Abstract
Earth’s biota produces vast quantities of polymerized silica at ambient temperatures and pressures by mechanisms that are not understood. Silica spicules constitute 75% of the dry weight of the sponge Tethya aurantia, making this organism uniquely tractable for analyses of the proteins intimately associated with the biosilica. Each spicule contains a central protein filament, shown by x-ray diffraction to exhibit a highly regular, repeating structure. The protein filaments can be dissociated to yield three similar subunits, named silicatein α, β, and γ. The molecular weights and amino acid compositions of the three silicateins are similar, suggesting that they are members of a single protein family. The cDNA sequence of silicatein α, the most abundant of these subunits, reveals that this protein is highly similar to members of the cathepsin L and papain family of proteases. The cysteine at the active site in the proteases is replaced by serine in silicatein α, although the six cysteines that form disulfide bridges in the proteases are conserved. Silicatein α also contains unique tandem arrays of multiple hydroxyls. These structural features may help explain the mechanism of biosilicification and the recently discovered activity of the silicateins in promoting the condensation of silica and organically modified siloxane polymers (silicones) from the corresponding silicon alkoxides. They suggest the possibility of a dynamic role of the silicateins in silicification of the sponge spicule and offer the prospect of a new synthetic route to silica and siloxane polymers at low temperature and pressure and neutral pH.
Biosilicification, the biological formation of opal-like amorphous hydrated silica, occurs on a globally vast scale in a wide variety of organisms, including diatoms, sponges, mollusks, and higher plants (1–3). The exquisite species-specific control of silica architecture from the nanoscale to macroscopic dimensions has focused attention on the possible roles of the occluded biopolymers and membranes of surrounding structures, but details of the biosynthetic mechanisms controlling silica polymerization in living systems remain unknown (4–7). It is thought that elucidation of these mechanisms may help resolve the unexplained requirement for silicon in the formation of the mammalian skeleton (8) and may lead to the development of new low-temperature routes to the synthesis and patterning of high-performance composite materials based on silicon (9–14).
Siliceous sponges deposit silica in needle-like spicules that support the organism and provide defense against predation. These spicules, produced in membrane-enclosed vesicles within specialized cells known as sclerocytes (15), each contain an axial filament of protein (16). Because the protein filaments can be produced during silica deprivation, and unsilicified filaments can be found in sclerocytes (17), it was suggested that the axial filaments may serve as a template or otherwise direct silica deposition.
To help understand the molecular mechanisms controlling biosilicification, it is essential to characterize the proteins that are occluded within biologically silicified structures. We report here the isolation and characterization of proteins, which we have named silicateins (for silica proteins), from the occluded axial protein filaments of the siliceous spicules of the marine sponge Tethya aurantia, and the cloning and sequence analysis of the cDNA encoding silicatein α, the most abundant of the silicateins. The results reveal that the axial filaments are composed of three similar silicatein subunits in a regular repeating array. Silicatein α is shown to belong to the cathepsin L class of the papain-like cysteine protease superfamily, supporting the suggestion that enzymatic activity may be involved in biosilicification in sponges.
MATERIALS AND METHODS
Isolation of Spicules and Axial Filaments.
Specimens of the sponge Tethya aurantia was collected at 17 m depth off Santa Barbara County, California (latitude 34°25.331′N, longitude 119°57.142′W) in February to June, 1997. Sponges (about 20 g wet weight) were washed with seawater, diced into 1-cm cubes, and bleached with sodium hypochlorite solution until the fragments were colorless. Bleached material was washed with Milli-Q (Millipore) -purified water five times and then soaked in HNO3/H2SO4 (1:4) overnight. Acid-insoluble material (consisting principally of the silica spicules) was rinsed with Milli-Q water until the pH was above 6 and then washed twice with acetone. After air-drying, this material was treated to dissolve the silica with 100 ml of 2 M HF/8 M NH4F (pH 5) until no sediment was observed. The solution was dialyzed against 4 liters of Milli-Q water at 4°C for 4 h per dialysis, and the outer solution was changed 9 times. The dialysate was centrifuged at 10,000 × g for 20 min at 4°C. The sedimented protein filaments were suspended in Milli-Q water and stored at 4°C.
Electron Microscopy and X-Ray Diffraction.
