Abstract
Bacteria can defend themselves against β-lactam antibiotics through the expression of class B β-lactamases, which cleave the β-lactam amide bond and render the molecule harmless. There are three subclasses of class B β-lactamases (B1, B2, and B3), all of which require Zn2+ for activity and can bind either one or two zinc ions. Whereas the B1 and B3 metallo-β-lactamases are most active as dizinc enzymes, subclass B2 enzymes, such as Aeromonas hydrophila CphA, are inhibited by the binding of a second zinc ion. We crystallized A. hydrophila CphA in order to determine the binding site of the inhibitory zinc ion. X-ray data from zinc-saturated crystals allowed us to solve the crystal structures of the dizinc forms of the wild-type enzyme and N220G mutant. The first zinc ion binds in the cysteine site, as previously determined for the monozinc form of the enzyme. The second zinc ion occupies a slightly modified histidine site, where the conserved His118 and His196 residues act as metal ligands. This atypical coordination sphere probably explains the rather high dissociation constant for the second zinc ion compared to those observed with enzymes of subclasses B1 and B3. Inhibition by the second zinc ion results from immobilization of the catalytically important His118 and His196 residues, as well as the folding of the Gly232-Asn233 loop into a position that covers the active site.
Class B β-lactamases (also called zinc β-lactamases or metallo-β-lactamases) play a key role in bacterial resistance to β-lactam antibiotics by efficiently catalyzing the hydrolysis of the β-lactam amide bond. The existence of such enzymes is a particular concern because they are effective against most β-lactam antibiotics (including the carbapenems), the corresponding genes are easily transferred between bacteria, and there are no clinically useful inhibitors. On the basis of the known sequences, three different lineages, identified as subclasses B1, B2, and B3, can be characterized (13, 14). All class B β-lactamases possess two potential zinc-binding sites and share a small number of conserved motifs bearing some of the residues that coordinate the zinc ion(s), notably His/Asn/Gln116-Xaa-His118-Xaa-Asp120 and Gly/Ala195-His196-Ser/Thr197 (14). Structural analysis of subclass B1 enzymes shows that one zinc ion has a tetrahedral coordination sphere involving His116, His118, His196, and a water molecule or OH− ion (histidine site), whereas the other has a trigonal-pyramidal coordination sphere involving Asp120, Cys221, His263, and two water molecules (cysteine site) (Table 1). One water molecule/hydroxide serves as a ligand for both metal ions (3). In the mononuclear structures of B1 enzymes (BcII, VIM-2, SPM-1, and VIM-4), the sole metal ion was found to be located in the histidine site (5, 16, 26; P. Lassaux, unpublished data). In some monozinc B1 structures, the Cys221 residue is found under an oxidized form. In subclass B3, the histidine site is similar to that found in subclass B1, but Cys221 is replaced by a serine residue and the second zinc ion is coordinated by Asp120, His121, His263, and the nucleophilic water molecule, which again forms a bridge between the two metal ions (Table 1) (33).
TABLE 1.
Composition of the two metal-binding sites in the three metallo-β-lactamase subclasses
| Enzyme | Histidine site | Cysteine site |
|---|---|---|
| B1 MβL | His116-His118-His196 | Asp120-Cys221-His263 |
| B2 MβL | His118-His196 | Asp120-Cys221-His263 |
| B3 MβL | His116-His118-His196 | Asp120-His121-His263 |
| B3 MβL GOB | (Gln116)-His118-His196a | Asp120-His121-His263 |
This histidine site is still putative, since the three-dimensional structure of the GOB enzyme is not available.
Aeromonas hydrophila CphA is a subclass B2 metallo-β-lactamase characterized by a uniquely narrow specificity. CphA efficiently hydrolyzes only carbapenems and shows very poor activity against penicillins and cephalosporins, in contrast to subclass B1 and B3 enzymes, which hydrolyze nearly all β-lactam compounds, with the exception of monobactams (11, 30). In contrast to subclass B1 and B3 enzymes, which are more active as dizinc species, subclass B2 is inhibited in a noncompetitive manner by the binding of a second zinc ion. For CphA, the dissociation constant of the second zinc ion (Kd2) is 46 μM at pH 6.5 (17). In agreement with extended X-ray absorption fine-structure studies (18) and site-directed mutagenesis (34), the crystallographic structure of CphA shows that the first zinc ion is in the cysteine site (15). However, the second binding site in subclass B2 enzymes remains to be determined because Garau and colleagues could not produce crystals of the dizinc form, despite the presence of zinc concentration well above Kd2 (15). The structure of Sfh-1, a subclass B2 enzyme from Serratia fonticola, has been solved recently and once again involves only one zinc ion in the active site (12). In subclass B2, the His116 residue found throughout the metallo-β-lactamase superfamily (with the exception of the B3 GOB enzymes in which Gln is present) is replaced by an asparagine (9, 13, 14). It has previously been shown that the Asn116 residue has no role in the binding of the zinc ions in CphA (34). On the basis of spectroscopic studies with the Aeromonas veronii cobalt-substituted ImiS enzyme, reports from the laboratory of Crawford and coworkers postulated that the second metal binding site was not the traditional histidine site but a site, remote from the active site, which involves both His118 and Met146 as zinc ligands (7, 8). However, a recent study of potential zinc ligand mutants contradicts this hypothesis and indicates that the position of the second zinc ion in CphA is probably equivalent to the histidine site observed with subclass B1 and B3 enzymes, with His118 and His 196 involved in the binding of this second zinc, with perhaps Cys221 (or Asn116 or Asp120) as the third ligand (4).
Since crystals of the wild-type CphA protein could not be obtained directly, single-site mutants were engineered by site-directed mutagenesis and overproduced. These mutants were selected in order to introduce residues that are conserved in either subclass B1 or B3, or both (2). Among these mutants, the N220G mutant (2) easily yielded crystals which were used as seeds to grow wild-type CphA crystals (15). The kinetic parameters of the wild-type and N220G mutant CphA enzymes were similar, indicating strong conservation of enzymatic properties in the mutant (2). However, the N220G mutation results in a slightly higher Kd2 value (86 μM) (2). We therefore set out to solve the crystallographic structures of the dizinc forms of the wild-type CphA enzyme and its N220G mutant. We confirmed that the second zinc ion occupies a slightly modified histidine site, in which the conserved His118 and His196 residues act as metal ligands. The implication of this discovery for what is known about the coordination of zinc ions by metallo-β-lactamases is discussed. We propose an explanation for the inhibition by the second zinc ion. Moreover, the role of the Asn233 residue in this inhibition phenomenon is also apprehended based on a site-directed mutagenesis study. In this work, a structural role for the zinc ions is also investigated.
