordano dataset (104 genes). Functional enrichments are shown for five identified gene clusters on the right, followed by p-values and the genes involved. The black text represents Level 3 Gene Ontology terms; the blue text represents canonical Ingenuity pathways. Gene profiles of the differential genes reflect the class similarities observed. WT: wild type tissue; PTC: papillary thyroid carcinoma; FTA: macrofollicular thyroid adenoma; FTC: follicular thyroid carcinoma; OTA: oncocytic thyroid adenoma; and OTC: oncocytic thyroid carcinoma.
Table 5. Cross validated differential genes from the two main datasets (n = 104).
| Cluster | Symbol | UGCluster | Clone ID | P-value | P-value |
| (Fontaine et al. 2008) | (Giordano et al. 2006) | ||||
| 1 | CASP10 | Hs.5353 | 241481 | 1,00E-07 | 1,00E-07 |
| 1 | CLGN | Hs.86368 | 1049033 | 1,00E-07 | 1,00E-07 |
| 1 | DEFB1 | Hs.32949 | 665086 | 1,00E-07 | 1,00E-07 |
| 1 | NTRK2 | Hs.494312 | 2048801 | 1,00E-07 | 1,00E-07 |
| 1 | SDHA | Hs.440475 | 40304 | 1,00E-07 | 1,00E-07 |
| 1 | HSPC159 | Hs.372208 | 365045 | 1,00E-07 | 3,00E-07 |
| 1 | ALDH1A3 | Hs.459538 | 486189 | 1,00E-07 | 8,00E-07 |
| 1 | PLCG2 | Hs.413111 | 201467 | 1,00E-07 | 9,00E-07 |
| 1 | ARVCF | Hs.370408 | 40633 | 1,00E-07 | 1,50E-06 |
| 1 | ELK4 | Hs.497520 | 236155 | 1,00E-07 | 1,50E-06 |
| 1 | DCI | Hs.403436 | 667892 | 1,00E-07 | 1,70E-06 |
| 1 | DIO1 | Hs.251415 | 296702 | 1,00E-07 | 3,50E-06 |
| 1 | CKS2 | Hs.83758 | 725454 | 2,00E-07 | 9,00E-07 |
| 1 | ID1 | Hs.504609 | 1087348 | 2,00E-07 | 3,90E-06 |
| 1 | MRPS11 | Hs.111286 | 471574 | 4,00E-07 | 1,00E-07 |
| 1 | ST6GALNAC2 | Hs.592105 | 823590 | 6,00E-07 | 8,00E-07 |
| 1 | GFPT2 | Hs.696497 | 485085 | 9,00E-07 | 1,00E-07 |
| 1 | PITPNC1 | Hs.591185 | 364436 | 9,00E-07 | 2,00E-07 |
| 1 | HRK | Hs.87247 | 767779 | 1,00E-06 | 1,00E-07 |
| 1 | MAPRE2 | Hs.532824 | 383868 | 1,20E-06 | 1,00E-07 |
| 1 | DOK5 | Hs.656582 | 25664 | 1,30E-06 | 1,00E-07 |
| 1 | RPS6KA5 | Hs.510225 | 258966 | 1,40E-06 | 1,00E-07 |
| 1 | DCXR | Hs.9857 | 724596 | 3,00E-06 | 1,00E-07 |
| 1 | ODAM | Hs.143811 | 364706 | 3,30E-06 | 1,00E-07 |
| 1 | KLK2 | Hs.515560 | 1102600 | 5,00E-06 | 1,30E-06 |
| 1 | F5 | Hs.30054 | 433155 | 6,60E-06 | 1,00E-07 |
| 1 | MFAP3L | Hs.593942 | 726821 | 7,60E-06 | 1,00E-07 |
| 1 | GALNT7 | Hs.548088 | 381854 | 1,10E-05 | 1,00E-07 |
| 2 | AHNAK | Hs.502756 | 238840 | 1,00E-07 | 1,00E-07 |
| 2 | APOD | Hs.522555 | 838611 | 1,00E-07 | 1,00E-07 |
| 2 | CAMK2N1 | Hs.197922 | 173820 | 1,00E-07 | 1,00E-07 |
| 2 | CFD | Hs.155597 | 666128 | 1,00E-07 | 1,00E-07 |
| 2 | FCGBP | Hs.111732 | 154172 | 1,00E-07 | 1,00E-07 |
| 2 | FOXO1 | Hs.370666 | 151247 | 1,00E-07 | 1,00E-07 |
| 2 | GLT8D2 | Hs.631650 | 365271 | 1,00E-07 | 1,00E-07 |
| 2 | MATN2 | Hs.189445 | 28584 | 1,00E-07 | 1,00E-07 |
| 2 | PRUNE | Hs.78524 | 364324 | 1,00E-07 | 1,00E-07 |
| 2 | SLC4A4 | Hs.5462 | 787938 | 1,00E-07 | 1,00E-07 |
| 2 | PAX8 | Hs.469728 | 545475 | 1,00E-07 | 3,00E-07 |
| 2 | HSD17B6 | Hs.524513 | 471641 | 1,00E-07 | 1,30E-06 |
| 2 | HBB | Hs.523443 | 469549 | 1,00E-07 | 2,60E-06 |
| 2 | APOE | Hs.654439 | 1870594 | 1,00E-07 | 3,70E-06 |
| 2 | CRABP1 | Hs.346950 | 739193 | 1,00E-07 | 4,20E-06 |
| 2 | ZNF76 | Hs.388024 | 745003 | 1,00E-07 | 4,70E-06 |
| 2 | C20orf19 | Hs.187635 | 366032 | 1,00E-07 | 5,00E-06 |
| 2 | DEPDC6 | Hs.112981 | 669318 | 2,00E-07 | 1,00E-07 |
| 2 | CDH16 | Hs.513660 | 726763 | 5,00E-07 | 1,00E-07 |
| 2 | HBA2 | Hs.654744 | 469647 | 6,00E-07 | 1,80E-06 |
| 2 | SASH1 | Hs.193133 | 31120 | 1,40E-06 | 1,00E-07 |
| 2 | HEXIM1 | Hs.15299 | 342551 | 2,20E-06 | 1,00E-07 |
| 2 | GJA1 | Hs.74471 | 839101 | 2,30E-06 | 1,40E-06 |
| 2 | TIMP3 | Hs.644633 | 501476 | 4,30E-06 | 4,80E-06 |
