Abstract
Influenza A virus infects many species, and amantadine is used as an antiviral agent. Recently, a substantial increase in amantadine-resistant strains has been reported, most of which have a substitution at amino acid position 31 in the M2 gene. Understanding the mechanism responsible for the emergence and spread of antiviral resistance is important for developing a treatment protocol for seasonal influenza and for deciding on a policy for antiviral stockpiling for pandemic influenza. The present study was conducted to identify the existence of drug pressure on the emergence and spread of amantadine-resistant influenza A viruses. We analyzed data on more than 5,000 virus sequences and constructed a phylogenetic tree to calculate selective pressures on sites in the M2 gene associated with amantadine resistance (positions 26, 27, 30, and 31) among different hosts. The phylogenetic tree revealed that the emergence and spread of the drug-resistant M gene in different hosts and subtypes were independent and not through reassortment. For human influenza virus, positive selection was detected only at position 27. Selective pressures on the sites were not always higher for human influenza virus than for viruses of other hosts. Additionally, selective pressure on position 31 did not increase after the introduction of amantadine. Although there is a possibility of drug pressure on human influenza virus, we could not find positive pressure on position 31. Because the recent rapid increase in drug-resistant virus is associated with the substitution at position 31, the resistance may not be related to drug use.
Influenza virus, a common cause of respiratory infections worldwide, infects humans and avian, swine, and equine species. The virus has a negative-sense, single-stranded RNA genome, which is comprised of eight segments that comprise 12 genes (42). Influenza A viruses cause epidemics and pandemics by antigenic drift and antigenic shift, respectively (42). Antigenic drift is due to an accumulation of point mutations leading to minor and gradual antigenic changes. Antigenic shift involves major antigenic changes by introduction of new hemagglutinin (HA) and/or neuraminidase subtypes into the human population. Since the majority of humans do not have immunity to such novel subtypes, the morbidity and mortality impacts of pandemic influenza can be much higher than those of seasonal influenza.
Amantadine and rimantadine are antiviral agents used for influenza A infection. Both inhibit virus replication by blocking the acid-activated ion channel formed by the virion-associated M2 protein encoded by the M gene (41). The M gene (1,027 bp) encodes two proteins, M1 (at nucleotide positions 26 to 784) and M2 (at positions 26 to 51 and 740 to 1007) (23). The M2 protein comprises 97 amino acids and has ion channel activity (27). Mutations of the M2 gene associated with amantadine (and rimantadine) resistance include mutations at amino acid positions 26, 27, 30, 31, and 34 (1, 12). Amantadine-resistant strains of influenza A virus are commonly isolated from clinical samples (14, 34), and they can be generated easily in vitro by culturing the viruses in the presence of amantadine (3, 12). Resistant strains can replicate as efficiently as sensitive ones, and they can also transmit efficiently (1, 13). Recently, a significant worldwide increase in resistant strains has been reported, not only among seasonal influenza viruses in humans (H1N1 and H3N2) (5, 31), but also in H5N1 avian influenza viruses (7, 15). Most of the resistant strains have a serine-to-asparagine substitution at amino acid position 31 (S31N) in the M2 gene. However, controversies exist regarding the implication of drug pressure (i.e., increasing use of the drug) in increasing resistance (5, 10, 17, 30, 35). It is believed that excess use of amantadine leads to an increase in amantadine-resistant viruses (5, 17, 30), but the drug pressure alone may not be able to explain the recent rapid and significant increase in amantadine resistance (10, 35), and such resistance may be totally unrelated to increasing drug use. For example, oseltamivir-resistant influenza A virus (H1N1) is increasing worldwide (22), yet it is unclear if the increasing oseltamivir resistance is associated with use of the drug, since resistance emerged in northern Europe, where oseltamivir is not widely used (11, 20). Understanding the mechanism responsible for the emergence and spread of antiviral resistance is important for developing a treatment protocol for seasonal and pandemic influenza.
We conducted the present study to clarify whether drug pressure affects the evolution of the M gene by analyzing large numbers of sequences of influenza A viruses from different hosts. The main purpose of the study was to understand the emergence and spread of amantadine resistance among different hosts.
MATERIALS AND METHODS
Sequence data.
All data were obtained from the influenza virus sequence database (Influenza Virus Resource [http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html], accessed on 21 July 2008 [4]). All sequencing data for the strains with a full-length M gene and any subtypes of influenza A virus from different host species, including avian, canine, equine, human, and swine viruses, were included. Sequences derived from laboratory strains and different sequences from the same strains, verified by the strain name, were excluded. A total of 5,489 sequences were obtained (the accession numbers are listed in the supplemental material). The sequences containing ambiguous nucleotides, minor insertions, minor deletions (data for full-length coding regions were used), or premature termination codons were excluded. As a result, 5,060 sequences were used for analysis. The sequencing data were obtained together with information about the host, subtype, isolation year, and isolation location. The numbers of sequences of viruses in each host are given in Table 1. A multiple-sequence alignment of the nucleotide sequences, which did not contain any gaps, was constructed using ClustalW. Among all 5,060 sequences, the number of strains with the amantadine resistance mutation was determined.