MATERIALS AND METHODS
Protein purification and crystallization.
Site-directed mutagenesis, protein expression, and purification of the proteins were carried out as described previously (2), with the addition of a third purification step consisting of a size-exclusion chromatography (Hiload 16/60 Superdex 75 prep-grade column; Pharmacia Biotech). The purified enzyme solution was dialyzed against 15 mM sodium cacodylate (pH 6.5). Monodispersity of the protein solutions and the hydrodynamic radii of the dissolved protein molecules were checked by dynamic light scattering. The crystallization conditions used previously (15) were not easy to reproduce because the crystallization drop comprised two phases. Therefore, a screen was carried out to find new conditions. Initial screens were carried out at 8°C using commercial Hampton crystal screens 1 and 2, grid screen ammonium sulfate, and grid screen PEG 6000 (Hampton Research, CA). N220G CphA (10 mg/ml) was crystallized from 2.2 to 2.4 M ammonium sulfate and 0.1 M morpholineethanesulfonic acid (pH 6.0 to 6.5), using the sitting-drop method with new crystallization plates designed by Taorad GmbH (Aachen, Germany). The reservoir solution (1 μl) was mixed with the protein solution (1 μl), and the mixture was left to equilibrate against the solution reservoir. Typically, orthorhombic crystals of the N220G mutant grew within a few days to dimensions of 80 by 100 by 100 μm and belonged to space group C2221 (a = 42.68 Å, b = 101.06 Å, and c = 116.82 Å). Crystals of the wild-type CphA enzyme were obtained under similar conditions using mutant microcrystals as starting seeds and showed similar orthorhombic symmetry in space group C2221 (a = 42.80 Å, b = 101.50 Å, and c = 116.49 Å). Before data collection, 1 μl of 100 mM ZnCl2 was added to the drops containing the wild-type and the mutant crystals, and the crystals were soaked for 1 day.
Data collection and processing.
For data collection, wild-type and mutant crystals were transferred to a cryoprotectant solution containing reservoir solution supplemented with 30% (vol/vol) glycerol. The mounted crystals were flash-frozen in a liquid nitrogen stream. Near-complete X-ray data sets were collected using a Bruker FR591 rotating anode X-ray generator and a Mar345dtb detector. Diffraction data were processed with XDS (21) and scaled with SCALA from the CCP4 suite (6).
Structure determination and refinement.
The structures of dizinc variants from the wild-type and N220G mutant enzymes were solved using a molecular replacement approach with Molrep (6) and the monozinc structure of CphA (Protein Data Bank [PDB] number 1X8G) as a starting model. The resulting model was rebuilt with ARP/wARP (29). The structures were refined in a cyclic process, including manual inspection of the electron density with Coot (10) and refinement with Refmac (27). The refinement to convergence was carried out with isotropic B values and using a translation/libration/screw parameter (28). The positions of the zinc, sulfate, and chloride ions were verified by calculating anomalous maps, and the occupancies of the ions were determined by the disappearing of the difference electron density. Alternative conformations were modeled for a number of side chains, and occupancies were adjusted to yield similar B values for the disordered conformations.
CD spectroscopy.
Circular dichroism (CD) spectra for the apo-, mono-, and dizinc forms of the wild-type and N220G enzymes (0.3 mg/ml) were obtained using a JASCO J-810 spectropolarimeter. The apoenzymes were obtained in the presence of 10 mM EDTA. The dizinc forms were obtained in the presence of 500 μM Zn2+. The spectra were scanned at 25°C with 1-nm steps from 200 to 250 nm (far UV). Thermal stability was assessed for the apo-, mono-, and dizinc forms of the wild-type and N220G proteins using CD spectroscopy at 220 nm with increasing temperature (25°C to 85°C). Urea denaturation studies of the wild-type and N220G enzymes were also carried out in the presence and absence of 500 μM Zn2+. The mono- and dizinc enzymes were incubated for 18 h in increasing concentrations of urea (0 to 8 M) at 4°C. Stability was assessed for the mono- and dizinc forms of both proteins using CD spectroscopy at 220 nm, with increasing concentrations of urea.
Thermal shift assay.
Thermal shift assays were carried out using a LightCycler real-time PCR instrument (Roche Diagnostics). Briefly, 10 μl of wild-type CphA enzyme or the N220G mutant (0.3 mg/ml) was mixed with 10 μl of 5,000× Sypro Orange (Molecular Probes) diluted 1:500 in 15 mM cacodylate (pH 6.5). Samples were heat denatured from 25 to 90°C at a rate of 0.5°C per minute. Protein thermal unfolding curves were monitored by detecting changes in Sypro Orange fluorescence. The inflection point of the fluorescence-versus-temperature curves was identified by plotting the first derivative over the temperature, and the minima were referred to as the melting temperature (Tm). Buffer fluorescence was used as the control.
Differential scanning calorimetry.
Thermal denaturation was investigated using an NDSC 6100 differential scanning calorimeter (Calorimetry Sciences Corp., Lindon, UT) after dialysis of the N220G enzyme sample (1 mg/ml) against 15 mM cacodylate (pH 6.5). The dialysis buffer was used as a reference. The data were collected and analyzed with an MCS Observer software package, assuming a two-state folding mechanism.
Analysis of the N233A mutant.
The QuikChange site-directed mutagenesis kit (Stratagene Inc., La Jolla, CA) was used to introduce the N233A mutation into CphA, using pET9a-CphA wild type as the template, forward primer (5′-GGAGAAGCTGGGCGCCCTGAGCTTTGCCG-3′), and reverse primer (5′-CGGCAAAGCTCAGGGCGCCCAGCTTCTCC-3′). The vector was then introduced into Escherichia coli strain BL21(DE3) pLysS Star. Overexpression and purification of the mutant protein; CD spectroscopy; electrospray ionization-mass spectrometry; and the determination of metal content, kinetic parameters, and residual activity in the presence of increasing concentrations of zinc were carried out as previously described (2, 4, 34).