Spicules and filaments were air dried, sputter coated with gold, and observed with a JEOL JSM63000F scanning electron microscope. The diffraction pattern was obtained from purified filaments oriented parallel to a glass substrate by using a Scintag x-ray diffractometer.
Protein Analyses.
Protein subunits from the filaments were analyzed by SDS/PAGE (18). Filaments (10 μg of protein) were suspended in sample buffer, incubated at 95°C for 5 min, and then applied to a 4–20% gradient polyacrylamide gel. After electrophoresis at 125 V for 2 h, gels were stained with Coomassie brilliant blue R250 to visualize the resolved proteins. For amino acid analyses, the intact filaments and the subunits separated by SDS/PAGE and electrophoretically transferred to poly(vinylidene difluoride) membranes (19) were hydrolyzed in 6 M HCl at 110°C for 24 h in vacuo and analyzed with an automated amino acid analyzer. Similarity of amino acid compositions was determined by the method of Cornish-Bowden (20). Amino-terminal and internal peptide sequences of silicatein α were obtained as follows: After negative staining of the SDS/PAGE gel containing 100 μg of filament protein with CuCl2 (21), the region corresponding to silicatein α was excised and destained, and the protein was recovered by electroelution with a Bio-Rad electro-eluter. Samples of the eluted and concentrated protein were incubated with endoproteinase Asp-N, Glu-C, or Lys-C (Sigma) for 18 h at 37°C. The resulting proteolytic fragments and the undigested protein were separated with SDS/PAGE using Tris/Tricine buffer, electrophoretically transferred onto polyvinylidene fluoride membranes, and analyzed for N-terminal sequences by using a gas-phase micro-Edman peptide sequencer.
Cloning and Characterization of cDNA.
To promote tissue regeneration and silicatein mRNA synthesis, live sponge was torn from the attached rock, and the fragments of tissue remaining attached to the rock were maintained in running seawater for 2 weeks. RNA was extracted from the regenerating sponge tissue by using TRIZOL solution (GIBCO/BRL). Reverse transcription and digestion of RNA with RNase H were performed with the Superscript preamplification system (GIBCO/BRL) with 5 μg of RNA according to the manufacturer’s instructions. The degenerate oligonucleotides CA(A/G) GG(A/C/G/T) GA(C/T) TG(C/T) GG(A/C/G/T) GC(A/C/G/T) TC and CC (A/C/G/T)GT (A/G/T)AT (A/C/G/T)AC CAT (A/C/G/T)GC (A/G)TG were synthesized from the partial protein sequences, QGDCGAS and HAMVITG, respectively, then were used for PCR amplification. PCR products containing the 3′ and 5′ regions of the silicatein α cDNA were obtained by rapid amplification of cDNA ends (RACE)–PCR (22). PCR products were blunt-ended, phosphorylated, and ligated into the SmaI site of plasmid pUC18 by standard methods, and both strands of the cloned inserts were sequenced by the dideoxynucleotide method (23). Northern hybridization was carried out using the Northern Max kit (Ambion) according to the manufacturer’s protocol, as follows: An EcoRI–PstI fragment of the cloned silicatein α sequence was subcloned into pGEM3Z; an antisense digoxigenin (DIG)-labeled RNA probe then was prepared with SP6 RNA polymerase from the plasmid linearized by EcoRI treatment. Hybridized RNA was detected by using the DIG detection system (Boehringer Mannheim).