TABLE 1.
Frequencies of amantadine-resistant strains by host and time
| Host | Time | Total no. | No. of strains with resistance mutation at amino acid position:
|
||||
|---|---|---|---|---|---|---|---|
| 26 | 27 | 30 | 31 | Double mutation of 27 and 31a | |||
| Human | Total | 2,763 | 5 | 54 | 1 | 368 | 14 |
| Before 1920 (1918-) | 1 | 0 | 0 | 0 | 0 | 0 | |
| 1920- | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1930- | 8 | 0 | 2 | 0 | 7 | 2 | |
| 1940- | 8 | 0 | 0 | 0 | 1 | 0 | |
| 1950- | 37 | 0 | 0 | 0 | 0 | 0 | |
| 1960- | 103 | 3 | 1 | 0 | 1 | 1 | |
| 1970- | 85 | 0 | 1 | 0 | 2 | 1 | |
| 1980- | 91 | 0 | 0 | 0 | 1 | 0 | |
| 1990- | 468 | 0 | 2 | 0 | 7 | 1 | |
| 2000- | 1962 | 2 | 48 | 1 | 349 | 9 | |
| Avianb | Total | 2009 (540) | 3 (0) | 24 (4) | 20 (14) | 197 (152) | 5 (0) |
| Unknown | 1 (1) | 0 | 0 | 0 | 0 | 0 | |
| Before 1960 (1902-) | 12 (1) | 0 | 0 | 0 | 0 | 0 | |
| 1960- | 11 (0) | 0 | 0 | 0 | 1 (0) | 0 | |
| 1970- | 143 (0) | 0 | 0 | 0 | 0 | 0 | |
| 1980- | 242 (1) | 0 | 0 | 0 | 0 | 0 | |
| 1990- | 340 (12) | 0 | 5 (0) | 0 | 11 (0) | 0 | |
| 2000- | 1260 (525) | 3 (0) | 19 (4) | 20 (14) | 185 (152) | 5 (0) | |
| Swine | Total | 201 | 3 | 7 | 3 | 31 | 5 |
| Before 1980 (1930-) | 71 | 0 | 1 | 0 | 0 | 0 | |
| 1980- | 38 | 0 | 1 | 0 | 3 | 1 | |
| 1990- | 26 | 2 | 1 | 0 | 10 | 1 | |
| 2000- | 66 | 1 | 4 | 3 | 18 | 3 | |
| Canine/Equine | Total | 87 | 0 | 0 | 0 | 0 | 0 |
| Total for all hosts | 5,060 | 11 | 85 | 24 | 596 | 24 | |
Strains with double mutations at positions 27 and 31 were counted three times in the positions and double-mutation column.
Numbers of H5N1 avian strains are shown in parentheses.
Phylogenetic-tree analysis.
A phylogenetic tree was inferred by RAxML (37) with all 5,060 sequences. The data used were the sequences for the coding region only, i.e., at nucleotide positions 26 to 1007. The basic sequential algorithm of RAxML is outlined elsewhere (8). RAxML is one of the fastest and most accurate sequential phylogeny programs (38). In this method, a rapid bootstrap search was combined with a rapid maximum-likelihood search of the original alignment. The tree was constructed using the Web server RAxML BlackBox (http://phylobench.vital-it.ch/raxml-bb/) (37). The M genes with amantadine resistance mutations were colored by FigTree (version 1.1.2).
Data sets for each influenza virus host.
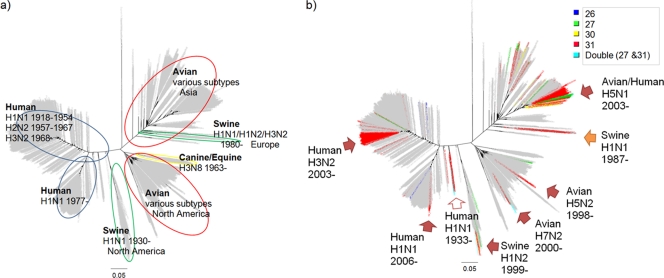
Data sets for each host (avian, canine/equine, human, and swine) were constructed. Only sequences from the host-specific lineage in the phylogenetic tree were used. For example, the data set for the human influenza viruses consists of sequences of human influenza A viruses that are in the human lineage in the phylogenetic tree (Fig. 1a). H5N1 influenza viruses that infect humans were excluded from the analyses because humans were accidental hosts infected with viruses in an avian lineage. These accidental infections should not reflect host-specific evolution. Also, sequences with identical nucleotides in the same data set were removed because the data set should not include identical sequences to analyze selective pressure. The profile of sequences we analyzed is in Table S1 in the supplemental material.