| 2 | IL11RA | Hs.591088 | 1101773 | 5,50E-06 | 7,00E-07 |
| 3 | APLP2 | Hs.695920 | 549054 | 1,00E-07 | 1,00E-07 |
| 3 | DCN | Hs.694789 | 666410 | 1,00E-07 | 1,00E-07 |
| 3 | FRMD4B | Hs.371681 | 669564 | 1,00E-07 | 1,00E-07 |
| 3 | HMGB2 | Hs.434953 | 884365 | 1,00E-07 | 1,00E-07 |
| 3 | IGF2BP2 | Hs.35354 | 743774 | 1,00E-07 | 1,00E-07 |
| 3 | LOC400451 | Hs.27373 | 667174 | 1,00E-07 | 1,00E-07 |
| 3 | NPAL3 | Hs.523442 | 738970 | 1,00E-07 | 1,00E-07 |
| 3 | PBXIP1 | Hs.505806 | 366042 | 1,00E-07 | 1,00E-07 |
| 3 | PDGFRL | Hs.458573 | 139242 | 1,00E-07 | 1,00E-07 |
| 3 | PDLIM1 | Hs.368525 | 135689 | 1,00E-07 | 1,00E-07 |
| 3 | PSAT1 | Hs.494261 | 366388 | 1,00E-07 | 1,00E-07 |
| 3 | SF3B1 | Hs.632554 | 739247 | 1,00E-07 | 1,00E-07 |
| 3 | TIAM1 | Hs.517228 | 23612 | 1,00E-07 | 1,00E-07 |
| 3 | TSHR | Hs.160411 | 565317 | 1,00E-07 | 1,00E-07 |
| 3 | FAM111A | Hs.150651 | 137454 | 1,00E-07 | 4,00E-07 |
| 3 | CA11 | Hs.428446 | 282587 | 3,00E-07 | 1,00E-07 |
| 3 | HNRPA3 | Hs.516539 | 365349 | 5,00E-07 | 2,60E-06 |
| 4 | ABCC3 | Hs.463421 | 208097 | 1,00E-07 | 1,00E-07 |
| 4 | CDH3 | Hs.461074 | 359051 | 1,00E-07 | 1,00E-07 |
| 4 | CITED1 | Hs.40403 | 265558 | 1,00E-07 | 1,00E-07 |
| 4 | CLDN1 | Hs.439060 | 594279 | 1,00E-07 | 1,00E-07 |
| 4 | DPP4 | Hs.368912 | 343987 | 1,00E-07 | 1,00E-07 |
| 4 | DUSP5 | Hs.2128 | 33285 | 1,00E-07 | 1,00E-07 |
| 4 | NOTCH2 | Hs.487360 | 1641901 | 1,00E-07 | 1,00E-07 |
| 4 | QPCT | Hs.79033 | 711918 | 1,00E-07 | 1,00E-07 |
| 4 | S100A4 | Hs.654444 | 868577 | 1,00E-07 | 1,00E-07 |
| 4 | SCEL | Hs.534699 | 668239 | 1,00E-07 | 1,00E-07 |
| 4 | SLPI | Hs.517070 | 378813 | 1,00E-07 | 1,00E-07 |
| 4 | TESC | Hs.525709 | 745490 | 1,00E-07 | 1,00E-07 |
| 4 | TIMP1 | Hs.522632 | 162246 | 1,00E-07 | 1,00E-07 |
| 4 | MGAT3 | Hs.276808 | 731060 | 1,00E-07 | 8,00E-07 |
| 4 | MDK | Hs.82045 | 309009 | 2,00E-07 | 1,00E-07 |
| 4 | BID | Hs.591054 | 128065 | 6,00E-07 | 1,00E-07 |
| 4 | NRIP1 | Hs.155017 | 38775 | 6,00E-07 | 1,00E-07 |
| 4 | COL8A2 | Hs.353001 | 486204 | 1,00E-06 | 1,00E-07 |
| 4 | MRC2 | Hs.7835 | 235882 | 1,60E-06 | 9,00E-07 |
| 4 | STAT1 | Hs.699271 | 110101 | 2,10E-06 | 1,00E-07 |
| 4 | ARMCX3 | Hs.592225 | 251452 | 2,40E-06 | 1,00E-07 |
| 4 | LAMB3 | Hs.497636 | 1103402 | 9,90E-06 | 1,00E-07 |
| 4 | CD55 | Hs.527653 | 627107 | 1,30E-05 | 1,00E-07 |
| 5 | DUS4L | Hs.97627 | 726904 | 1,00E-07 | 1,00E-07 |
| 5 | ESRRG | Hs.444225 | 44064 | 1,00E-07 | 1,00E-07 |
| 5 | ISG20 | Hs.459265 | 740604 | 1,00E-07 | 1,00E-07 |
| 5 | ROR2 | Hs.644776 | 1089025 | 1,00E-07 | 1,00E-07 |
| 5 | S100A11 | Hs.417004 | 143957 | 1,00E-07 | 1,00E-07 |
| 5 | SEMA4D | Hs.655281 | 210587 | 1,00E-07 | 1,00E-07 |
| 5 | CD58 | Hs.34341 | 2103105 | 1,00E-07 | 2,00E-07 |
| 5 | IRF9 | Hs.1706 | 724588 | 1,10E-06 | 3,20E-06 |
| 5 | MCOLN1 | Hs.631858 | 726156 | 2,00E-06 | 2,00E-07 |
| 5 | ITGB7 | Hs.654470 | 1337232 | 1,09E-05 | 2,90E-06 |
Independent validation of selected genes by real-time RT-PCR
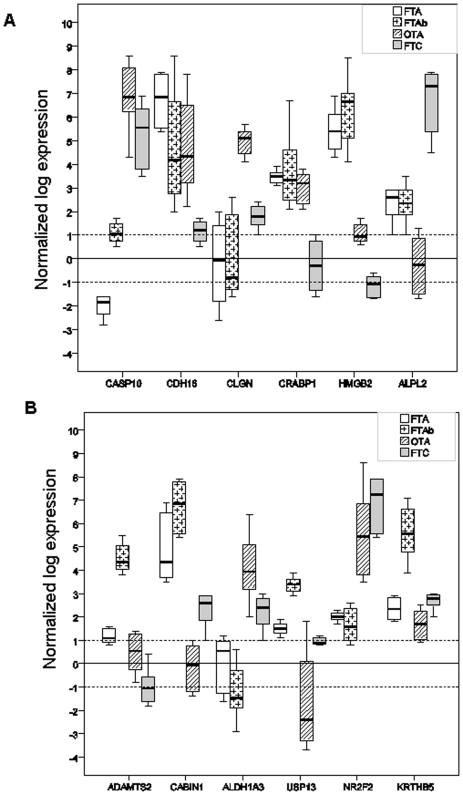
To test the relevance of some markers selected by cross-validation between the two main datasets, we measured the expression levels of six differential genes, CASP10, CDH16, CLGN, CRABP1, HMGB2 and ALPL2, on 32 new follicular tumor samples, including eight samples each of FTA, FTAb, OTA and FTC (Figure 4A). For all six genes, the expression levels differentiated between follicular adenomas (macro- and micro-follicular) and follicular carcinomas (t-test, p<0.05). In all cases except one (CASP10, F-test, p<0.05), the gene expression level differentiated not only between macro- and micro-follicular adenomas but also between follicular adenomas and oncocytic adenomas, as in the case of CDH16 and CRABP1 genes.