RESULTS
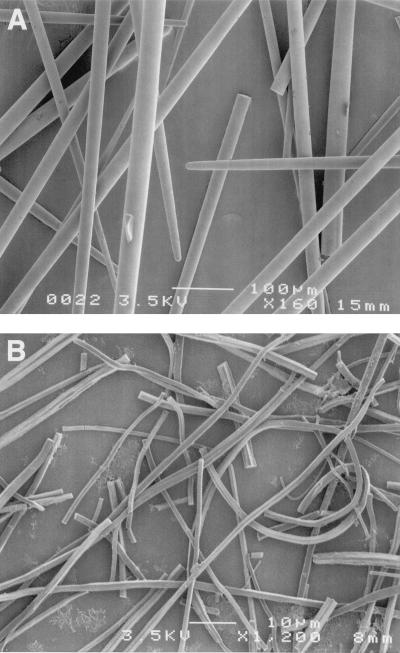
Silica spicules (Fig. 1A) were purified from the shallow-water Pacific sponge Tethya aurantia by centrifugation following sequential digestion with bleach and concentrated acids. This treatment removed all detectable protein from the outside of the spicules; no proteins were detectable when 10 mg of acid-cleaned spicules were boiled in SDS and the solution was analyzed by SDS/PAGE. The intact axial filaments then were purified (Fig. 1B) by dissolution of the silica with buffered HF, followed by centrifugation. The purified filaments contained no detectable remaining Si (≤0.002% by weight of protein detectable with molybdate assays). Dimensions of the silica spicules are typically about 2 mm length × 30 μm diameter, and the filaments are of the same length and 1 μm in diameter. The typical yields of spicules, filaments, and filament protein from the sponge are shown in Table 1. Silica spicules constitute 75% of the dry weight of the sponge, while the axial filament contributes only 0.1% of the mass of each spicule. Amino acid analyses after acid hydrolysis revealed that about 91% of the filament is protein.
Figure 1.
Scanning electron micrographs of isolated silica spicules (×130) (A) and axial filaments (×1,000) (B) from Tethya aurantia.
Table 1.
Yields of silica spicules, filaments, and filament protein from Tethya aurantia
| Material | Weight, g | Yield, %* |
|---|---|---|
| Sponge (wet) | 20.0 | |
| Sponge (dry) | 7.3 | 100 |
| Spicules | 5.5 | 75.3 |
| Filaments | 0.0058 | 0.11 |
| Protein | 0.0053 | 0.10 |
Yield was calculated relative to sponge dry weight as 100%. (Dry weight of the sponge was ca. 35% of the live wet weight).
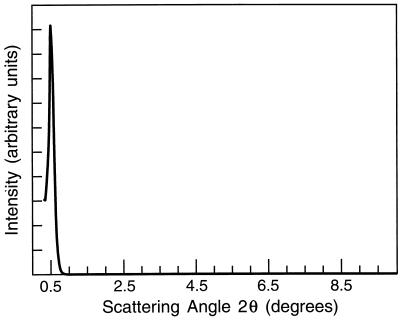
The filament proteins were dissociated and separated by SDS/PAGE (Fig. 2). When 10 μg of protein was analyzed electrophoretically, three bands, 29, 28, and 27 kDa, were observed; these were designated silicatein α, β, and γ, respectively. Densitometric analyses showed these proteins to be present in the relative proportions α:β:γ = 12:6:1. Amino acid analyses demonstrate that the compositions of the three proteins are highly similar (Table 2). Small-angle x-ray diffraction data (Fig. 3) reveal evidence for a highly regular, repeating structure within the filament (periodicity = 17.2 nm).
Figure 2.
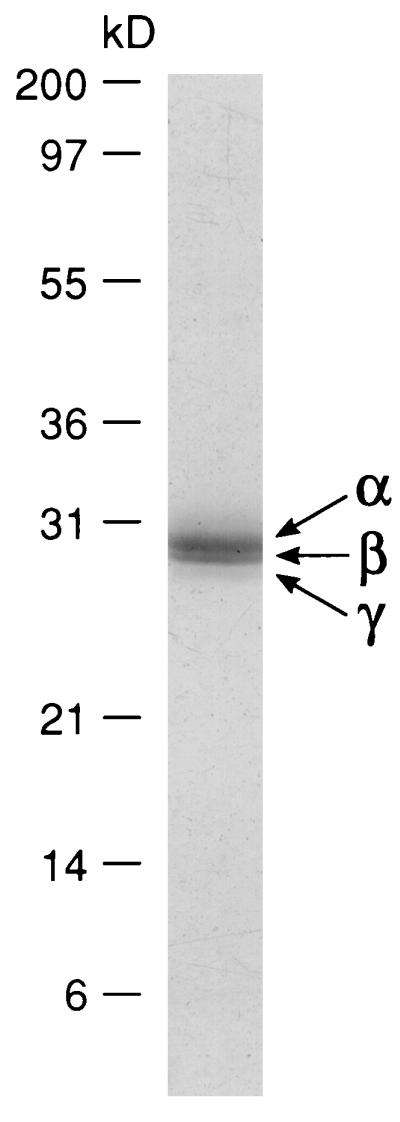
Electrophoretic profile of proteins from axial filaments. Filaments (10 μg of protein) were subjected to SDS/PAGE on a 4–20% gradient polyacrylamide gel followed by staining with Coomassie brilliant blue R250. Silicatein α, β, and γ are indicated by arrows. Numbers indicate molecular masses of standard proteins.