FIG. 1.
Phylogenetic trees for the M gene. Shown are phylogenetic trees constructed using RAxML. The scale bars show evolutionary distances inferred by the RAxML algorithm. The trees are marked with host-specific lineages and their profiles (a) and amantadine resistance mutations shaded in colors by mutation positions (b). The arrows in panel b indicate major clusters of amantadine-resistant strains. These results suggest that viruses with amantadine resistance mutation(s) occurred independently by point mutation in the M gene in each host and/or subtype and that they were not acquired by reassortment with the M genes with resistance mutations from viruses in other lineages.
The number of base substitutions per site from averaging over all sequence pairs was calculated to define the diversity of sequences in a data set (see Tables S1 and S2 in the supplemental material) using the maximum composite likelihood method in MEGA (ver. 4) (21).
Evaluation of pressure.
Selective pressures were calculated for each data set for each influenza virus host. Phylogenetic trees for each data set were constructed with the maximum-likelihood method implemented in PhyML-aLRT (2) using the General Time Reversible model (four rate categories, with all parameters estimated from the data).
Selective pressure among host populations was calculated using the trees. Selective pressure was analyzed with HyPhy (29). All analyses in HyPhy were conducted after identifying the best-fit model out of every possible time-reversible model (e.g., F81 and HKY85) by Akaike's information criterion (24, 33).
Relative rates of nonsynonymous (dN) and synonymous (dS) substitutions were calculated. Positive selection sites in human influenza virus were detected by two methods, single-likelihood ancestor counting (SLAC) and fixed-effects likelihood (FEL). The relative rates of nonsynonymous and synonymous substitutions were compared. Sites where dN/dS was >1 and where dN/dS was <1 were inferred as positively and negatively selected, respectively. The details of the two methods are described elsewhere (6, 19, 33). Briefly, in the SLAC method, the nucleotide and codon model parameter estimates are used to reconstruct the ancestral codon sequences at internal nodes of the tree. The single most likely ancestral sequences are then fixed as known variables and applied to infer the expected number of nonsynonymous or synonymous substitutions that have occurred along each branch for each codon position. SLAC is a substantially modified and improved derivative of the Suzuki-Gojobori method (40). The FEL method is based on maximum-likelihood estimates. The FEL method estimates the ratio of nonsynonymous to synonymous substitutions on a site-by-site basis for the entire tree (eFEL) or only the interior branch (iFEL). iFEL is essentially the same as eFEL, except that selection is tested only along internal branches of the phylogeny (28).
Separate analyses were conducted by testing hypotheses for the entire tree, the internal branch, and the terminal branch: the SLAC (for the entire tree [eSLAC], internal branches [iSLAC], and terminal branches [tSLAC]) and FEL (for the entire tree [eFEL] and internal branches [iFEL]) methods. Pond et al. (28) revealed that many recent nonsynonymous substitutions, i.e., those in the terminal branches of the tree, were not represented on internal branches. At codons where internal substitutions are seen, the strength of selection along terminal branches is high.
Comparison of pressures.
The differential of evolutionary pressures was analyzed by HyPhy. HyPhy tests whether the dN/dS ratios at a given site differ between two data sets along the entire tree (eFEL) or only with interior sequences (iFEL). The details are described elsewhere (28, 33). The differential between hosts was tested. In addition, the human data set that was constructed as described above was divided by time (before 1965 and after 1966, and before 1999 and after 2000) to create new datasets (see Table S2 in the supplemental material). The differentials between them were also tested.
RESULTS
Frequency of drug resistance.
The numbers of strains with amantadine resistance mutation(s) among different hosts are shown in Table 1. There were no amantadine resistance mutations at position 34 of M2 in any of the strains except laboratory strains. We therefore conducted the analyses focusing on positions 26, 27, 30, and 31 as sites for amino acid substitutions associated with naturally occurring amantadine resistance. Canine/equine influenza virus had no amantadine-resistant strains. Amantadine-resistant mutations were detected at all four sites (positions 26, 27, 30, and 31) in human influenza virus, as well as avian and swine influenza viruses. We also found strains with double resistance mutations (positions 27 and 31) in these hosts. Amantadine resistance mutations were detected most frequently at position 31, followed by position 27, in all hosts except canine/equine. We found more amantadine-resistant strains in human influenza virus after 2000 (Table 1) because of an increase in the resistance mutation at position 31 in both H3N2 and H1N1. Since 2000, amantadine-resistant strains of avian influenza virus have also been found more frequently, and many of these (170 out of 227) were H5N1 viruses. The most common resistance mutation was S31N in human, avian, and swine influenza viruses.