Protein structure accession numbers.
Coordinates and structure factors were deposited in the PDB using accession numbers 3F90 (dizinc form of wild-type CphA) and 3FAI (dizinc form of the N220G mutant).
RESULTS
Structure determination.
The crystal structures of the zinc-saturated wild-type and N220G CphA enzymes were solved. Stereochemical parameters were calculated by PROCHECK (22) and WHAT_CHECK (19) and fell within the range expected for a structure with similar resolution. The crystallographic and model statistics for the dizinc form of the wild-type and N220G mutant structures are shown in Table 2.
TABLE 2.
X-ray data collection and structure refinement
| Parameter | Value(s)
|
|
|---|---|---|
| CphA WT dizinca | CphA N220G dizinc | |
| Data statistics | ||
| Resolution range (Å) | 19.7-2.03 | 19.6-1.70 |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Unit cell parameters | ||
| Space group | C2221 | C2221 |
| a (Å) | 42.80 | 42.68 |
| b (Å) | 101.05 | 101.06 |
| c (Å) | 116.49 | 116.83 |
| Rsym | 0.062 | 0.032 |
| No. of observed reflections | 235,065 | 206,898 |
| No. of unique reflections | 16,754 | 28,046 |
| Completeness (%) | 99.7 | 99.7 |
| I/σ(I) | 34.4 | 42.1 |
| Multiplicity | 14 | 7.4 |
| B (Wilson)b | 15.69 | 11.96 |
| Refinement statistics | ||
| No. of reflections used | 16,754 | 28,046 |
| No. of molecules/asymmetric unit | 1 | 1 |
| Rworking | 0.1568 | 0.1388 |
| Rfree | 0.2020 | 0.16024 |
| No. of residues in disallowed region | 1 | 1 |
| RMSD from ideal | ||
| Bonds (Å) | 0.011 | 0.011 |
| Angles (°) | 1.217 | 1.550 |
| Mean B factor | 9.81 | 7.84 |
| No. of atoms (non-H) | 2,090 | 2,198 |
| No. of atoms (protein) | 1,850 | 1,882 |
| No. of H20 molecules | 202 | 252 |
| Other molecules/ions (no.) | ||
| Zn2+ | 3 | 3 |
| SO4 | 4 | 7 |
| CO3 | 2 | |
| Cl− | 7 | 7 |
| Glycerol | 3 | |
WT, wild type.
Value resulting from the Wilson plot.
Dizinc form of the wild-type CphA metallo-β-lactamase.
The crystal structure of the dizinc form of the wild-type CphA enzyme was solved by molecular replacement using the structure of the monozinc form (PDB number 1X8G) as the starting model. The structure was refined to a resolution of 2.03 Å. The Rwork and Rfree values for the refined structure were 0.1589 and 0.2020, respectively. The crystals adopted a C2221 space group, with one molecule in the asymmetric unit. Only one residue (Ala195) was found in a disallowed region of the Ramachandran plot. Ala195 (ψ = 105°; φ = 155°) is located on the loop between strands β8 and β9, with His196 at its apex.
The model of the wild-type dizinc structure includes 226 amino acid residues (41 to 312, according to the BBL numbering [14]). The last serine residue at the C terminus is missing. The electron density was good enough to identify three zinc ions (with two in the active site and one on the surface) and a sulfate ion in the active site. The composition of the visible solvent sphere is mentioned in Table 2. The Gly232-Phe236 mobile loop region had a low electron density, and the main chain for the residues was modeled in two conformations. The Phe236 side chain was not visible in the electron density map, and other side chains were modeled with alternative conformations.
CphA shows the typical αββα fold of the metallo-β-lactamase superfamily. The overall fold is not altered by the presence of the second zinc. The dizinc form has an average root mean square deviation (RMSD) value of 0.272 Å for backbone atoms with respect to the monozinc form.
The positions of the zinc ions were verified at the level of the electron density and by using anomalous signals. Both zinc ions were refined to a 100% occupancy and show an increased mobility. This is indicated by the B factors (17.68 and 20.63 for the first and second zinc ions, respectively) lying over the mean B factor (9.81). The first zinc ion is found in the Asp120-Cys221-His263 site or cysteine site as previously observed with the monozinc form (15) (Fig. 1). The distances between the zinc ion and the Asp120 carboxyl oxygen atom, the Cys221 sulfur atom, and the His263 side chain nitrogen atom are 2.00, 2.26, and 2.09 Å, respectively. In the monozinc form, these distances were 1.96, 2.20, and 2.05 Å, respectively (15). The tetrahedral coordination sphere is completed by a sulfate ion from the crystallization solution at a distance of 2.04 Å.
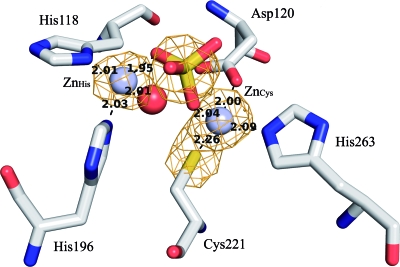
FIG. 1.
Structure of the wild-type CphA metal-binding sites. Zinc ions and water molecule are shown as gray and red spheres, respectively. ZnHis is the zinc ion that binds in the histidine site, while ZnCys is the zinc ion that binds in the cysteine site. The anomalous map at a contour level of 4 σ is shown in orange. Relevant distances between zinc ions and ligands are indicated.
The second zinc ion is coordinated by the His118 and His196 residues. Both histidine residues belong to the conserved histidine site in metallo-β-lactamases. The distances between the zinc ion and the His118 ND1 and the His196 NE2 are 2.01 and 2.03 Å, respectively. In CphA, the third ligand His116 is not conserved and is replaced by an asparagine residue, which is not involved in the binding of the second zinc (the distance is more than 4 Å). A sulfate ion (1.95 Å) and a water molecule (2.01Å) complete the vacant coordination positions of the zinc ion in the histidine site, forming a tetrahedral coordination sphere. The sulfate ion acts as a bridging agent between the zinc ions (Fig. 1). The distances between the sulfate ion and zinc in the cysteine site and zinc in the histidine site are 2.04 and 1.95 Å, respectively. The distance between both zinc ions is 4.08 Å.