Figure 4. Gene expression of selected markers.
Differential expression of 12 genes measured by real-time quantitative RT-PCR on 32 new follicular thyroid tumors (8 FTA, 8 FTAb, 8 OTA and 8 FTC). The upper and the lower limits of each box represent the upper and the lower quartiles, respectively. The bold lines represent medians. The expression of ADAMTS2, CABIN1, ADLH1A3, USP13, NR2F2, KRTHB5, CASP10, CDH16, CLGN, CRABP1, HMGB2 and ALPL2 genes was referred to the β-globin expression level. Significances of differential expression between the classes were assessed by t-tests and those over the classes by F-tests. FTA: macrofollicular thyroid adenoma; FTAb: microfollicular thyroid adenoma; FTC: follicular thyroid carcinoma; and OTA: oncocytic thyroid adenoma.
To test the relevance of our microarray analysis in identifying subclasses of adenomas, we measured the expression levels of six differential genes from the microfollicular adenoma subclass (ADAMTS2, CABIN1, ALDH13, USP13, NR2F2 and KRTHB5), on the same 32 follicular thyroid samples mentioned above (Figure 4B). Except for NR2F2 (p<0.954), all genes were differentially expressed not only between the three subclasses of adenomas (t-test, p<0.05), but also between the four groups of follicular tumors (F-tests, p<0.05 for ADAMTS2, CABIN1, ALDH13, USP13; p = 0.06 for KRTHB5).
Mutational status of follicular tumors
The mutational status of three genes (TSHR, GNAS and NRAS) was investigated on 59 solitary follicular adenomas (43 from the microarray study, and 16 from the RT-PCR study) to compare these to the expression profile of FTA samples and evaluate their functional status. To identify the role of the PAX8-PPARG translocation in the expression profile of the FTC class, the rearrangement status for PAX8-PPARG was explored on 11 FTC samples (three samples from the microarray study, and eight samples from the RT-PCR study).
In the follicular adenoma samples, we identified mutations in the TSHR gene in seven of the 59 samples (12%): six of which were macrofollicular adenomas from the microarray study and one was a macrofollicular adenoma from the RT-PCR study. All the mutations were situated in the seventh transmembrane domain of the TSH receptor (from codon 619 to codon 633). Our results are in accordance with the mutational status of the TSH receptor gene described in the literature, especially concerning the macrofollicular subtype [19]. One macrofollicular adenoma presented a mutation in codon 201 of the GNAS gene. None of the microfollicular adenomas showed TSHR and GNAS mutations in the four exons explored. Five microfollicular adenomas (four from the microarray study, and one from the RT-PCR study) were positive for the NRAS mutation in codon 61.
No PAX8-PPARG translocation was found in any of the FTC samples studied.
Protein expression of selected differential genes in independent samples
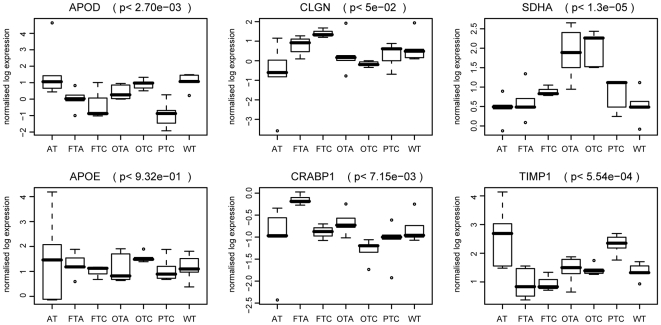
We collected 49 new tissue samples to measure the protein expression of six selected differential genes in the classes common to the two main datasets (FTA, FTC, OTA, OTC, PTC and WT), and also for the AT class as a non-tumoral control (Figure 5). There was significant differential expression between the classes (F-tests, p<0.05) for five proteins (SDHA, CLGN, APOD, CRABP1 and TIMP1). The APOE protein was not differentially expressed (p<0.932). Among the differentially expressed proteins, three had profiles similar to their gene profiles (SDHA, APOD and TIMP1), whereas two proteins (CLGN and CRABP1) showed some differences, as reported in previous studies of selected classes [20], [21]. The expression of the CLGN protein was higher in the FTA and FTC samples, but not in the oncocytic samples, contrary to the gene profile observed. The CRABP1 protein was differentially expressed between the FTA samples compared to the PTC and FTC samples, with a decreased expression of both RNA and protein in the malignant tumors. Considering the appurtenance of this gene to Cluster 2 (Figure 3), the expression of CRABP1 protein could serve to distinguish between the benign and malignant groups of thyroid tissue.
Figure 5. Protein expression of selected genes.
The protein expression of six selected differential genes was measured on 49 new tissue samples. These independent samples represent six tissue classes that are common to the two main datasets (WT, PTC, FTA, FTC, OTA, and OTC). The AT class was added to compare the expression levels with a non-tumoral lesion. Seven samples were used for each class. Protein expression profiles of APOD, CLGN, SDHA, APOE, CRABP1 and TIMP1 are shown by box-plots for the different classes of tissue. Significances of differential expression over the classes were assessed by F-tests. P-values are shown between brackets. WT: wild type tissue; PTC: papillary thyroid carcinoma; FTA: macrofollicular thyroid adenoma; FTC: follicular thyroid carcinoma; OTA: oncocytic thyroid adenoma; and OTC: oncocytic thyroid carcinoma.