Phylogenetic trees.
Phylogenetic trees for all M gene sequence data are shown in Fig. 1a. The features of the tree were described in detail under subtypes, hosts, and temporal and geographical distribution in our previous study (9). The analysis revealed seven host-specific lineages: (i) a human influenza virus lineage that consisted of H1N1 between 1918 and 1954, H2N2 between 1957 and 1967, and H3N2 after 1968; (ii) a human influenza virus lineage that comprised H1N1 after 1977; (iii) an avian lineage that included viruses mainly from Asia, but also from other regions; (iv) an avian lineage that included viruses mostly from North America; (v) a swine lineage that was between the human and avian lineages and mainly included viruses from North America; (vi) a swine lineage that diverged from an avian lineage and consisted of swine viruses after 1980, mainly from Europe; and (vii) a canine/equine lineage that diverged from an avian-lineage root.
Strains with mutations associated with amantadine resistance were identified in all lineages except the canine/equine lineage (Fig. 1b). Amantadine resistance mutations appeared across different subtypes, different hosts, and different geographic regions.
The viruses with mutations at positions 26, 27, and 30 were found sporadically, but those viruses did not become dominant strains and disappeared. There were eight major clusters of resistant strains (indicated in Fig. 1b). All of these major resistant clusters had the S31N mutation. Such a cluster was found in the 1930s (human H1N1 1933 in Fig. 1b). They were not laboratory strains (e.g., A/Melbourne/35, CY009325, and A/Alaska/1935, CY019956). Most major resistant clusters were found after the late 1990s.
Site-by-site pressures for human influenza virus.
We analyzed selective pressures on human influenza virus. The data on the M2 sites associated with amantadine resistance are shown in Table 2. “dN/dS” indicates the ratio of nonsynonymous and synonymous substitutions at each codon. When the pressure on a codon is significantly larger than 1, the site is regarded as under significant positive selection. When the pressure on a codon is significantly smaller than 1, the site is regarded as under significant negative selection (33, 40).
TABLE 2.
Selective pressures (dN/dS) on sites associated with amantadine resistance for human influenza virus
| Position | dN/dS | Selective pressure
|
||||
|---|---|---|---|---|---|---|
| eSLAC | iSLAC | tSLAC | eFEL | iFEL | ||
| 26 | 1.35 | 0.57 | 1 | 0.35 | 0.72 | 1 |
| 27 | 4.42 | 0.054 | 0.59 | 0.039a | 0.082 | 0.2 |
| 30 | 0.21 | 1 | 1 | 1 | 1 | 1 |
| 31 | 0.83 | 0.46 | 1 | 0.74 | 1 | 1 |
Statistically significant positive selection (P < 0.05).
The selective pressure on the entire sequence of M2 was 0.45. Position 26 had a dN/dS ratio greater than 1 (1.35), but it was not statistically significant (P > 0.05 for all tests). Position 27 had a much larger dN/dS ratio (4.42) and was under significant positive selection based on tSLAC (P = 0.039). Position 30 had a dN/dS ratio smaller than 1, though significant negative selection was not found in any tests. Position 31 showed results similar to those of position 30.
Differences in selective pressures between hosts.
We analyzed the differences in selective pressures on sites associated with amantadine resistance between hosts. The site-specific selective pressures (dN/dS) for each host are shown in Fig. 2a. The dN/dS ratio of avian influenza virus was calculated, excluding sequences of chicken viruses (discussed below).
FIG. 2.
