Abstract
Over the past several years, significant advances have been made in the molecular genetics of the Mollicutes (the simplest cells that can be grown in axenic culture). Nevertheless, a number of basic molecular tools are still required before genetic manipulations become routine. Here we describe the development of a new dominant selectable marker based on the enzyme puromycin-N-acetyltransferase from Streptomyces alboniger. Puromycin is an antibiotic that mimics the 3′-terminal end of aminoacylated tRNAs and attaches to the carboxyl terminus of growing protein chains. This stops protein synthesis. Because puromycin conscripts rRNA recognition elements that are used by all of the various tRNAs in a cell, it is unlikely that spontaneous antibiotic resistance can be acquired via a simple point mutation—an annoying issue with existing mycoplasma markers. Our codon-optimized cassette confers pronounced puromycin resistance on all five of the mycoplasma species we have tested so far. The resistance cassette was also designed to function in Escherichia coli, which simplifies the construction of shuttle vectors and makes it trivial to produce the large quantities of DNA generally necessary for mycoplasma transformation. Due to these and other features, we expect the puromycin marker to be a widely applicable tool for studying these simple cells and pathogens.
The mycoplasmas, which is the generic name for members of the class Mollicutes, are cell wall-less bacteria that are often important pathogens in a variety of species (2, 20). In addition to their importance as pathogens, their small genomes and limited metabolic complexity make them platform organisms for studies on the universal elements of life itself. For example, transposon mutagenesis of the simplest mycoplasma (Mycoplasma genitalium) showed that more than 100 of the 485 protein-encoding genes could be individually disrupted under laboratory conditions (5, 7, 18). Although it is unknown if the disrupted genes are simultaneously dispensable, the remaining 370 or so undisturbed genes provide an initial map of the minimum genetic requirements for life.
The simplicity of mycoplasmas also introduces some technical complications. For example, auxotrophic markers may be difficult to establish in organisms that utilize ill-defined media and that cannot synthesize most biological building blocks. Similarly, antibiotics that target the cell wall are not useful because this structure is completely absent. The use of other markers is not advisable because they are clinically relevant and the simplicity of mycoplasmas naturally restricts clinical options in the first place. In our studies, fairly reliable markers include the tetracycline resistance protein (TetM) and, to a lesser extent, chloramphenicol acetyltransferase and aminoglycoside resistance protein (AacA/AphD). The TetM-tetracycline combination is usually preferable because of the potent antimycoplasma activity of tetracyclines and the low levels of spontaneous tetracycline resistance. In comparison, spontaneous resistance frequently arises with the other markers (presumably via a point mutation in an rRNA). Such resistance is rarely insurmountable, but screening numerous colonies for authentic transformants wastes valuable resources unnecessarily.
In spite of the limitations imposed by mycoplasma biology, the characteristics and mode of action of puromycin suggested that it was a likely candidate for marker development. Puromycin is readily available, is relatively inexpensive, has no clinical value, and is a potent inhibitor of translation in both prokaryotes and eukaryotes. To our knowledge, no example of rRNA-based resistance to the antibiotic exists. Most likely, this is a consequence of the universality of the acceptor end of tRNA and the mechanism of peptidyl transferase—the two translational features puromycin exploits as a nearly perfect tRNA mimic (25). Moreover, the puromycin N-acetyltransferase (PAC) gene from Streptomyces alboniger encodes a small protein which provides resistance to the antibiotic. This suggested that a codon-optimized version of the enzyme might be efficiently expressed. The resistance genes for the usual markers have not been codon optimized for production in mycoplasmas, and expression issues likely contribute to the use of low antibiotic concentrations and thus to the establishment of spontaneous resistance. Here we describe the use of the PAC gene as a new selectable marker in several mycoplasma species.
MATERIALS AND METHODS
Mycoplasma strains.