The third observed zinc ion was found on the surface, away from the active site (about 17 Å). It is coordinated by His289 (2.09 Å from His NE), and the coordination sphere is completed by three chloride ions (2.19, 2.25, and 3.17 Å).
Dizinc form of the N220G mutant.
The crystal structure of the dizinc form of N220G was solved by molecular replacement based on the monozinc structure (PDB number 1X8G). The structure was refined to 1.70 Å with Rwork and Rfree values of 0.1308 and 0.1602, respectively.
The model of the N220G dizinc structure included all the 227 residues of the protein (41 to 313 according to the BBL numbering [14]), three zinc ions (two in the active site and one on the surface), and one sulfate ion located in the active site. The complete content of the solvent sphere is described in Table 2. The C-terminal residues were highly flexible. Ala195 was found in a disallowed region of the Ramachandran plot. The electron density for the Gly232-Phe236 mobile loop region allowed a well-defined model to be constructed.
The active site of the N220G mutant with two bonded zinc ions had a conformation nearly identical to that of the wild-type dizinc enzyme. The RMSD for all main chain atoms was 0.216 Å. In contrast to the zinc ion that occupied two sites 1.5 Å apart in the monozinc form of the N220G mutant (15), in the dizinc form of the enzyme described here, the zinc ion was in the classical cysteine site, with distances to ligands identical to those in the wild-type structure. Also, a sulfate ion and a water molecule (both with 50% occupancy) were included in the coordination sphere at distances of 2.04 and 2.27 Å, respectively. Under the new crystallization conditions described above, a monozinc form of the N220G mutant, in which a fully occupied zinc ion was present in the canonical cysteine site, was also obtained (data not shown).
The second zinc ion could not be modeled with an occupancy higher than 0.8. This second zinc ion shows a B factor of 9.85, nearly the same as the mean B factor, 7.84, and in the same range as the B factor of the zinc ion in the cysteine site, 10.50. This zinc ion was coordinated by His118 and His196. The coordination sphere was completed by a water molecule (2.02 Å) and the sulfate ion (2.15 Å). An additional water molecule was visible at a distance of 2.54 Å.
The third zinc ion was found in nearly the same position as that in the wild-type structure. Here, the zinc ion and the coordinating chloride ions exhibited 50% occupancy. The coordination sphere was affected by a water molecule and a disordered side chain from Glu292.
Comparison between the mono- and dizinc structures of CphA.
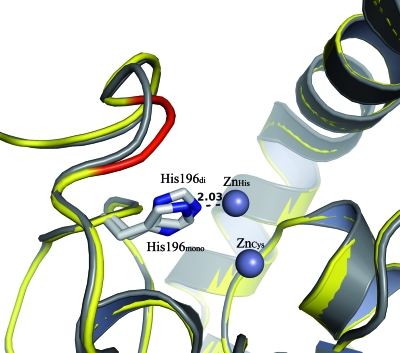
The binding of the second zinc ion in the active site has no influence on the global structure of CphA. This is confirmed by the low RMSD values obtained by superposition of the mono- and dizinc forms. The differences between the mono- and dizinc structures are limited to the Gly232-Asn233 loop region (Fig. 2). This loop, located at the entrance to the active site, is known to be stabilized upon the binding of the substrate (15) or inhibitors (20, 23). For the wild-type dizinc structure, this loop can exist in two conformations (both half occupied), corresponding, respectively, to the open form (as in the monozinc structures 1X8G and 1X8H) and the closed form (as in structures with substrate or inhibitor in the active site, 1X8I, 2QDS, and 2GKL). In the dizinc form of the N220G mutant, this loop is found in the closed form (Fig. 2). The closed conformation is stabilized by additional H bonds between the Asn233 side chain and the Ser235 side-chain oxygen and backbone nitrogen.
FIG. 2.
Superposition of the mono- (gray) and dizinc (yellow) forms of the N220G CphA enzyme. The Gly232-Asn233 loop region of the dizinc form is represented in red to underline the conformational change. The positions of the His196 residue in the monoform (His196mono) and in the dizinc form (His196di) are represented. The zinc ions in the histidine site (ZnHis) and in the cysteine site (ZnCys) are shown as gray spheres. The distance between His196di and the zinc ion located in the histidine site is indicated.
The active site of the dizinc forms can be superimposed nearly perfectly over all the previously available CphA structures (PDB numbers 1X8G, 1X8H, 1X8I, 2QDS, and 2GKL). For example, the RMSD is 0.56 Å for the superposition of the cysteine and histidine sites over their counterparts in 1X8G. The positions of the zinc in the cysteine sites are nearly identical, with the exception of that of the monozinc N220G mutant, in which a disordered zinc ion was observed (15). The coordinating His118 residue in the histidine site is not affected by the binding of the second zinc. In contrast, the His196 residue displays a slight flexibility. It is noticeable that the orientation of the imidazole ring of His196 in all X-ray structures is selected to achieve an optimal pattern of contacts in the coordination sphere (H bonds and zinc binding). Therefore, in dizinc structures, the His196 imidazole ring is rotated 180° compared to its position in the monozinc structures (Fig. 2).
Comparison with the active sites in B1 and B3 metallo-β-lactamases.