Table 2.
Amino acid compositions of total filament proteins and the purified silicateins
| Residue | Composition, mol %
|
|||
|---|---|---|---|---|
| Filament proteins | Silicatein α | Silicatein β | Silicatein γ | |
| Asx | 11.8 | 12.5 | 12.4 | 12.7 |
| Thr | 4.8 | 6.7 | 4.8 | 4.3 |
| Ser | 10.9 | 13.0 | 12.3 | 11.8 |
| Glx | 8.8 | 8.8 | 10.2 | 10.0 |
| Pro | 3.2 | 2.1 | 2.2 | 2.5 |
| Gly | 12.7 | 15.2 | 14.8 | 13.7 |
| Ala | 9.2 | 9.6 | 10.0 | 10.6 |
| Val | 6.4 | 4.1 | 5.7 | 6.3 |
| Met | 2.3 | 1.6 | 1.2 | 1.0 |
| Ile | 4.3 | 4.0 | 3.9 | 4.0 |
| Leu | 6.0 | 7.0 | 6.2 | 7.4 |
| Tyr | 7.9 | 6.3 | 6.6 | 7.8 |
| Phe | 2.8 | 2.4 | 2.5 | 2.2 |
| His | 1.3 | 0.8 | 0.5 | 0.4 |
| Lys | 4.4 | 3.3 | 3.7 | 3.8 |
| Arg | 3.1 | 2.6 | 2.9 | 1.6 |
Amino acid compositions (mol % exclusive of cysteine and tryptophan) were determined after acid hydrolysis (6 M HCl; 24 h at 110°C).
Figure 3.
X-ray diffraction of axial protein filaments. The diffraction pattern was obtained from filaments oriented parallel to a glass substrate by using a Scintag x-ray diffractometer. The peak at 2θ = 0.51° represents a periodicity of 17.245 nm.
To facilitate cloning and sequence analysis of the cDNA coding for silicatein α, the protein was purified and partial amino acid sequences were obtained from the N terminus and five purified proteolytic fragments as described in Materials and Methods. These sequences then were used to design degenerate oligonucleotide primers for amplification of the corresponding cDNA by standard reverse transcription–PCR and RACE methods, starting from mRNA purified from regenerating sponge tissue that was rapidly producing new silica spicules and silicatein mRNA. The amino acid sequence of silicatein α deduced from the full-length 1360-bp cDNA is shown in Fig. 4A. The N-terminal peptide and five internal sequences obtained directly from the purified protein are completely contained in this deduced sequence. The sequence contains 112 amino acid residues upstream from the N terminus of the mature protein; these include a typical 17-residue signal peptide that is presumed to mediate secretion, followed by a propeptide. Northern blot analysis of the sponge mRNA (Fig. 4B) shows the single silicatein α transcript to be 1.4 kb in length, demonstrating that the PCR products sequenced cover the entire cDNA.
Figure 4.
(A) Amino acid sequence of silicatein α deduced from cDNA. The putative signal peptidase cleavage site is marked with an arrow; the observed boundary between the propeptide and mature peptide is shown by an arrowhead. Amino acid sequences obtained from peptide sequencing are underlined as follows: solid line, N-terminal sequence; long-dashed, endoproteinase Asp-N fragments; short dashed, endoproteinase Glu-C fragment; and dotted, endoproteinase Lys-C fragment. (B) Northern hybridization analysis of silicatein α transcript. Total RNA prepared from regenerating sponge tissue was subjected to Northern blot analysis with digoxigenin-labeled RNA probe as described in the text.