Discussion
The clinical diagnosis of thyroid tumors is subject to significant inter- and intra-observer variations [22], [23]. Since 2001, various molecular markers for thyroid tumors have been proposed on the basis of microarray analysis. However, none of these markers has proved satisfactory in clinical practice, possibly because only a small number of thyroid tumor classes were initially taken into account. In our study, we analyzed six publicly available microarray datasets containing 347 thyroid tissue samples and defining as many as 12 classes of thyroid tumors. One of these datasets, the Fontaine dataset generated by our laboratory, comprised about half the total number of samples, including all the differentiated follicular thyroid lesions.
With regard to the 12 classes of thyroid tumor defined, we found that more than half of the 104 relevant and cross-validated differential genes were also related to other classes, such as AT, GD, TUM, FTAb, which were not shared by the two main datasets, i.e. the Giordano dataset and the Fontaine dataset. This may have affected the relevance of the signature in the classification of the follicular pathologies, explaining the low percentage of genes common to the two classifiers. Although the genes were selected from different sets of classes, discrepancies of some gene profiles, for example in Cluster 5, may be related to incomplete microarray platform compatibility or to differential cellular environment. Interestingly, most of the genes in Cluster 5 were involved in the process of vascular endothelial cell expansion[24]–[26], which may represent the stromal reaction. Despite the small number of FTC and OTC samples and the microarray platform being restricted to 9,216 probes, our classifier was able to validate the classification of PTC, FTC, FTA and OTC on five other datasets (Table 3). This demonstrated the relevance of our approach to the molecular classification of the main types of differentiated follicular thyroid pathology.
Unsupervised analysis of the class signatures showed a separation into three groups corresponding to benign, malignant, and oncocytic thyroid tumors (Figure 1). The pairwise distinctions of benign/normal and malignant tumors were consistent with published reports, as in the case of PTC and FTA [14]. The surprising proximity of the FTC and FTA classes in the Giordano dataset has already been discussed [4]. This could be related to the high homogeneity in the expression profiles of the FTC samples, which were positive for the PAX8-PPARG rearrangement and significantly related to the malignant profile of the PTC samples. Thus, the clustering of the FTC- and FTA classes appears to be related more to their heterogeneous expression profiles rather than to their tendency to tumoral behavior. These results strongly suggest the existence of molecular subclasses for the FTA and FTC- samples. Interestingly, the FTC samples in the Fontaine dataset, included in the malignant group, were negative for the PAX8-PPARG translocation.
Although the histologic diagnosis of oncocytic follicular tumors presents few problems, the two latest WHO classifications have debated whether these tumors should be considered as an independent class of tumors or as a variant of follicular tumors. This question is still relevant in view of the recent results of miRNA profiling that clearly revealed the segregation of oncocytic tumors from other conventional follicular tumors [27]. According to their specific gene-expression profiles as well as their mutational status for RAS and PAX8-PPARγ genes [7], oncocytic tumors should be considered as distinct from the other follicular tumors. The role of the “oncocytic group” signature in the appropriate classification of benign and malignant tumors is debatable. Considering the mitochondrial richness of follicular thyroid tumors, this signature may highlight the metabolic changes during tumorigenesis. Little is known about the etiology of oncocytomas but their signature is composed of numerous mitochondrial and metabolic genes, and the role played by these genes in tumor aggressivity has not yet been established [7], [20]. Interestingly, we found that microfollicular adenomas (FTAb) were related by their molecular profiles to oncocytic adenomas as well as to carcinomas. Relationships between thyroid metabolism and microfollicular lesions have been explored [28], but never in the context of mitochondrial functions. However, in a recent study, the galectin-3 profile associated with FTAb was reported to be related to its role in the regulation of mitochondrial stability [29]. Galectin-3 has been also involved in thyroid tumorigenesis through its anti-apoptotic activity [30]. Thus, the malignant potential of FTAb is still under discussion [31].
Evaluation of the performance of the classifiers used showed that poor performance was associated with the FTA subclasses in the two main datasets. In our study, the functional status of the FTA samples may have influenced the performance of the classifiers in discriminating true FTA from hyperfunctioning nodules. Only 12% of macrofollicular adenomas presented mutations in the TSH receptor or GNAS genes. These results correlated on microarray analysis with the expression profile of several genes previously associated with the hyperfunctional status of nodules [32]. Even though our FTA group represents a mix of adenomas in terms of functional status, the signature of follicular adenoma defined by our classifier are involved in other processes than iodine uptake and thyroid hormone secretion. As the selected genes are mainly associated to cell metabolism (CABIN1, CAMK2A, DAP, PDK4), signal transduction (ASGR1, PLXNB2, TBC1, PLCG1) and cell adhesion (COL18A1), we believe that the FTA signature we propose can discriminate true FTA from other follicular thyroid tumors. The relevance of constructing subclasses of follicular adenomas was tested on six cross-validated markers and six specific markers from our own analysis (Figure 4). Five specific markers enabled us to segregate micro- and macro-follicular adenomas from oncocytic adenomas and minimally invasive follicular carcinomas, while only one of the six cross-validated markers showed the same performance. Distinguishing between subclasses of adenoma allowed the identification of relevant molecular markers of follicular adenomas. These markers had never been differentially selected during previous microarray analyses of thyroid tumors. However, three of the markers (ADAMTS2, ALDH1A3 and CABIN1) had been proposed as prognostic markers for the recurrence of epithelial tumors [33]–[35].