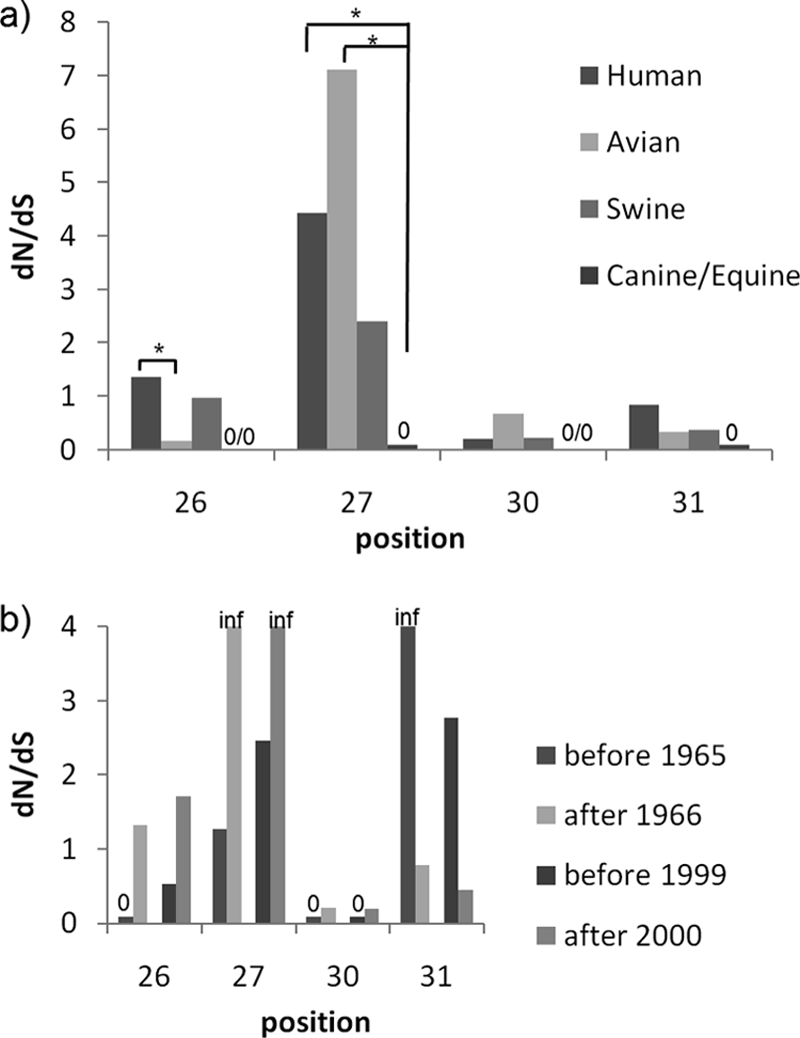
Differentials of selective pressures on sites associated with amantadine resistance. (a) Differentials of selective pressures on sites associated with amantadine resistance between hosts. Selective pressures for human viruses are higher than for the viruses of other hosts at positions 26 and 31. Significantly higher selective pressures for human viruses were found only when compared to avian viruses at position 26 and only when compared to canine/equine viruses at position 27. The significant differences were observed only by eFEL. No significant differences were found by iFEL. The dN/dS ratios at positions 26 and 30 for canine/equine influenza virus could not be calculated because both the denominator and the numerator were zero. The dN/dS ratios at positions 27 and 31 for canine/equine influenza viruses are zero, as only the numerators were zero. *, significant differences were found by eFEL (P < 0.05). (b) Differentials of selective pressures on sites associated with amantadine resistance in human influenza virus by time. The dN/dS ratios for positions 26, 27, and 30 have become larger since 1966 (the introduction of amantadine) and 2000 (the beginning of the recent surge of amantadine-resistant strains), though there are no significant differences. In contrast, the dN/dS ratio for position 31 became smaller rather than larger. “inf” means infinity, as the denominator was zero. The dN/dS ratios at position 26 before 1965, at position 30 before 1965, and at position 30 before 1999 were zero, as only the numerators were zero. No significant differences were found by either eFEL or iFEL.
We found higher selective pressures for human viruses than for viruses of the other hosts at positions 26 and 31 (Fig. 2a). Significantly higher selective pressures for human viruses were found only when compared to avian viruses at position 26 and to canine/equine viruses at position 27. The significant differences were observed only by a test of the entire tree (eFEL) (Fig. 2a). iFEL, which is a test for internal branches, did not detect significant differences (see Table S3 in the supplemental material). We could not find any significant differences between human and swine viruses. Furthermore, human influenza virus was under lower selective pressure at positions 27 and 30 than the avian virus, though the difference was not significant.
Change in selective pressure with time.
We divided the human data set by the year of isolation: before 1965 versus after 1966 and before 1999 versus after 2000. In 1966, amantadine was approved as a drug for influenza virus infection in the United States (25), and in 2000, the escalating trend of circulating amantadine-resistant viruses in humans began (5). We found that the entire selective pressure for the M gene (both M1 and M2) became smaller with time (data not shown).
The dN/dS ratio for positions 26, 27, and 30 associated with amantadine resistance has increased since 1966. The selective pressures were also higher after 2000 than before 1999 (Fig. 2b), although there were no significant differences. In contrast, the dN/dS ratio for position 31 became smaller rather than larger in both analyses (before 1965 versus after 1966 and before 1999 versus after 2000) (Fig. 2b).
DISCUSSION
Amantadine-resistant strains were found in avian and swine, as well as human, influenza viruses. The recent spread of amantadine-resistant viruses is caused by the emergence of viruses with the S31N mutation in the M2 gene. The phylogenetic tree suggests that viruses with an amantadine resistance mutation(s) occurred independently by point mutation in the M gene in each lineage (Fig. 1). The emergence of the S31N mutation was not caused by acquiring the M gene from other hosts/subtypes by reassortment.