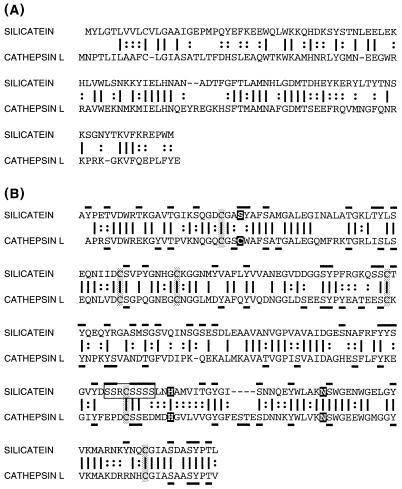
Analyses of sequence similarity revealed that silicatein α is a novel member of the cathepsin L subfamily of papain-like cysteine proteases (24). Alignment of silicatein α with human cathepsin L (Fig. 5) reveals that 45% of the corresponding amino acids are identical in the two full-length proteins, which differ in length by only three residues. When the regions corresponding to the mature protease and silicatein α are compared (after removal of the signal and propeptides), the sequence identity is 52%; correspondence of residues with biochemically identical or similar side chains is 75%. The same degree of similarity was found between silicatein α and the recently sequenced cathepsin L from the marine sponge Geodia cydonium (50% identity; 78% similarity) (25). Silicatein α showed less similarity to other members of the papain-like protease family (24), including papain (39% identity) and cathepsin B (28% identity).
Figure 5.
Alignment and comparison of silicatein α with human cathepsin L. (A) Propeptide regions. (B) Mature proteins. Identical amino acids are indicated by vertical bars; similar residues indicated by colons. Cysteine residues involved in disulfide bonds in cathepsin L are shaded. Catalytic triad amino acids of the active site of cathepsin L and corresponding amino acids in silicatein α are highlighted. Hydroxy amino acid residues in silicatein α and cathepsin L are overlined and underlined, respectively. Silicatein α-specific hydroxy amino acid cluster is boxed.
DISCUSSION
The high relative mass of silica spicules in the marine sponge Tethya aurantia made it possible for us to purify the spicules, the occluded axial protein filaments, and their constituent silicatein subunits in sufficient quantities for characterization of the proteins and determination of the sequence of silicatein α from the cloned cDNA. Results of SDS/PAGE and amino acid analyses demonstrate that the molecular weights and compositions of the silicateins α, β, and γ are highly similar, suggesting that the filaments are composed primarily of protein subunits of a single family. Small-angle x-ray diffraction revealed a regular, repeating structure within the filament (periodicity = 17.245 nm), as would be predicted if a simple repeating subunit structure underlies the macroscopic filament. This finding also is consistent with the electron micrographic evidence for paracrystallinity of the protein filaments from silica spicules of another sponge observed earlier (16).
Analyses of sequence similarity revealed that silicatein α is a novel member of the cathepsin L subfamily of papain-like cysteine proteases (24). Identities include the six cysteine residues that form intramolecular disulfide bridges in the protease, making it likely that the three-dimensional structure of silicatein α is similar to that of cathepsin L. The propeptides of silicatein α and the human and sponge cathepsin L molecules also are highly similar, further suggesting a common evolutionary origin of these proteins. Cathepsin L is a typical lysosomal proteolytic enzyme in both human and sponge tissues (25, 26), whereas silicatein α is presumably localized in the silicalemma, which forms the membrane-enclosed vesicle in which silica deposition occurs in the sponge sclerocytes (15). The relationship between these proteins thus apparently includes the nature of their subcellular localizations as well as their molecular structures, further supporting the suggestion of a common evolutionary ancestry.
Interestingly, the catalytic cysteine (sulfhydryl) residue at the active site of the proteases is replaced in silicatein α with a serine (hydroxyl). Consistent with this replacement, we have found that the silicateins do not display esterase activity when tested with synthetic chromogenic substrates. Similarly, a mutant form of cathepsin L in which the catalytic cysteine is replaced by serine also is inactive as a protease (27).
Although the calculated pIs of silicatein α and cathepsin L are identical (≈pH 5), there are fewer charged amino acids in silicatein α and somewhat more hydroxy amino acids than in the protease (20.3% in the mature silicatein vs. 17.2% in mature cathepsin L). This observation is consistent with our finding that the silicateins are released from the filament by treatment with SDS or urea, indicating that the subunits associate by means of nonionic, noncovalent interactions. The assembly of macroscopic filaments exhibiting a highly ordered nanostructure from protein subunits that are small and presumably globular appears to be a distinctive feature of the silicateins, not previously reported for cathepsin L or related proteases.