The observation of six protein profiles highlights the difficulty of proposing immunomarkers for clinical practice. However, some markers may be relevant for the classification of differentiated follicular pathologies (FTA, FTC, PTC, OTA and OTC). It has been suggested that APOD, which has a specific protein profile for FTC and PTC, may modify the proliferative activity of cancer cells [36]. The differential expression of the folding protein calmegin (CLGN) with FTA and FTC samples may be associated with a difference in the abundance of the protein, as recently described in thyroid tumors [37]. Furthermore, increasing the number of thyroid tumor classes has revealed new aspects of some of the markers. Thus, TIMP1 and CRABP1 were not selected by our classifier but figured in the intersecting differential gene lists. Our data clearly questioned the relevance of TIMP1 as a marker of thyroid cancer. Since high protein levels were observed in the AT and PTC classes, TIMP1 expression may be more closely related to the frequently described lymphocytic infiltration in PTC [38]. Quantitative RT-PCR analysis on new follicular samples confirmed the upregulation of CRABP1 expression in all the adenomas as compared to follicular carcinomas. Underexpression of CRABP1 mRNA was relevant in distinguishing the FTC as well as PTC classes, as reported previously [39], [40]. However, our results differed at the protein level compared to a study on cold thyroid nodules where both the expression of CRABP1 mRNA and the protein were downregulated compared to normal surrounding tissue [41]. As our FTA samples were mostly non-functioning nodules, we postulate that this difference in protein level may be due to the histological class studied, macrofollicular adenomas in the Fontaine dataset, as opposed to the mix of follicular adenomas and adenomatous nodules in the other study. While all these markers can be used to complement current markers [42], the exploration of a large panel of protein markers to improve the differential diagnosis may rapidly become more cumbersome and less relevant than the study of the expression profile of selected genes, especially in the case of suspicious nodular thyroid lesions.
In conclusion, diagnostic tools defined on the basis of thyroid microarray data are more relevant when many samples and tissue classes are used, especially with the inclusion of oncocytic tumors. Our study has revealed additional potentially useful gene and protein markers allowing the classification of several thyroid pathologies. However, to identify early events of transformation, these markers should be viewed in the context of the biological processes in which they participate. We therefore believe that dedicated gene-expression profiling offers the most powerful method of identifying gene groups that correlate to nodular growth, thyroid dedifferentiation or malignancy, taking into account the potential contamination by other cell types such as activated lymphocytes. In this context, the molecular classification of microfollicular adenomas on fine needle aspiration biopsies should have a direct impact in the management of such tumors.
Materials and Methods
Samples and microarray datasets
We analyzed gene-expression data from six microarray datasets containing up to 12 classes of differentiated thyroid pathologies and normal thyroid tissue. The FTA samples were divided into two subclasses according to their follicular architecture, FTA (macrofollicular) and FTAb (microfollicular). The dominant nodule within the multinodular goiter (MNG) was investigated to explore the incidence of genetic or environmental events on the molecular signature of the class of follicular adenoma. When possible, the FTC class was also divided into two subclasses, FTC+ and FTC-, defined respectively by the presence or absence of the PAX8/PPARγ translocation in the samples. Atypical (TUM) and oncocytic features (OT) were individualized on criteria specified elsewhere [15], [20]. We also explored non-tumoral lesions such as those in Graves' disease (GD) and autoimmune thyroiditis (AT) to identify the immunological environment of the tumoral samples. This study is based on the Fontaine dataset generated by our laboratory, which comprises 12 subclasses containing a total of 166 anonymized human thyroid tissues. The five other datasets, particularly the Giordano dataset with 7 subclasses, were used to cross-validate the results of the molecular classification, the classifiers, and the functional analysis of differential genes [5], [43]–[45]. One dataset (Reyes et al.) has not yet associated citation. Details of the six datasets, containing a total of 347 samples, are summarized in Table 1.
The Fontaine and the Giordano datasets are referred as the main datasets. All the datasets, generated by various microarray platforms (Table 1), are publicly available. The Fontaine dataset (GSE6339), the He dataset (GSE3467) and the Reyes dataset (GSE3678) are hosted by the Gene Expression Omnibus (GEO) database of the U.S. National Center for Biotechnology Information. The Weber dataset was downloaded from European Bioinformatics Institute ArrayExpress database (E-MEXP-97). The two other datasets were downloaded from the authors' websites: the Giordano dataset (http://dot.ped.med.umich.edu:2000/pub/PPARG/index.html) and the Jarzab dataset (http://www.genomika.pl/thyroidcancer/PTCCancerRes.html).
Data analysis
For the Fontaine dataset generated in our laboratory, hierarchical clustering of the data was computed on log-transformed, median gene-centered and normalized data as described elsewhere [15]. Normalized data from the Giordano and Jarzab datasets were downloaded from the authors' websites. For the remaining datasets, raw data were downloaded from the GEO or Array Express databases and normalized with BRB Array Tools v3.7.0b1 developed by Dr. Richard Simon. Since the data originated from three different but one-color platforms, global normalization to the median was used to center the gene-expression values in each array. Genes showing minimal variation across the set of arrays were excluded from the analysis (log-intensity variation P-value>0.15). Hierarchical clustering was done on centered genes by using the centered correlation distance and an average linkage method. Robustness and discrepancy indices were computed from 10,000 data perturbations.
For the two largest datasets [5], [15], we identified genes that were differentially expressed among the classes by using a multivariate 10,000-permutation test, to provide 95% confidence that the number of false discoveries did not exceed 1. Although F-statistics were used for each gene, the multivariate permutation test is non-parametric and does not require the assumption of Gaussian distributions. Functional enrichments in gene clusters were defined at 5% risk for Level 3 Gene Ontology terms by Fatigo tools (adjusted P-values, www.fatigo.bioinfo.cipf.es [46]) as compared to the whole genome, and for canonical pathways by Ingenuity Pathway Analysis tools as compared to the Ingenuity Pathways Knowledge database (www.ingenuity.com).
Classifiers were defined for each pathological class and trained either versus the wild type class (binary training) or the entire dataset (complete training). Twenty genes were selected by the Greedy Pairs algorithm, and Diagonal Linear Discriminant Analysis (DLDA) was used for training and sample [47]. Intra- and inter-dataset cross-validations were done by the 0.632+ method and DLDA respectively [48]. Since the classical accuracy coefficient is biased in large datasets containing unbalanced class sizes, we preferred to use the positive accuracy coefficient defined as the geometric mean of the sensitivity (i.e. the proportion of true positives in the samples belonging to the targeted class in the entire datasets) and the positive predictive value (i.e. the proportion of true positives in the samples belonging to the targeted class) [49].
Quantitative RT-PCR analysis
An independent set of 32 follicular tumors, comprising 8 minimally invasive FTC and 24 follicular adenoma (8 OTA, 8 FTA and 8 FTAb), were subjected to real-time quantitative RT-PCR to test the relevance of several selected markers. The anonymized samples were obtained from the Department of Pathology of the University Hospital of Angers, France.