The mycoplasma strains used were Mycoplasma mycoides subsp. mycoides large colony GM12, which has recently been renamed M. mycoides subsp. capri GM12 (15), Mycoplasma capricolum subsp. capricolum California kid (ATCC 27343), M. genitalium G37, Mycoplasma gallisepticum (ATCC 15302), Mycoplasma pneumoniae Eaton (ATCC 15531), and M. pneumoniae M129-B170 (ATCC 29343). M. mycoides and M. capricolum were grown at 37°C in SP4 medium (8, 9, 23). M. genitalium and the M. pneumoniae strains were grown at 37°C in the presence of 5% CO2 in SP4 medium. M. gallisepticum was grown in Hayflick's medium (23) in the presence of 5% CO2 at 37°C.
PAC gene construction.
The 597-bp PAC gene was synthesized with overlapping oligonucleotides as described previously (22). Briefly, 5′-phosphorylated oligonucleotides encoding both strands of a codon-optimized version (for expression in M. genitalium) were ordered from IDT (Coralville, IA). The oligonucleotides were 48 bases long with an overlap of 24 bases. The top-strand and bottom-strand oligonucleotides were mixed, heated to 95°C, and slowly cooled to allow annealing of the overlaps. The reaction products were ligated for 12 h and used as the template for PCR. The PCR amplicon was cloned into pGEM-3Zf(+) (Promega, Madison, WI) and sequenced to identify correct PAC clones. The optimized PAC gene was then cloned under the control of the Spiroplasma citri spiralin promoter (Ps) (12) and used to replace the tetM gene in a derivative of Mini-Tn4001tet (17, 19) (GenBank accession no. GQ420676), as well as the tetM gene in pMyco1 (11). The new plasmid (Mini-Tn4001PsPuro; GenBank accession no. FJ872396) was used to transform M. genitalium, M. gallisepticum, and M. pneumoniae, while the pMyco1 derivative (pMycoPuro; GenBank accession no. GQ420675) was used to transform M. mycoides and M. capricolum.
Mycoplasma transformations.
M. genitalium and M. pneumoniae were transformed by electroporation as previously described (1, 6, 21). Polyethylene glycol-mediated transformation was used for M. mycoides, M. gallisepticum, and M. capricolum (8, 9, 13, 17).
Puromycin inhibitory concentration determination.
Wild-type M. capricolum cells and cells bearing the pMycoPuro plasmid were grown in SP4 medium containing various concentrations of puromycin to determine bacteriostatic activity. After 24 h of exposure to the antibiotic, aliquots were removed, diluted 10,000-fold, and plated without antibiotic to evaluate bactericidal activity. A similar procedure was used for the other organisms, but bactericidal activity was not determined.
RESULTS AND DISCUSSION
All five mycoplasma species were grown in SP4 or Hayflick's medium supplemented with various concentrations of puromycin. All of the wild-type mycoplasmas that were tested were unable to grow in the presence of puromycin when the concentration exceeded 1 to 3 μg/ml. Thus, distantly related mycoplasmas from both the mycoides group and the pneumoniae group are sensitive to the antibiotic. This sensitivity may be commonplace within the Mollicutes, as they have streamlined genomes unlikely to contain the sophisticated efflux pumps or the modifying enzymes typically required to establish puromycin resistance.
Mini-Tn4001PsPuro (Fig. 1) established resistance in M. genitalium, M. gallisepticum, and M. pneumoniae by transposition. The pMycoPuro plasmid replicates as a free DNA in M. capricolum, while in M. mycoides the plasmid integrates by single crossover at the oriC gene of the M. mycoides genome (data not shown). The puromycin constructs have not been used for directed mutagenesis, although we presume it would work in mycoplasmas, where such mutagenesis has been successful with a tetracycline marker. In general, expression of the PAC gene confers wild-type growth at concentrations beyond 50 μg/ml (M. capricolum had wild-type growth at 500 μg/ml). In practice, we typically selected for transformants on solid medium at lower concentrations (3 μg/ml for M. genitalium, M. gallisepticum, and M. pneumoniae and 8 μg/ml for M. capricolum and M. mycoides). Isolated colonies of M. capricolum or M. mycoides were subsequently grown in liquid culture at 8 μg/ml for the first passage and 50 μg/ml for later passages. M. genitalium, M. gallisepticum, and M. pneumoniae were maintained at 3 μg/ml during passage. As shown in Fig. 2, the inhibitory concentration for M. capricolum is approximately 3 μg/ml.