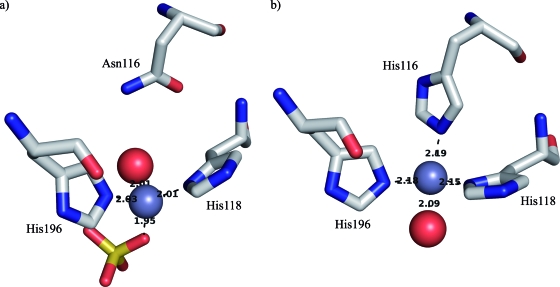
The active sites of all three metallo-β-lactamase subclasses show a high degree of correlation (RMSD values between 0.5 Å and 1.6 Å). In subclass B2, the coordination sphere of the zinc in the histidine site is altered by the substitution of His116 by an asparagine residue. In subclasses B1 and B3, the His116 residue is involved in the binding of the zinc ion in the histidine site (Fig. 3). Removing one coordinating residue causes this zinc ion to shift by 1.12, 1.03, 1.03, and 0.92 Å compared to the BcII (PDB number 1BVT) (Fig. 3), VIM-2 (PDB number 1KO3), VIM-2 (PDB number 1KO2), and L1 (PDB number 2FM6) structures, respectively. The position of the zinc in the cysteine site is nearly unchanged compared to the available structures of subclass B1 enzymes.
FIG. 3.
The histidine site in CphA (a) and in BcII (PDB number 1BVT) (b). Zinc ions and water molecules are represented as gray and red spheres, respectively.
Stability study.
As already reported (17), the helical contents of the apo-, mono-, and dizinc forms of the wild-type CphA enzyme are similar. The apparent Tm values determined by CD spectroscopy were 46.9, 58.7, and 61.6°C for the apo-, mono-, and dizinc forms, respectively. Monozinc wild-type CphA is also less stable than the dizinc form with urea as a denaturing reagent. Transitions between native and denatured states occurred at 3.4 and 4.4 M urea for the mono- and dizinc forms, respectively (data not shown).
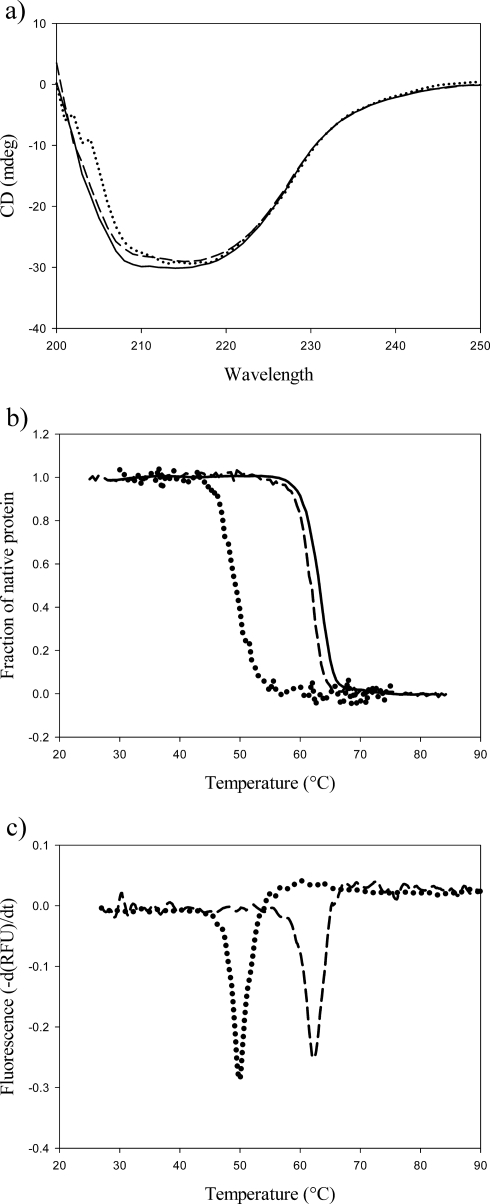
The far-UV CD spectra of the apo-, mono-, and dizinc forms of the N220G mutant exhibited small but significant differences (Fig. 4a). However, as observed for the wild-type enzyme, the helical content did not vary significantly. Thermal denaturation was irreversible in all cases. The apparent Tm values determined by CD spectroscopy were 49.2, 61.7, and 62.9°C for the apo-, mono-, and dizinc forms, respectively (Fig. 4b). Also, the apparent Tm value determined for the monozinc form of N220G using a thermal shift assay (Fig. 4c) and by differential scanning calorimetry was 62°C but decreased to 50°C in the presence of 10 mM EDTA (Fig. 4c). The monozinc form of the N220G mutant was slightly less stable than the dizinc form with urea as the denaturing reagent. Transitions between native and denatured states occurred at 4.1 and 4.7 M urea for the mono- and dizinc forms, respectively (data not shown).
FIG. 4.
Structural studies of the N220G mutant. (a) Circular dichroism spectra of the apo-enzyme (dotted line), monozinc (dashed line), and dizinc forms (solid line) of the N220G enzyme (0.3 mg/ml in both cases). (b) Thermal denaturation of the N220G mutant (0.5 mg/ml); fraction of native protein versus temperature (°C). Apo-N220G (dotted line), monozinc N220G (dashed line), dizinc N220G (solid line). (c) Thermal shift assay for N220G (0.3 mg/ml). N220G plus 10 mM EDTA (dotted line) and monozinc N220G (dashed line).
Is the closed position of the Gly232-Asn233 loop important for inhibition by the second zinc ion? Analysis of the N233A mutant.
An Asn233 residue is present in all metallo-β-lactamases, with the exception of the L1, BlaB, and IND-1 enzymes (14). In CphA, the binding of a carbapenem substrate or inhibitor modifies the Asn233 ψ angle so that its side chain closes the entrance of the active site in a conformation stabilized by the formation of an H bond between the oxygen of the Asn233 side chain and the hydroxyl of Ser235 (15, 20, 23). In the structure of the dizinc form of CphA, the loop adopts the closed position, even in the absence of substrate, thus hindering access to the active site (Fig. 2). To explore the role of the Asn233 residue in the inhibition phenomenon, the N233A mutant was expressed in Escherichia coli BL21(DE3) pLysS Star, purified to homogeneity, and characterized. The mass of the protein was verified by electrospray ionization mass spectrometry. Within experimental error, the mutant was found to exhibit the expected mass (25,144 Da measured versus 25,146 Da expected). The far-UV CD spectrum of the mutant enzyme showed the same α/β ratio as that of the wild type. The enzyme was stored in 15 mM sodium cacodylate (pH 6.5) with <0.4 μM free zinc. Under these conditions, the N233A protein contained one zinc ion per molecule, as reported for the wild-type β-lactamase. The activity of the mutant was measured in the absence of added Zn2+ (<0.4 μM) with imipenem, nitrocefin, and CENTA (Table 3). The N233A mutant showed an increased Km value for imipenem. In contrast to the wild-type enzyme, the N233A mutant was able to hydrolyze (albeit rather poorly) both cephalosporins, with quite low Km values. The hydrolysis of imipenem by N233A was inhibited by the binding of the second zinc ion, but the Kd2 value decreased to 11 ± 2 μM compared to 46 μM for the wild-type enzyme.