Total RNA was isolated using the TRIzol reagent (Invitrogen, Paisley, UK). RNA integrity was determined using a Bio-Analyzer 2100 (Agilent Technologies, Waldbronn, Germany). Reverse transcription was performed on 1 µg of RNA with Advantage RT-for-PCR kit (Clontech Laboratory, Palo Alto, CA, USA) following the manufacturer's recommendations. Real-time quantification was performed in a 96-well plate using the IQ SYBR Green supermix and Chromo4 (Biorad, Hercules, CA, USA) with the recommended protocol. Twelve genes were explored for their expression levels: CASP10, CDH16, CLGN, CRABP1, HMGB2, ALPL2, ADAMTS2, CABIN1, ALDH1A3, USP13, NR2F2 and KRTHB5. Data were normalized to the β-globin expression level. The significance of differential expression between classes was assessed by t-tests and that over the classes by F-tests. The primer sequences used in this study are specified in Table 6.
Table 6. Primer sequences used for real-time quantitative RT-PCR.
| Gene | Forward Sequence | Reverse Sequence |
| ADAMTS2 | 5′-GGG-AAG-GAG-CAC-GTA-CAG-AA-3′ | 5′-CGT-GAT-CGT-GGT-ATT-CAT-CG-3′ |
| ALDH1A3 | 5′-TCT-CGA-CAA-AGC-CCT-GAA-GT-3′ | 5′-GTC-CGA-TGT-TTG-AGG-AAG-GA-3′ |
| APLP2 | 5′-GCT-GTC-GTT-CCG-GTT-ATG-TT-3′ | 5′-GAG-GGT-CTC-TCA-CGT-GCT-TC-3′ |
| CABIN1 | 5′-AGG-CCC-TGG-AGG-TGT-ACT-TT-3′ | 5′-CCA-GCT-GAA-GAG-TGG-GAG-TC-3′ |
| CASP10 | 5′-TCT-CAG-GAT-CAC-TGG-GCT-CT-3′ | 5′-ATG-AAG-GCG-TTA-ACC-ACA-GG-3′ |
| CDH16 | 5′-CAA-GTC-ATG-AGG-TGG-TGG-TG-3′ | 5′-GAT-GGT-CAG-CAG-GAA-AGA-GC-3′ |
| CLGN | 5′-CAA-TGG-ACC-TGG-AAG-AGG-AA-3′ | 5′-TGA-CTT-TAT-CGG-CCC-ATC-TC-3′ |
| CRABP1 | 5′-CAG-GAC-GGG-GAT-CAG-TTC-TA-3′ | 5′-CGC-CAA-ACG-TCA-GGA-TAA-GT-3′ |
| HMGB2 | 5′-AGA-GGC-TGA-GGA-TTG-CGT-TA-3′ | 5′-GGG-GTC-TCC-TTT-ACC-CAT-GT-3′ |
| KRTHB5 | 5′-AGC-TCT-CAG-GGA-CAA-GAC-CA-3′ | 5′-AAC-AGG-TTA-GCC-CAG-AAG-CA-3′ |
| NR2F2 | 5′-TGC-CTG-TGG-TCT-CTC-TGA-TG-3′ | 5′-ATA-TCC-CGG-ATG-AGG-GTT-TC-3′ |
| USP13 | 5′-GAC-CTG-CGA-GAA-AAC-CTC-TG-3′ | 5′-CAG-GAG-TGA-TGG-TTC-CCA-GT-3′ |
| β-GLOBIN | 5′-CAA-CTT-CAT-CCA-CGT-TCA-CC-3′ | 5′-ACA-CAA-CTG-TGT-TCA-CTA-GC-3′ |
Detection of mutations and PAX8-PPARG rearrangement
The mutational status for the TSHR, GNAS and NRAS genes was determined for all the FTA and FTAb explored in studies by microarray (26 FTA and 17 FTAb) and RT-PCR (8 FTA and 8 FTAb). DNA was isolated from frozen tissues during the guanidium isothiocyanate procedure (TRIzol Reagent, Invitrogen Life Technologies). Exon 2 of the NRAS gene was amplified using primer sequences described elsewhere[50]. Exons 9 and 10 of the TSH receptor gene, and exons 8 and 9 of the Gs alpha gene were amplified using primer sequences previously specified [51]. PCR were performed on 5 µl DNA with the HotGoldstar DNA polymerase (Eurogentec, Seraing, Belgium) according to the manufacturer's recommendations. Amplified fragments were purified and directly sequenced on a CEQ 8000 apparatus, using the CEQ DTCS Quick Start Kit (Beckman Coulter, Fullerton, CA, USA) following the manufacturer's instructions.
The presence of the PAX8-PPARG translocation was evaluated as described elsewhere [52], on the three FTC samples in the microarray study and the eight FTC samples in the RT-PCR study. Quantification, degradation and DNA contamination of RNA were assessed using an RNA 6000 Nano Assay kit (Agilent Technologies, Palo Alto, CA, USA). After reverse transcription, we looked for the translocation in 5 µl cDNA, using primer sequences previously specified [5] and the HotGoldstar DNA polymerase (Eurogentec, Seraing, Belgium) according to the manufacturer's recommendations.
Dot blot analysis
Two micrograms of protein corresponding to the TRIzol fraction of 49 independent and anonymized thyroid tissues were spotted onto nitrocellulose membranes at room temperature, using a Dot Blot apparatus (Biorad, Hercules, CA, USA) following the manufacturer's recommendations. These tissues, provided by the Department of Pathology of the University Hospital of Angers, differed from those used for the microarray and RT-PCR studies. Six groups of thyroid pathologies were defined according to WHO cytological criteria: AT, FTA, FTC, PTC, OTA and OTC and compared to normal thyroid tissue (WT). Seven samples were tested from each group and the assays were performed in duplicate. Seven primary antibodies (all from Abcam, Cambridge, UK) were used at specific dilutions: 1/750 for TIMP1, 1/1000 for APOD, SDHA, CLGN, CRABP1 and APOE, and 1/5000 for α-tubulin as a control. Peroxidase anti-rabbit or anti-mouse secondary antibodies were revealed using the ECL Plus reagent kit (ECL, Amersham, Chalfont, UK). Spots were quantified on a GelDoc XRS apparatus using Quantity One software (Biorad) and expressed in terms of the control value. Significances of differential expression over the classes were assessed by F-tests.