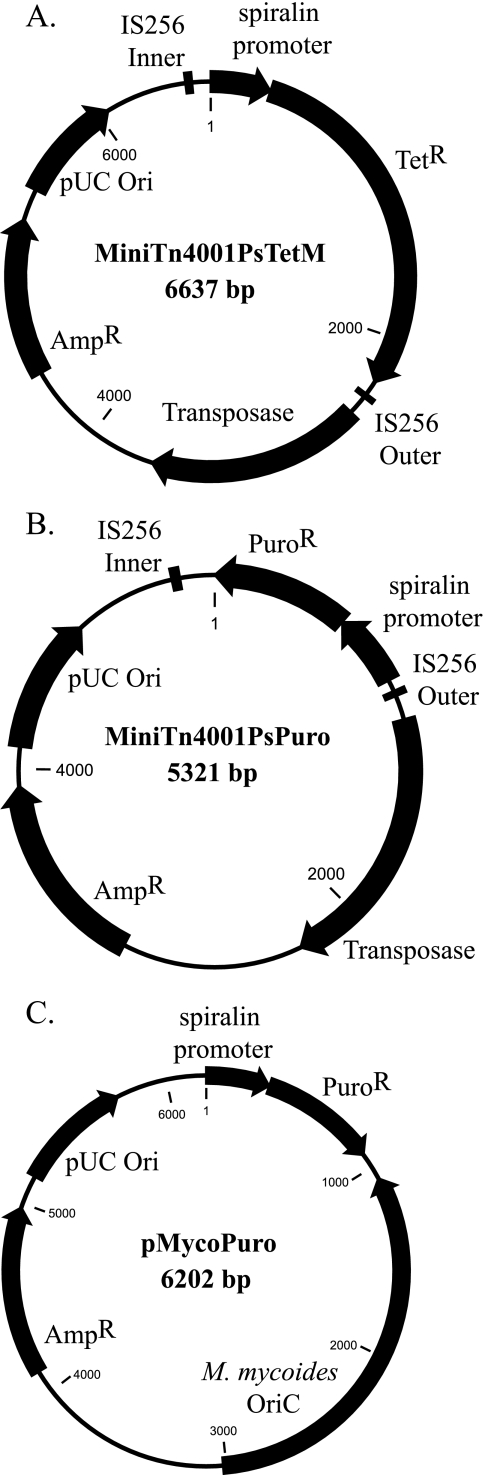
FIG. 1.
Maps of plasmids used in mycoplasmas. (A) Map of the Mini-Tn4001PsTetM plasmid. The spiralin promoter is upstream of the TetM open reading frame. The IS256 elements flank the tetracycline resistance cassette with the transposase gene outside of the repeats. (B) Map of the Mini-Tn4001PsPuro plasmid. The features are identical to those of the Mini-Tn4001PsTetM plasmid, except that the antibiotic resistance cassette is in the opposite orientation and the TetM open reading frame has been replaced with the puromycin N-acetlytransferase open reading frame. After the spiralin promoter was cloned upstream of the PAC gene in pGEM-3Zf(+), the entire cassette was cloned into the backbone of the Mini-Tn4001PsTetM plasmid that had the promoter and TetM open reading frame removed by digestion with PstI and BamHI. (C) Map of the pMycoPuro plasmid. The puromycin resistance unit was cloned into pMyco1, which had the promoter and TetM open reading frame removed by digestion with PstI. M. mycoides oriC allows propagation in M. mycoides. The pUC ori and β-lactamase-encoding genes are used for propagation in E. coli.
FIG. 2.