Clusters of the hydroxy amino acids—serine, tyrosine, and (to a lesser extent) threonine—constitute one of the most distinctive features of silicatein α. Serine-rich sequences including Ser-Ser-Cys-Thr-Tyr, Ser-Ser-Arg-Cys-Ser-Ser-Ser-Ser, two Ser-Xaa-Ser-Xaa-Ser sequences, and the Ser-Tyr sequence at the site of the serine-replaced active site of the protease are notable examples of the high localized concentrations of hydroxyls in this protein (Fig. 5).
The mechanism of biosilicification remains poorly understood, although Perry and Lu demonstrated that hydroxyl-rich polysaccharides and oligosaccharides that had been occluded in plant biosilicas are capable of accelerating silica polymerization from silicon catecholates (28). Hydroxyls also are abundant in the proteins associated with the silicified cell walls of diatoms (5, 6, 29), and these groups long have been proposed to participate in the organization of silicic acid precursors, either by hydrogen bonding or by covalent condensation with the elimination of water (4). Modeling of a multistep reaction sequence involving condensation of silicic acid with the hydroxyls of an organizing protein suggests thermodynamic favorability (30). A family of cDNAs coding for silica-associated proteins in the diatom cell wall recently has been cloned (29). One of these proteins contains 16.5% serine and threonine, primarily in regular structural domains, although the function of this protein is not yet known and silicatein α shows no significant similarity to it. A family of genes coding for silicon transporters also has been cloned from a diatom (31), and the encoded protein has been demonstrated to mediate the cellular uptake of dissolved silicon, presumably as silicic acid. It remains to be determined, however, whether the proximate precursor of intracellular polymerization of silica is the free silicic acid or an as-yet-unidentified organic conjugate as some have suggested (32–35). Because the biological synthesis of silica by the condensation of silicic acid monomers (or their conjugates such as alkoxides or esters) is formally similar to the reverse of the hydrolysis reaction catalyzed by the proteases, the structural similarities between the silicatein and protease may help explain the mechanism of biosilicification. We recently have found that the sponge silicatein filaments and subunits catalyze the polymerization of silicas and organosiloxanes (silicones) from silicon alkoxide precursors at neutral pH (unpublished results). This finding, in conjunction with the apparent conservation of structural features of their enzyme counterparts reported here, supports the possibility of a dynamic role of the silicateins in silicification of the sponge spicule, and raises the possibility of a new synthetic route to silica and siloxane polymers at low temperature and pressure and neutral pH.
Acknowledgments
We thank M. Brzezinski for helpful discussions and results of the molybdate assay. This work was supported by grants from the U.S. Army Research Office Multidisciplinary University Research Initiative (DAAH04-96-1-0443), the U.S. Office of Naval Research (N00014-93-1-0584), the Materials Research Division of the National Science Foundation (MCB-9202775), the NOAA National Sea Grant College Program, U.S. Department of Commerce, under Grant NA36RG0537, Project E/G-2, through the California Sea Grant College System, the Materials Research Science and Engineering Centers Program of the National Science Foundation under award DMR-96-32716 to the University of California Santa Barbara Materials Research Laboratory, and a donation from the Dow Corning Corporation.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF032117).