Acknowledgments
We thank Dominique Couturier and Anne Coutoleau for technical help. We thank Marja Steenman and Kanaya Malkani for the critical reading of this paper.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the French Ministry of Research, the Institut National de la Recherche Medicale (INSERM), the University Hospital of Angers and the University of Angers (PHRC 03-10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hundahl SA, Fleming ID, Fremgen AM, Menck HR. A National Cancer Data Base report on 53,856 cases of thyroid carcinoma treated in the U.S., 1985-1995. Cancer. 1998;83:2638–2648. doi: 10.1002/(sici)1097-0142(19981215)83:12<2638::aid-cncr31>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Yeh MW, Demircan O, Ituarte P, Clark OH. False-negative fine-needle aspiration cytology results delay treatment and adversely affect outcome in patients with thyroid carcinoma. Thyroid. 2004;14:207–215. doi: 10.1089/105072504773297885. [DOI] [PubMed] [Google Scholar]
- 4.Lacroix L, Lazar V, Michiels S, Ripoche H, Dessen P, et al. Follicular thyroid tumors with the PAX8-PPARgamma1 rearrangement display characteristic genetic alterations. Am J Pathol. 2005;167:223–231. doi: 10.1016/s0002-9440(10)62967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordano TJ, Au AY, Kuick R, Thomas DG, Rhodes DR, et al. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clin Cancer Res. 2006;12:1983–1993. doi: 10.1158/1078-0432.CCR-05-2039. [DOI] [PubMed] [Google Scholar]
- 6.Barden CB, Shister KW, Zhu B, Guiter G, Greenblatt DY, et al. Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clin Cancer Res. 2003;9:1792–1800. [PubMed] [Google Scholar]
- 7.Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW, et al. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88:2318–2326. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- 8.Aldred MA, Huang Y, Liyanarachchi S, Pellegata NS, Gimm O, et al. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J Clin Oncol. 2004;22:3531–3539. doi: 10.1200/JCO.2004.08.127. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci U S A. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarzab B, Wiench M, Fujarewicz K, Simek K, Jarzab M, et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 11.Weber F, Shen L, Aldred MA, Morrison CD, Frilling A, et al. Genetic classification of benign and malignant thyroid follicular neoplasia based on a three-gene combination. J Clin Endocrinol Metab. 2005;90:2512–2521. doi: 10.1210/jc.2004-2028. [DOI] [PubMed] [Google Scholar]
- 12.Eszlinger M, Wiench M, Jarzab B, Krohn K, Beck M, et al. Meta- and reanalysis of gene expression profiles of hot and cold thyroid nodules and papillary thyroid carcinoma for gene groups. J Clin Endocrinol Metab. 2006;91:1934–1942. doi: 10.1210/jc.2005-1620. [DOI] [PubMed] [Google Scholar]
- 13.Griffith OL, Melck A, Jones SJ, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol. 2006;24:5043–5051. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- 14.Finley DJ, Zhu B, Fahey TJ,, III Molecular analysis of Hurthle cell neoplasms by gene profiling. Surgery. 2004;136:1160–1168. doi: 10.1016/j.surg.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 15.Fontaine JF, Mirebeau-Prunier D, Franc B, Triau S, Rodien P, et al. Microarray analysis refines classification of non-medullary thyroid tumours of uncertain malignancy. Oncogene. 2008;27:2228–2236. doi: 10.1038/sj.onc.1210853. [DOI] [PubMed] [Google Scholar]
- 16.Stolf BS, Abreu CM, Mahler-Araujo MB, Dellamano M, Martins WK, et al. Expression profile of malignant and non-malignant diseases of the thyroid gland reveals altered expression of a common set of genes in goiter and papillary carcinomas. Cancer Lett. 2005;227:59–73. doi: 10.1016/j.canlet.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 17.Yukinawa N, Oba S, Kato K, Taniguchi K, Iwao-Koizumi K, et al. A multi-class predictor based on a probabilistic model: application to gene expression profiling-based diagnosis of thyroid tumors. BMC Genomics. 2006;7:190. doi: 10.1186/1471-2164-7-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eszlinger M, Krohn K, Hauptmann S, Dralle H, Giordano TJ, et al. Perspectives for improved and more accurate classification of thyroid epithelial tumors. J Clin Endocrinol Metab. 2008;93:3286–3294. doi: 10.1210/jc.2008-0201. [DOI] [PubMed] [Google Scholar]
- 19.Corvilain B, Van SJ, Dumont JE, Vassart G. Somatic and germline mutations of the TSH receptor and thyroid diseases. Clin Endocrinol (Oxf) 2001;55:143–158. [PubMed] [Google Scholar]
- 20.Baris O, Savagner F, Nasser V, Loriod B, Granjeaud S, et al. Transcriptional profiling reveals coordinated up-regulation of oxidative metabolism genes in thyroid oncocytic tumors. J Clin Endocrinol Metab. 2004;89:994–1005. doi: 10.1210/jc.2003-031238. [DOI] [PubMed] [Google Scholar]
- 21.Wasenius VM, Hemmer S, Kettunen E, Knuutila S, Franssila K, et al. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: a cDNA and tissue microarray study. Clin Cancer Res. 2003;9:68–75. [PubMed] [Google Scholar]
- 22.Franc B, de la SP, Lange F, Hoang C, Louvel A, et al. Interobserver and intraobserver reproducibility in the histopathology of follicular thyroid carcinoma. Hum Pathol. 2003;34:1092–1100. doi: 10.1016/s0046-8177(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 23.Lloyd RV, Erickson LA, Casey MB, Lam KY, Lohse CM, et al. Observer variation in the diagnosis of follicular variant of papillary thyroid carcinoma. Am J Surg Pathol. 2004;28:1336–1340. doi: 10.1097/01.pas.0000135519.34847.f6. [DOI] [PubMed] [Google Scholar]