The pMycoPuro plasmid confers puromycin resistance. M. capricolum was grown in SP4 medium with a range of puromycin concentrations in the presence or absence of the pMycoPuro plasmid. Without the plasmid, M. capricolum grew slowly at a puromycin concentration of 1.2 μg/ml and no growth was observed at 3.7 μg/ml. In this color-changing unit assay, the change of the pH indicator phenol red in the medium from red to yellow was used as an indicator of growth. In the absence of cell growth, the SP4 medium does not change color over the time course of the experiment (24 h). M. capricolum cells containing the vector were resistant to puromycin at concentrations of at least 300 μg/ml.
We wanted to determine if the antibiotic action of puromycin on these mycoplasma species was bactericidal or bacteriostatic. After M. capricolum cells were exposed to 300 μg/ml puromycin for 24 h, they could recover when plated in the absence of the antibiotic (data not shown). Apparently, puromycin is not lethal, even at high concentrations. It is possible that the stringent response, which is active in the organism, induces a persistent state and thereby death is avoided (4, 10, 24). If this supposition is correct, the presence of stringent-response proteins (RelA/SpoT family proteins) in most sequenced mycoplasmas suggests that these bacteria might also survive puromycin exposure even if they are incapable of growth. From a practical standpoint, this issue does not affect the usefulness of the marker. For example, puromycin has become our marker of choice when working with the very-slow-growing organism M. genitalium. Over the 3-week period required to produce clones, no special procedures are required to prevent the appearance of untransformed colonies.
There are several minor features of the expression cassette that were designed to broaden its usefulness. Transcription of the PAC gene is driven by the promoter for spiralin (Ps), which is an abundant protein from S. citri. We have found this promoter to be widely applicable. The efficacy of the spiralin promoter in the range of mycoplasmas examined here further documents its usefulness. Because the promoter also functions in E. coli (16), we designed the codon-optimized PAC gene so that it is free of TGA codons (TGA codes for tryptophan in most mycoplasmas, rather than a stop codon, as in the standard genetic code). In the mycoplasmas that use TGA as a tryptophan codon, its use is heavily preferred over the other tryptophan codon (TGG). Coding the Trp residues in the PAC gene with only TGG is a minor violation of codon optimization rules, but together with the Ps promoter, it allows for expression in E. coli. This simplifies the construction of shuttle vectors, as only a single marker is required. Puromycin sensitivity in E. coli begins at 10 to 50 μg/ml, and the cassette provides resistance beyond 200 μg/ml. We typically use 125 μg/ml puromycin for selection with E. coli. For puromycin expression, as well as the expression of numerous other proteins, we have used a large region upstream of the spiralin start codon (324 nucleotides). In the past, such a large fragment could complicate its movement into various constructs. Now, with the advent of new DNA assembly methods, this is no longer a concern as we can seamlessly join 2 to 10 sequences without regard to restriction sites (3, 14).
Our main objective with this study was to identify a more consistent marker than tetracycline for our work on genome transplantation in M. genitalium (2). The finding that the puromycin-PAC gene combination is broadly applicable for mollicute research suggests that it may be generally preferable to the current antibiotic generally used by molecular mycoplasmologists, tetracycline. This is in part due to the recombinant DNA guidelines that regulate the insertion into pathogens of resistance genes for antibiotics in clinical or veterinary use. Tetracyclines remain a clinical option for some mycoplasma infections and are additives in some animal feeds. The efficacy of puromycin also introduces the possibility of generating double knockouts to further refine the minimal gene content able to support replication and life itself.
Acknowledgments
We thank Radha Krishnakumar for critically reading the manuscript.