TABLE 3.
Kinetic parameters of the wild-type and N233A CphA enzymesa
| β-Lactam antibiotic | WT
|
N233A
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | |
| Imipenem | 1,200 | 340 | 3,500,000 | >700 | >1,000 | 700,000 |
| Nitrocefin | 0.008 | 1,300 | 6 | 0.03 | 55 | 520 |
| CENTA | NH | NH | NH | 0.0035 | 14 | 250 |
All the experiments were performed at 30°C in 15 mM cacodylate (pH 6.5) with <0.4 μM Zn2+. Standard errors were below 10%. NH, no observed hydrolysis; WT, wild type.
DISCUSSION
There are three subclasses of metallo-β-lactamases (B1, B2, and B3), all of which require Zn2+ for activity and can bind either one or two zinc ions (13, 14). Subclass B2 metallo-β-lactamases are active only in the monozinc form, because the binding of a second zinc ion inhibits the enzyme in a noncompetitive manner (17). This behavior contrasts with that of the B1 and B3 subclasses, in which the presence of a second zinc ion in the active site has either little effect or facilitates enzyme activity. Since it has thus far proven difficult to determine the structure of the dizinc form of a subclass B2 β-lactamase to investigate the basis of this inhibitory effect, we set out to crystallize the dizinc form of Aeromonas hydrophila CphA and solve its structure.
Previous attempts to determine the crystal structure of dizinc A. hydrophila CphA have not been successful, even with a high concentration of zinc in the mother liquor (15). It was proposed that this might reflect the conditions used to grow crystals and/or the presence of a carbonate ion in the active site. Also, it was suggested that the binding of the second zinc ion required a change of active site conformation impossible to obtain in the crystal. Indeed, nuclear magnetic resonance measurements indicate major conformation changes upon binding of the second zinc ion (C. Damblon, personal communication). However, CD spectra showed that the secondary structures of the monozinc forms of the wild-type (17) and N220G CphA enzymes are only slightly different from those of the dizinc forms. By modifying the crystallization conditions, we succeeded in obtaining CphA crystals with two zinc ions in the active site. Moreover, the overall αββα conformation of the enzyme was not affected by the presence of a second zinc ion. The dizinc form of CphA was obtained by soaking the crystal in a solution containing a zinc concentration well above the Kd2 value. In the crystal, major movements leading to the significant unfolding of the protein as observed by nuclear magnetic resonance are impossible.
In the monozinc form, a carbonate ion was present in the active site (15). In our structures, a sulfate ion from the crystallization solution was bound to both zinc ions.
As previously postulated (4, 34), the second inhibitory zinc ion is located in a slightly modified histidine binding site, with the two conserved His118 and His196 residues as ligands. The Met146 residue, proposed as one of the inhibitory zinc ion ligands together with His118 in the ImiS metallo-β-lactamase (7), was 12.54 Å from this ion in CphA, and the sulfur atom points in the opposite direction. Also, the space between the His118 and Met146 residues is occupied by the backbone of the Asn114-His118 loop, connecting strand β6 and helix α2, and thus does not allow the accommodation of a zinc ion. The sequence of ImiS is 96% identical to that of CphA, so it would be remarkable if these few substitutions produced such a radically different mechanism for the binding of the inhibitory zinc ion.
This atypical coordination sphere with only two of the three conserved histidine residues (Fig. 3) could probably explain the rather high value of the dissociation constant for the histidine site in CphA (46 μM) compared to that in subclasses B1 and B3. The value of the dissociation constant for the histidine site is 1.8 nM for BcII (a subclass B1 enzyme), and subclass B3 enzymes have high affinity constants for both binding sites (Kd < 6 nM) (35).
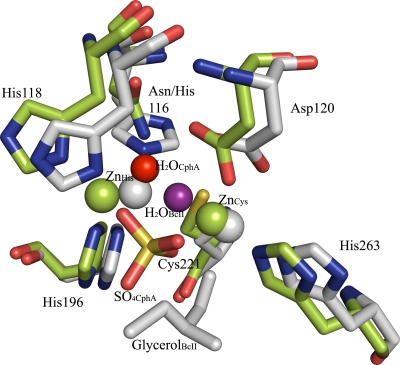
Vanhove et al. proposed an explanation for the inhibition of subclass B2 enzymes by the second zinc ion. The side chain of the Cys221, which is essential for binding the first zinc ion, would be displaced by the binding of the second zinc ion (34). However, a comparison between mono- and dizinc structures showed that this residue was not displaced. On the basis of the results obtained here, we can confirm that the binding of the inhibitory zinc ion to the catalytically important His118 and His196 residues prevents them from playing their catalytic roles in the hydrolysis of carbapenems (4, 15), in which His118 is the general base which activates the hydrolytic water molecule and His196 contributes to the oxyanion hole by forming an H bond with the carbonyl oxygen of the β-lactam bond (15). This model is supported by the very weak activities of the H118A and H196A mutants (4). This conclusion reinforces the hypothesis that the nucleophile is not adequately metal activated in B2 β-lactamases (31, 36). Superimposition of the structures of the dizinc forms of BcII (B1) and CphA shows that in the latter, the water molecule does not bridge the two zinc ions but is in interaction only with that in the histidine site (Fig. 5). It is possible that an interaction with this sole zinc ion does not sufficiently decrease the water pKa, so the concentration of OH− ions remains very low. Moreover, His118, which now serves as a zinc ligand, can no longer play the role of a general base. The closure of the Gly232-Asn233 loop in the dizinc form could also help to inhibit the enzyme, but Asn233 probably plays only a minor role since the N233A mutant is still inhibited by the binding of the second zinc ion. Finally, a third zinc ion is bound to a superficial histidine residue, but this is unlikely to be involved in the inhibition phenomenon. Its binding is probably due to the high zinc concentration utilized.