- 24.Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci U S A. 2008;105:7821–7826. doi: 10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sierra JR, Corso S, Caione L, Cepero V, Conrotto P, et al. Tumor angiogenesis and progression are enhanced by Sema4D produced by tumor-associated macrophages. J Exp Med. 2008;205:1673–1685. doi: 10.1084/jem.20072602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor KL, Leaman DW, Grane R, Mechti N, Borden EC, et al. Identification of interferon-beta-stimulated genes that inhibit angiogenesis in vitro. J Interferon Cytokine Res. 2008;28:733–740. doi: 10.1089/jir.2008.0030. [DOI] [PubMed] [Google Scholar]
- 27.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nucera C, Mazzon E, Caillou B, Violi MA, Moleti M, et al. Human galectin-3 immunoexpression in thyroid follicular adenomas with cell atypia. J Endocrinol Invest. 2005;28:106–112. doi: 10.1007/BF03345351. [DOI] [PubMed] [Google Scholar]
- 29.Fukumori T, Oka N, Takenaka Y, Nangia-Makker P, Elsamman E, et al. Galectin-3 regulates mitochondrial stability and antiapoptotic function in response to anticancer drug in prostate cancer. Cancer Res. 2006;66:3114–3119. doi: 10.1158/0008-5472.CAN-05-3750. [DOI] [PubMed] [Google Scholar]
- 30.Savin S, Cvejic D, Isic T, Paunovic I, Tatic S, et al. Thyroid peroxidase and galectin-3 immunostaining in differentiated thyroid carcinoma with clinicopathologic correlation. Hum Pathol in press. 2008 doi: 10.1016/j.humpath.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Schmid KW, Farid NR. How to define follicular thyroid carcinoma?. Virchows Arch. 2006;448:385–393. doi: 10.1007/s00428-006-0162-0. [DOI] [PubMed] [Google Scholar]
- 32.Wattel S, Mircescu H, Venet D, Burniat A, Franc B, et al. Gene expression in thyroid autonomous adenomas provides insight into their physiopathology. Oncogene. 2005;24:6902–6916. doi: 10.1038/sj.onc.1208849. [DOI] [PubMed] [Google Scholar]
- 33.Carinci F, Lo ML, Piattelli A, Rubini C, Chiesa F, et al. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol. 2005;18:513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- 34.Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, et al. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10:2429–2440. doi: 10.1158/1078-0432.ccr-0398-3. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe T, Kobunai T, Sakamoto E, Yamamoto Y, Konishi T, et al. Gene expression signature for recurrence in stage III colorectal cancers. Cancer. 2009;115:283–292. doi: 10.1002/cncr.24023. [DOI] [PubMed] [Google Scholar]
- 36.Soiland H, Soreide K, Janssen EA, Korner H, Baak JP, et al. Emerging concepts of apolipoprotein D with possible implications for breast cancer. Cell Oncol. 2007;29:195–209. doi: 10.1155/2007/487235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Netea-Maier RT, Hunsucker SW, Hoevenaars BM, Helmke SM, Slootweg PJ, et al. Discovery and validation of protein abundance differences between follicular thyroid neoplasms. Cancer Res. 2008;68:1572–1580. doi: 10.1158/0008-5472.CAN-07-5020. [DOI] [PubMed] [Google Scholar]
- 38.Delys L, Detours V, Franc B, Thomas G, Bogdanova T, et al. Gene expression and the biological phenotype of papillary thyroid carcinomas. Oncogene. 2007;26:7894–7903. doi: 10.1038/sj.onc.1210588. [DOI] [PubMed] [Google Scholar]
- 39.Hawthorn L, Stein L, Varma R, Wiseman S, Loree T, et al. TIMP1 and SERPIN-A overexpression and TFF3 and CRABP1 underexpression as biomarkers for papillary thyroid carcinoma. Head Neck. 2004;26:1069–1083. doi: 10.1002/hed.20099. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, de la CA, Pellegata NS. Hypermethylation, but not LOH, is associated with the low expression of MT1G and CRABP1 in papillary thyroid carcinoma. Int J Cancer. 2003;104:735–744. doi: 10.1002/ijc.11006. [DOI] [PubMed] [Google Scholar]
- 41.Eszlinger M, Krohn K, Berger K, Lauter J, Kropf S, et al. Gene expression analysis reveals evidence for increased expression of cell cycle-associated genes and Gq-protein-protein kinase C signaling in cold thyroid nodules. J Clin Endocrinol Metab. 2005;90:1163–1170. doi: 10.1210/jc.2004-1242. [DOI] [PubMed] [Google Scholar]
- 42.Barroeta JE, Baloch ZW, Lal P, Pasha TL, Zhang PJ, et al. Diagnostic value of differential expression of CK19, Galectin-3, HBME-1, ERK, RET, and p16 in benign and malignant follicular-derived lesions of the thyroid: an immunohistochemical tissue microarray analysis. Endocr Pathol. 2006;17:225–234. doi: 10.1385/ep:17:3:225. [DOI] [PubMed] [Google Scholar]
- 43.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarzab B, Wiench M, Fujarewicz K, Simek K, Jarzab M, et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 45.Weber F, Shen L, Aldred MA, Morrison CD, Frilling A, et al. Genetic classification of benign and malignant thyroid follicular neoplasia based on a three-gene combination. J Clin Endocrinol Metab. 2005;90:2512–2521. doi: 10.1210/jc.2004-2028. [DOI] [PubMed] [Google Scholar]
- 46.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudoit S, Fridlyand J. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol. 2002;3:1–21. doi: 10.1186/gb-2002-3-7-research0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Efron B, Tibshirani R. Improvments of cross-validation: the.632+ bootstrap method. 1997;92:548–560. [Google Scholar]
- 49.Kubat M. Decision trees can initialize radial-basis function networks. IEEE Trans Neural Netw. 1998;9:813–821. doi: 10.1109/72.712154. [DOI] [PubMed] [Google Scholar]
- 50.Vasko V, Ferrand M, Di CJ, Carayon P, Henry JF, et al. Specific pattern of RAS oncogene mutations in follicular thyroid tumors. J Clin Endocrinol Metab. 2003;88:2745–2752. doi: 10.1210/jc.2002-021186. [DOI] [PubMed] [Google Scholar]
- 51.Bourasseau I, Savagner F, Rodien P, Duquenne M, Reynier P, et al. No evidence of thyrotropin receptor and G(s alpha) gene mutation in high iodine uptake thyroid carcinoma. Thyroid. 2000;10:761–765. doi: 10.1089/thy.2000.10.761. [DOI] [PubMed] [Google Scholar]
- 52.Kroll TG, Sarraf P, Pecciarini L, Chen CJ, Mueller E, et al. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma. Science. 2000;289:1357–1360. doi: 10.1126/science.289.5483.1357. [DOI] [PubMed] [Google Scholar]