This work was supported by Synthetic Genomics, Inc.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Cao, J., P. A. Kapke, and F. C. Minion. 1994. Transformation of Mycoplasma gallisepticum with Tn916, Tn4001, and integrative plasmid vectors. J. Bacteriol. 176:4459-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fadiel, A., K. D. Eichenbaum, N. El Semary, and B. Epperson. 2007. Mycoplasma genomics: tailoring the genome for minimal life requirements through reductive evolution. Front. Biosci. 12:2020-2028. [DOI] [PubMed] [Google Scholar]
- 3.Gibson, D. G., G. A. Benders, C. Andrews-Pfannkoch, E. A. Denisova, H. Baden-Tillson, J. Zaveri, T. B. Stockwell, A. Brownley, D. W. Thomas, M. A. Algire, C. Merryman, L. Young, V. N. Noskov, J. I. Glass, J. C. Venter, C. A. Hutchison III, and H. O. Smith. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319:1215-1220. [DOI] [PubMed] [Google Scholar]
- 4.Glaser, G., A. Razin, and S. Razin. 1981. Stable RNA synthesis and its control in Mycoplasma capricolum. Nucleic Acids Res. 9:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 7.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 8.King, K. W., and K. Dybvig. 1991. Plasmid transformation of Mycoplasma mycoides subspecies mycoides is promoted by high concentrations of polyethylene glycol. Plasmid 26:108-115. [DOI] [PubMed] [Google Scholar]
- 9.King, K. W., and K. Dybvig. 1994. Transformation of Mycoplasma capricolum and examination of DNA restriction modification in M. capricolum and Mycoplasma mycoides subsp. mycoides. Plasmid 31:308-311. [DOI] [PubMed] [Google Scholar]
- 10.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 11.Lartigue, C., A. Blanchard, J. Renaudin, F. Thiaucourt, and P. Sirand-Pugnet. 2003. Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res. 31:6610-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lartigue, C., S. Duret, M. Garnier, and J. Renaudin. 2002. New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid 48:149-159. [DOI] [PubMed] [Google Scholar]
- 13.Lartigue, C., J. I. Glass, N. Alperovich, R. Pieper, P. P. Parmar, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2007. Genome transplantation in bacteria: changing one species to another. Science 317:632-638. [DOI] [PubMed] [Google Scholar]
- 14.Li, M. Z., and S. J. Elledge. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4:251-256. [DOI] [PubMed] [Google Scholar]
- 15.Manso-Silvan, L., E. M. Vilei, K. Sachse, S. P. Djordjevic, F. Thiaucourt, and J. Frey. 2009. Mycoplasma leachii sp. nov. as a new species designation for Mycoplasma sp. bovine group 7 of Leach, and reclassification of Mycoplasma mycoides subsp. mycoides LC as a serovar of Mycoplasma mycoides subsp. capri. Int. J. Syst. Evol. Microbiol. 59:1353-1358. [DOI] [PubMed] [Google Scholar]
- 16.Mouchès, C., T. Candresse, G. Barroso, C. Saillard, H. Wroblewski, and J. M. Bové. 1985. Gene for spiralin, the major membrane protein of the helical mollicute Spiroplasma citri: cloning and expression in Escherichia coli. J. Bacteriol. 164:1094-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papazisi, L., K. E. Troy, T. S. Gorton, X. Liao, and S. J. Geary. 2000. Analysis of cytadherence-deficient, GapA-negative Mycoplasma gallisepticum strain R. Infect. Immun. 68:6643-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pich, O. Q., R. Burgos, R. Planell, E. Querol, and J. Pinol. 2006. Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152:519-527. [DOI] [PubMed] [Google Scholar]
- 19.Pour-El, I., C. Adams, and F. C. Minion. 2002. Construction of mini-Tn4001tet and its use in Mycoplasma gallisepticum. Plasmid 47:129-137. [DOI] [PubMed] [Google Scholar]
- 20.Razin, S., and R. Herrmann. 2002. Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum, New York, NY.
- 21.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1996. Isolation and characterization of transposon Tn4001-generated, cytadherence-deficient transformants of Mycoplasma pneumoniae and Mycoplasma genitalium. FEMS Immunol. Med. Microbiol. 15:199-211. [DOI] [PubMed] [Google Scholar]
- 22.Smith, H. O., C. A. Hutchison III, C. Pfannkoch, and J. C. Venter. 2003. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 100:15440-15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tully, J. G., R. F. Whitcomb, H. F. Clark, and D. L. Williamson. 1977. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892-894. [DOI] [PubMed] [Google Scholar]
- 24.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youngman, E. M., J. L. Brunelle, A. B. Kochaniak, and R. Green. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117:589-599. [DOI] [PubMed] [Google Scholar]