FIG. 5.
Superimposition of the structures of the dizinc forms of BcII (PDB number 2BFK) and CphA. Zinc ligands of BcII and CphA are represented as gray and green sticks, respectively. Zinc ions of BcII and CphA are shown as gray and green spheres, respectively. ZnHis is the zinc ion that binds in the histidine site, while ZnCys is the zinc ion that binds in the cysteine site. Water molecules of BcII and CphA are shown as purple and red spheres, respectively. The sulfate ion that bridges the two zinc ions in the CphA structure (SO4CphA) is also represented, as well as glycerol, the fifth ligand of ZnCys in BcII (glycerolBcII).
Whereas the N233S mutant of ImiS exhibits kinetic parameters similar to those of the wild-type enzyme (31), the N233A mutant of CphA shows an increased Km value for imipenem and thus seems to play a role in the binding of this substrate. Two-thirds of all sequenced metallo-β-lactamases have an Asn at position 233 (13, 14), and this residue was shown to be involved in substrate binding and activation by interacting electrostatically with the substrate β-lactam carbonyl (37). The same role for the Asn233 residue in substrate binding by CphA was already predicted by computational modeling (36) and is in agreement with our results.
In contrast to the zinc ion located in the cysteine site, the second zinc ion does not play an important structural role. Indeed, the binding of the first zinc ion stabilizes the wild-type and N220G CphA enzymes significantly, whereas the second zinc ion has a much more limited impact on stability. Both the mono- and dizinc forms of the N220G mutant are somewhat more stable than their wild-type counterparts, perhaps explaining why crystals of the N220G mutant are obtained more easily than those of the wild-type enzyme.
In conclusion, the catalytic metal ion is located in the cysteine site of CphA, and the second inhibitory zinc ion binds to a slightly modified histidine site. The histidine site had first been considered the catalytic site in the mononuclear metallo-β-lactamases. Indeed, in the first reported monozinc structures (B1 enzymes, such as BcII, VIM-2, VIM-4, and SPM-1), the single metal ion was shown to be located in the histidine site (5, 16, 26). In contrast, a B3 enzyme called GOB was shown to be active as a monozinc enzyme with the metal ion located in the cysteine site (25). Recently, reports from the laboratory of Vila and coworkers proposed that B1 enzymes can be active in their mono- and dizinc forms and that in monocobalt BcII, the metal ion is localized in the cysteine site (24, 32). Two mononuclear mutants of BcII in which each of the metal binding sites was selectively removed produced inactive variants. They concluded that the mononuclear form can be active only if assisted by residues at the other binding site (1). According to the model proposed by Tioni et al. (32), the metal ion in the histidine site would activate the hydrolytic water molecule in the dizinc enzyme. Reports from the laboratory of Vila and coworkers proposed that this is facilitated by a net of H-bond interactions in the mononuclear enzyme (24, 32), including probably His118 or Asp120, as already proposed for CphA (4, 15, 36). However, Asp120 appears unlikely because it already coordinates a zinc ion. In subclass B2, as previously postulated by Vanhove et al. (34), Asn116 is not involved in the binding of the second metal ion, but the N116H (34) and N116H-N220G (2) mutants could have a reconstituted His116-His118-His196 site and behave similarly to B1 enzymes. Indeed, the N116H-N220G mutant has an extended substrate spectrum, and its dizinc form is active (2).
The crystal structure of the subclass B2 CphA β-lactamase in its metal-inhibited form thus provides an answer to the structural characterization of the inhibitory site that has been elusive and controversial so far.
Acknowledgments
The work in Liège was supported by the Belgian Federal Government (PAI P5/33) and grants from the FNRS (Brussels, Belgium; FRFC grant no. 2.4511.06 and Lot. Nat. 9.4538.03). C.B. is an FRS/FNRS postdoctoral researcher.
Footnotes
Published ahead of print on 3 August 2009.
REFERENCES
- 1.Abriata, L. A., L. J. Gonzalez, L. I. Llarrull, P. E. Tomatis, W. K. Myers, A. L. Costello, D. L Tierney, and A. J. Vila. 2008. Engineered mononuclear variants in Bacillus cereus metallo-β-lactamase are inactive. Biochemistry 47:8590-8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebrone, C., C. Anne, K. De Vriendt, B. Devresse, J. Van Beeumen, J. M. Frère, and M. Galleni. 2005. Dramatic broadening of the substrate profile of the Aeromonas hydrophila CphA metallo-β-lactamase by site-directed mutagenesis. J. Biol. Chem. 17:180-188. [DOI] [PubMed] [Google Scholar]
- 3.Bebrone, C. 2007. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74:1686-1701. [DOI] [PubMed] [Google Scholar]
- 4.Bebrone, C., C. Anne, F. Kerff, G. Garau, K. De Vriendt, R. Lantin, B. Devreese, J. Van Beeumen, O. Dideberg, J. M. Frère, and M. Galleni. 2008. Mutational analysis of the zinc and substrate binding sites in the CphA metallo-β-lactamase from Aeromonas hydrophila. Biochem. J. 114:151-159. [DOI] [PubMed] [Google Scholar]
- 5.Carfi, A., S. Parès, E. Duée, M. Galleni, C. Duez, J. M. Frère, and O. Dideberg. 1995. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14:4914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project, Number 4. 1994. CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 7.Costello, A. L., N. P. Sharma, K. W. Yang, M. W. Crowder, and D. L. Tierney. 2006. X-ray absorption spectroscopy of the zinc-binding sites in the class B2 metallo-β-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry 45:13650-13658. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, P. A., K. W. Yang, N. Sharma, B. Bennett, and M. W. Crowder. 2005. Spectroscopic studies on cobalt(II)-substituted metallo-β-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry 44:5168-5176. [DOI] [PubMed] [Google Scholar]
- 9.Daiyasu, H., K. Osaka, Y. Ishino, and H. Toh. 2001. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 503:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 11.Felici, A., G. Amicosante, A. Oratore, R. Strom, P. Ledent, B. Joris, L. Fanuel, and J. M. Frère. 1993. An overview of the kinetic parameters of class B β-lactamases. Biochem. J. 291:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonseca, F., A. Correia, and J. Spencer. 2008. Structural and kinetic characterisation of Sfh-1, the B2 metallo-β-lactamase of Serratia fonticola, p. 46. In Proceedings of the 10th β-Lactamase Meeting, Eretria, Greece.