The tree shows that all major resistant clusters have the S31N mutation. Only M genes with the S31N mutation were maintained and could become dominant strains, indicating that strains with this mutation could efficiently transmit it to the next generation. It must be noted that strains with S31N appeared and were maintained in the human population in the 1930s, which was before amantadine was discovered and used (Fig. 1b). Furthermore, it is intriguing that clusters of M genes with mutations at position 31 have emerged separately since the late 1990s in different hosts and different subtypes (Fig. 1b). The influenza viruses of various hosts acquired the mutation at position 31 independently and almost simultaneously. Schmidtke et al. reported the emergence of amantadine-resistant strains of swine influenza virus in the 1980s and suggested that this might have been caused by a reassortment event and that further reassortment between these swine and human influenza viruses could cause an increase in the amantadine-resistant M gene in human influenza virus (32). Our results are contrary to this suggestion. Although amantadine resistance did increase in human and avian viruses after 2000 (Table 1), these resistant viruses did not acquire the M gene from swine viruses (Fig. 1b).
The next question was why amantadine resistance has increased so rapidly. One possible explanation is that there was drug pressure on the influenza viruses of various hosts that led to an amino acid change at position 31. Otherwise, S31N could be just a genetic variant in a diverse gene pool. It should also be noted that amantadine has been less frequently used since 2000 because neuraminidase inhibitors (oseltamivir and zanamivir) were licensed and became the most commonly prescribed drugs for influenza A and B virus infections in developed countries. However, there is a possibility of localized amantadine use in some countries even after 2000 (5, 35). We conducted further analysis to determine whether the drug pressure had any effect on the recent emergence and spread of viruses with the S31N mutation.
Analysis for selective pressure on human influenza virus indicates that the mutations at sites associated with amantadine resistance are not generally driven by external pressure affecting the entire tree, including drug pressure. If anything, the pressure affects only terminal branches without affecting internal branches at position 27, because only tSLAC, which is a test only for the terminal branches, found significant positive selection at position 27. iSLAC and iFEL, which are tests for internal branches, did not find significant positive selection. Although Suzuki showed that there was no positive pressure on the sites in human influenza virus (39), we found positive selection at position 27. This must be because we applied various methods of calculation and analyzed a data set that was 10 times larger than the data Suzuki used.
Although positive selection was not found in three of the four sites linked to amantadine resistance, we could not reject drug pressure on evolution of the M gene. In fact, the selective pressures on the sites, except position 30, were higher than the selective pressure for the entire M2 gene. Particularly at position 31, other substitutions apart from S31N must be under strong negative pressure, since we found only serine and asparagine at the site. The negative pressure might conceal drug pressure. Therefore, we compared selective pressures by hosts and time.
Amantadine is known to have been used to treat human disease and possibly in poultry, such as chickens (36). Even though veterinary use is possible, drug pressure on nonhuman influenza A viruses (except possibly in chickens) will not be stronger than on human viruses. If the drug pressure is exerted on M gene evolution, it must be stronger in the human population than in other hosts, even if the pressure is not significant positive selection. For the analysis, we removed the chicken virus data from the avian data set because chicken viruses may have been under selective pressure due to amantadine use. Hill et al. found positive selection at positions 27 and 31 in H5N1 avian influenza virus (16).
We found higher selective pressure for human viruses than for those of other hosts at positions 26 and 31, while human influenza virus was under lower selective pressure at positions 27 and 30 than avian virus (Fig. 2a). We could not find any significant differences between human and swine viruses, which are unlikely to be under drug pressure. In addition, significant differences in some combinations were observed only by testing the entire tree (eFEL) and not by using iFEL, which is a test for internal branches. That is, drug pressure, if any, was not strong enough to affect the interior branches. The results could not support the hypothesis that human influenza virus is under substantially higher drug pressure than viruses of other hosts.
In case of drug pressure on sites in M2, the significance of selective pressure (dN/dS) could become larger after the introduction of amantadine and/or the beginning of excess use of amantadine. It is said that the recent rapid increase of amantadine-resistant strains might be caused by excessive use of amantadine in Asian and adjacent countries, since amantadine is available as an over-the-counter formulation in those countries (5, 35).
We found that recent drug pressure might be stronger than before, although there were no significant differences (Fig. 2b). These results suggest that amantadine may be exerting pressure on human influenza viruses. However, selective pressure on position 31 did not increase even after the introduction of amantadine or the surge in drug-resistant viruses. Although selective pressure on position 31 has decreased, strains with the S31N mutation have increased (Table 1), suggesting that most of the resistant strains originated from a single or a few viruses in each lineage with the S31N mutation. This hypothesis is supported by the phylogenetic tree constructed in the present study (Fig. 1b). Most of the recent resistant strains have been derived from a single or a few strains and formed small clusters.
We used a mathematical-biological approach to determine if there was any selective pressure on amino acid positions associated with amantadine resistance. We could not find significant evidence for drug pressure on position 31 in human influenza virus. Shiraishi et al. showed that S31N, and also various mutants with amantadine-resistant mutations in M2, were detected in patients under treatment with amantadine (34). The reason why only S31N, which has weak selective pressure, has spread so rapidly remains unclear.