References
- 1.Lowenstam H A. Science. 1981;211:1126–1131. doi: 10.1126/science.7008198. [DOI] [PubMed] [Google Scholar]
- 2.Simpson T L, Volcani B E, editors. Silicon and Siliceous Structures in Biological Systems. New York: Springer; 1981. [Google Scholar]
- 3.Voronkov M G, Zelchan G I, Lukevits E J. Silicon and Life. 2nd Ed. Riga, Latvia: Zinatne; 1977. [Google Scholar]
- 4.Hecky R E, Mopper K, Kilham P, Degens E T. Mar Biol. 1973;19:323–331. [Google Scholar]
- 5.Swift D M, Wheeler A P. J Phycol. 1992;28:202–209. [Google Scholar]
- 6.Kröger N, Bergsdorf C, Sumper M. EMBO J. 1994;13:4676–4683. doi: 10.1002/j.1460-2075.1994.tb06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison C C. Phytochemistry. 1996;41:37–42. doi: 10.1016/0031-9422(95)00576-5. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz K. In: Biochemistry of Silicon and Related Problems. Bendz G, Lindqvist I, editors. New York: Plenum; 1977. pp. 207–230. [Google Scholar]
- 9.Mann S. Science. 1993;261:1286–1292. doi: 10.1126/science.261.5126.1286. [DOI] [PubMed] [Google Scholar]
- 10.Harrison C C, Loton N. J Chem Soc Faraday Trans. 1995;91:4287–4297. [Google Scholar]
- 11.Firouzi A, Kumar D,, Bull M, Besier T, Sieger P, Hyo Q, Walker S A, Zasadzinski J A, Glinka C, Nicol J, et al. Science. 1995;267:1138–1143. doi: 10.1126/science.7855591. [DOI] [PubMed] [Google Scholar]
- 12.Mann S. In: Biomimetic Materials Chemistry. Mann S, editor. New York: VCH; 1996. pp. 1–40. [Google Scholar]
- 13.Kuhn L T, Fink D J, Heuer A H. In: Biomimetic Materials Chemistry. Mann S, editor. New York: VCH; 1996. pp. 41–68. [Google Scholar]
- 14.Aksay I A, Staley J T, Prud’homme R K. In: Biomimetic Materials Chemistry. Mann S, editor. New York: VCH; 1996. pp. 361–378. [Google Scholar]
- 15.Simpson T L. The Cell Biology of Sponges. New York: Springer; 1984. [Google Scholar]
- 16.Garrone R. Phylogenesis of Connective Tissue. Basel: Karger; 1978. pp. 176–179. [Google Scholar]
- 17.Imsiecke G, Steffen R, Custodio M, Borojevic R, Müller W E G. In Vitro Cell Dev Biol. 1995;31:528–535. doi: 10.1007/BF02634030. [DOI] [PubMed] [Google Scholar]
- 18.Laemli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornish-Bowden A. Methods Enzymol. 1983;91:60–75. doi: 10.1016/s0076-6879(83)91011-x. [DOI] [PubMed] [Google Scholar]
- 21.Lee C, Levin A, Branton D. Anal Biochem. 1987;166:308–312. doi: 10.1016/0003-2697(87)90579-3. [DOI] [PubMed] [Google Scholar]
- 22.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanger F S, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berti P J, Storer A C. J Mol Biol. 1995;246:273–283. doi: 10.1006/jmbi.1994.0083. [DOI] [PubMed] [Google Scholar]
- 25.Krasko A, Gamulin V, Seack J, Steffen R, Schröder H C, Müller W E G. Mol Mar Biol Biotech. 1997;6:296–307. [PubMed] [Google Scholar]
- 26.Kirschke H, Langner J, Wiederanders B, Ansorge S, Bohley P. Eur J Biochem. 1977;74:293–301. doi: 10.1111/j.1432-1033.1977.tb11393.x. [DOI] [PubMed] [Google Scholar]
- 27.Coulombe R, Grochulski P, Sivaraman J, Ménard R, Mort J S, Cygler M. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 28.Perry C C, Lu Y. J Chem Soc Faraday Trans. 1992;88:2915–2921. [Google Scholar]
- 29.Kröger N, Lehmann G, Rachel R, Sumper M. Eur J Biochem. 1997;250:99–105. doi: 10.1111/j.1432-1033.1997.00099.x. [DOI] [PubMed] [Google Scholar]
- 30.Lobel K D, West J K, Hench L L. Mar Biol. 1996;126:353–360. [Google Scholar]
- 31.Hildebrand M, Volcani B E, Gassmann W, Schroeder J I. Nature (London) 1997;385:688–689. doi: 10.1038/385688b0. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya P, Volcani B E. Biochem Biophys Res Commun. 1983;114:365–372. doi: 10.1016/0006-291x(83)91636-4. [DOI] [PubMed] [Google Scholar]
- 33.Perry C C. In: Biomineralization: Chemical and Biochemical Perspectives. Mann S, Webb J, Williams R J P, editors. Weinheim, Germany: VCH; 1989. pp. 223–256. [Google Scholar]
- 34.Weiss A, Herzog A. In: Biochemistry of Silicon and Related Problems. Bendz G, Lindqvist I, editors. New York: Plenum; 1977. pp. 109–128. [Google Scholar]
- 35.Voronkov M G. In: Biochemistry of Silicon and Related Problems. Bendz G, Lindqvist I, editors. New York: Plenum; 1977. pp. 395–434. [Google Scholar]