- 13.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, J.-M. Frère, and the Metallo-β-Lactamases Working Group. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garau, G., I. Garcia-Saez, C. Bebrone, C. Anne, P. S. Mercuri, M. Galleni, J. M. Frère, and O. Dideberg. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garau, G., C. Bebrone, C. Anne, M. Galleni, J. M. Frère, and O. Dideberg. 2005. A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 345:785-795. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Saez, I., J. D. Docquier, G. M. Rossolini, and O. Dideberg. 2008. The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375:604-611. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez-Valladares, M., A. Felici., G. Weber, H. W. Adolph, M. Zeppezauer, G. M. Rossolini, G. Amicosante, J. M. Frère, and M. Galleni. 1997. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry 36:11534-11541. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Valladares, M., M. Kiefer, U. Heinz, R. Paul Soto, W. Meyer-Klaucke, H. Friederich Nolting, M. Zeppezauer, M. Galleni, J. M. Frère, G. M. Rossolini, G. Amicosante, and H. W. Adolph. 2000. Kinetic and spectroscopic characterization of native and metal-substituted β-lactamase from Aeromonas hydrophila AE036. FEBS Lett. 467:221-225. [DOI] [PubMed] [Google Scholar]
- 19.Hooft, R. W. W., G. Vriend, C. Sander, and E. E. Abola. 1996. Errors in protein structures. Nature 381:272. [DOI] [PubMed] [Google Scholar]
- 20.Horsfall, L. E., G. Garau, B. M. Liénard, O. Dideberg, C. J. Schofield, J. M. Frère, and M. Galleni. 2007. Competitive inhibitors of the CphA metallo-β-lactamase from Aeromonas hydrophila. Antimicrob. Agents Chemother. 51:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 22.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 23.Liénard, B. M. R., G. Garau, L. Horsfall, A. I. Karsisiotis, C. Damblon, P. Lassaux, C. Papamicael, G. C. K. Roberts, M. Galleni, O. Dideberg, J. M. Frère, and C. J. Schofield. 2008. Structural basis for the broad-spectrum inhibition of metallo-β-lactamases by thiols. Org. Biomol. Chem. 6:2282-2294. [DOI] [PubMed] [Google Scholar]
- 24.Llarull, L. I., M. F. Tioni, and A. J. Vila. 2008. Metal content and localization during turnover in Bacillus cereus metallo-β-lactamase. J. Am. Chem. Soc. 130:15842-15851. [DOI] [PubMed] [Google Scholar]
- 25.Morán-Barrio, J., J. M. González, M. N. Lisa, A. L. Costello, M. Dal Peraro, P. Carloni, B. Bennett, D. L. Tierney, A. S. Limansky, A. M. Viale, and A. J. Vila. 2007. The metallo-β-lactamase GOB is a mono-Zn(II) enzyme with a novel active site. J. Biol. Chem. 282:18286-18293. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, T. A., L. E. Catto, S. E. Halford, A. T. Hadfield, W. Minor, T. R. Walsh, and J. Spencer. 2006. Crystal structure of Pseudomonas aeruginosa SPM-1 provides insights into variable zinc affinity of metallo-β-lactamases. J. Mol. Biol. 357:890-903. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 28.Painter, J., and E. A. Merritt. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62:439-450. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 30.Segatore, B., O. Massida, G. Satta, D. Setacci, and G. Amicosante. 1993. High specificity of cphA-encoded metallo-β-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to β-lactam resistance. Antimicrob. Agents Chemother. 37:1324-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma, N. P., C. Hajdin, S. Chandrasekar, B. Bennett, K. W. Yang, and M. W. Crowder. 2006. Mechanistic studies on the mononuclear Zn(II)-containing metallo-β-lactamase ImiS from Aeromonas sobria. Biochemistry 45:10729-10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tioni, M. F., L. I. Llarull, A. A. Poeylaut-Palena, M. A. Marti, M. Saggu, G. R. Periyannan, E. G. Mata, B. Bennett, D. H. Murgida, and A. J. Vila. 2008. Trapping and characterization of a reaction intermediate in carbapenem hydrolysis by Bacillus cereus metallo-β-lactamase. J. Am. Chem. Soc. 130:15852-15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullah, J. H., T. R. Walsh, I. A. Taylor, D. C. Emery, C. S. Verma, S. J. Gamblin, and J. Spencer. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J. Mol. Biol. 284:125-136. [DOI] [PubMed] [Google Scholar]
- 34.Vanhove, M., M. Zakhem, B. Devreese, N. Franceschini, C. Anne, C. Bebrone, G. Amicosante, G. M. Rossolini, J. Van Beeumen., J. M. Frère, and M. Galleni. 2003. Role of Cys221 and Asn116 in the zinc-binding sites of the Aeromonas hydrophila metallo-β-lactamase. Cell. Mol. Life Sci. 60:2501-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wommer, S., S. Rival, U. Heinz, M. Galleni, J. M. Frère, N. Franceschini, G. Amicosante, B. Rasmussen, R. Bauer, and H. W. Adolph. 2002. Substrate-activated zinc binding of metallo-β-lactamases: physiological importance of mononuclear enzymes. J. Biol. Chem. 277:24142-24147. [DOI] [PubMed] [Google Scholar]
- 36.Xu, D., D. Xie, and H. Guo. 2006. Catalytic mechanism of class B2 metallo-β-lactamase. J. Biol. Chem. 281:8740-8747. [DOI] [PubMed] [Google Scholar]
- 37.Yanchak, M. P., R. A. Taylor, and M. W. Crowder. 2000. Mutational analysis of metallo-β-lactamase CcrA from Bacteroides fragilis. Biochemistry 39:11330-11339. [DOI] [PubMed] [Google Scholar]