It is possible that the S31N mutation has occurred naturally to some extent, irrespective of the use of amantadine. We showed that M genes with S31N appeared and were maintained in the 1930s before the development of amantadine. Other strains with the S31N mutation in the M2 gene also appeared sporadically in the period from the 1940s to the 1990s (Table 1). M genes with S31N might have increased by genetic drift, as in Kimura's neutral theory of molecular evolution (18, 26). In this theory, mutations that are not under selective pressure and are not advantageous or disadvantageous can predominate in a population by chance. Even if conversion from amantadine sensitivity to resistance caused by the S31N mutation occurs less commonly than mutations at other sites, such as position 27, the virus may be easily maintained once it occurs. Simonsen et al. proposed that a combination of S31N in the M2 gene and some specific amino acid substitutions in the HA genes was advantageous to the virus (35). However, our previous study revealed that strains with S31N without the same substitutions in the HA gene became prevalent after further reassortment (10). It is not known whether S31N alone or S31N together with other amino acid substitutions can change the growth characteristics of the virus.
In the present study, we showed that the recent rapid increase in drug-resistant virus was associated with a substitution at position 31, but the resistance may not be related to drug pressure. Further in silico, in vitro, and in vivo studies are needed to elucidate the mechanism responsible for the recent emergence of resistant strains with the S31N mutation.
Supplementary Material
Acknowledgments
We are indebted to Jianzhi George Zhang (University of Michigan) and Sergei L. Kosakovsky Pond (University of California, San Diego) for their kind advice regarding analyses.
This study was supported by the Acute Respiratory Infections Panels, United States-Japan Cooperative Medical Science Program (U.S. Department of Health and Human Services, U.S. Department of State; and Ministry of Foreign Affairs; Ministry of Health, Labor, and Welfare; and Ministry of Education, Culture, Sports, Science and Technology, Japan).
Footnotes
Published ahead of print on 3 August 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abed, Y., N. Goyette, and G. Boivin. 2005. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob. Agents Chemother. 49:556-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anisimova, M., and O. Gascuel. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539-552. [DOI] [PubMed] [Google Scholar]
- 3.Appleyard, G. 1977. Amantadine-resistance as a genetic marker for influenza viruses. J. Gen. Virol. 36:249-255. [DOI] [PubMed] [Google Scholar]
- 4.Bao, Y., P. Bolotov, D. Dernovoy, B. Kiryutin, L. Zaslavsky, T. Tatusova, J. Ostell, and D. Lipman. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bright, R. A., M.-J. Medina, X. Xu, G. Perez-Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, N. J. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175-1181. [DOI] [PubMed] [Google Scholar]
- 6.Campo, D. S., Z. Dimitrova, R. J. Mitchell, J. Lara, and Y. Khudyakov. 2008. Coordinated evolution of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 105:9685-9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, C.-L., J. M. Rayner, G. J. D. Smith, P. Wang, T. S. P. Naipospos, J. Zhang, K.-Y. Yuen, R. G. Webster, J. S. M. Peiris, Y. Guan, and H. Chen. 2006. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J. Infect. Dis. 193:1626-1629. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 9.Furuse, Y., A. Suzuki, T. Kamigaki, and H. Oshitani. 2009. Evolution of the M gene of the influenza A virus in different host species: large-scale sequence analysis. Virol. J. 6:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuse, Y., A. Suzuki, T. Kamigaki, M. Shimizu, N. Fuji, and H. Oshitani. 2008. Reversion of Influenza A (H3N2) from amantadine resistant to amantadine sensitive by further reassortment in Japan during the 2006-to-2007 influenza season. J. Clin. Microbiol. 47:841-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauge, S. H., S. Dudman, K. Borgen, A. Lackenby, and O. Hungnes. 2009. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 15:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayden, F. G., and A. J. Hay. 1992. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr.t Top. Microbiolo. Immunol. 176:119-130. [DOI] [PubMed] [Google Scholar]
- 14.Hayden, F. G., S. J. Sperber, R. B. Belshe, R. D. Clover, A. J. Hay, and S. Pyke. 1991. Recovery of drug-resistant influenza A virus during therapeutic use of rimantadine. Antimicrob. Agents Chemother. 35:1741-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, G., J. Qiao, C. Dong, C. He, L. Zhao, and Y. Tian. 2008. Amantadine resistance among H5N1 avian influenza viruses isolated in Northern China. Antivir. Res. 77:72-76. [DOI] [PubMed] [Google Scholar]
- 16.Hill, A. W., R. P. Guralnick, M. J. Wilson, F. Habib, and D. Janies. 2008. Evolution of drug resistance in multiple distinct lineages of H5N1 avian influenza. Infect. Genet. Evol. 9:169-178. [DOI] [PubMed] [Google Scholar]
- 17.Ilyushina, N. A., E. A. Govorkova, and R. G. Webster. 2005. Detection of amantadine-resistant variants among avian influenza viruses isolated in North America and Asia. Virology 341:102-106. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, M. 1968. Evolutionary rate at the molecular level. Nature 217:624-626. [DOI] [PubMed] [Google Scholar]
- 19.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 20.Kramarz, P., D. Monnet, A. Nicoll, C. Yilmaz, and B. Ciancio. 2009. Use of oseltamivir in 12 European countries between 2002 and 2007—lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses. Euro Surveill. 14:19112. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief.n Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lackenby, A., C. I. Thompson, and J. Democratis. 2008. The potential impact of neuraminidase inhibitor resistant influenza. Curr. Opin. Infect. Dis. 21:626-638. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, R. A., C. J. Lai, and P. W. Choppin. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. USA 78:4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanave, C., G. Preparata, C. Saccone, and G. Serio. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86-93. [DOI] [PubMed] [Google Scholar]
- 25.Maugh, T. H. 1976. Amantadine: an alternative for prevention of influenza. Science 192:130-131. [DOI] [PubMed] [Google Scholar]
- 26.Ohta, T., and J. H. Gillespie. 1996. Development of neutral and nearly neutral theories. Theor. Popul. Biol. 49:128-142. [DOI] [PubMed] [Google Scholar]
- 27.Pinto, L. H., L. J. Holsinger, and R. A. Lamb. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517-528. [DOI] [PubMed] [Google Scholar]
- 28.Pond, S. L. K., S. D. W. Frost, Z. Grossman, M. B. Gravenor, D. D. Richman, and A. J. L. Brown. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pond, S. L. K., S. D. W. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 30.Regoes, R. R., and S. Bonhoeffer. 2006. Emergence of drug-resistant influenza virus: population dynamical considerations. Science 312:389-391. [DOI] [PubMed] [Google Scholar]
- 31.Saito, R., Y. Suzuki, D. Li, H. Zaraket, I. Sato, H. Masaki, T. Kawashima, S. Hibi, Y. Sano, Y. Shobugawa, T. Oguma, and H. Suzuki. 2008. Increased incidence of adamantane-resistant influenza A(H1N1) and A(H3N2) viruses during the 2006-2007 influenza season in Japan. J. Infect. Dis. 197:630-632. (Author's reply, 197:632-633.) [DOI] [PubMed] [Google Scholar]
- 32.Schmidtke, M., R. Zell, K. Bauer, A. Krumbholz, C. Schrader, J. Suess, and P. Wutzler. 2006. Amantadine resistance among porcine H1N1, H1N2, and H3N2 influenza A viruses isolated in Germany between 1981 and 2001. Intervirology 49:286-293. [DOI] [PubMed] [Google Scholar]
- 33.Sergei, L., A. F. Y. P. Kosakovsky Pond, and S. D. W. Frost. 2007. Estimating selection pressures on alignments of coding sequences analyses using HyPhy. http://www.hyphy.org/pubs/hyphybook2007.pdf.
- 34.Shiraishi, K., K. Mitamura, Y. Sakai-Tagawa, H. Goto, N. Sugaya, and Y. Kawaoka. 2003. High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. J. Infect. Dis. 188:57-61. [DOI] [PubMed] [Google Scholar]
- 35.Simonsen, L., C. Viboud, B. T. Grenfell, J. Dushoff, L. Jennings, M. Smit, C. Macken, M. Hata, J. Gog, M. A. Miller, and E. C. Holmes. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol. Biol. Evol. 24:1811-1820. [DOI] [PubMed] [Google Scholar]
- 36.Sipress, A. 1 November 2005, posting date. Bird flu drug rendered useless. Washington Post Foreign Service http://www.washingtonpost.com/wp-dyn/content/article/2005/06/17/AR2005061701214.html.
- 37.Stamatakis, A., P. Hoover, and J. Rougemont. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758-771. [DOI] [PubMed] [Google Scholar]
- 38.Stamatakis, A. P., T. Ludwig, and H. Meier. 1 November 2008, posting date. Computing large phylogenies with statistical methods: problems and solutions. icwww.epfl.ch/∼stamatak/index-Dateien/publications/BGRS2004.PDF.
- 39.Suzuki, Y. 2006. Natural selection on the influenza virus genome. Mol. Biol. Evol. 23:1902-1911. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, Y., and T. Gojobori. 1999. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16:1315-1328. [DOI] [PubMed] [Google Scholar]
- 41.Wang, C., K. Takeuchi, L. H. Pinto, and R. A. Lamb. 1993. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67:5585-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.