Abstract
The hylEfm gene (encoding a putative hyaluronidase) has been found almost exclusively in Enterococcus faecium clinical isolates, and recently, it was shown to be on a plasmid which increased the ability of E. faecium strains to colonize the gastrointestinal tract. In this work, the results of mating experiments between hylEfm-containing strains of E. faecium belonging to clonal cluster 17 and isolated in the United States and Colombia indicated that the hylEfm gene of these strains is also carried on large plasmids (>145 kb) which we showed transfer readily from clinical strains to E. faecium hosts. Cotransfer of resistance to vancomycin and high-level resistance (HLR) to aminoglycosides (gentamicin and streptomycin) and erythromycin was also observed. The vanA gene cluster and gentamicin resistance determinants were genetically linked to hylEfm, whereas erm(B) and ant(6)-I, conferring macrolide-lincosamide-streptogramin B resistance and HLR to streptomycin, respectively, were not. A hylEfm-positive transconjugant resulting from a mating between a well-characterized endocarditis strain [TX0016 (DO)] and a derivative of a fecal strain of E. faecium from a healthy human volunteer (TX1330RF) exhibited increased virulence in a mouse peritonitis model. These results indicate that E. faecium strains use a strategy which involves the recruitment into the same genetic unit of antibiotic resistance genes and determinants that increase the ability to produce disease. Our findings indicate that the acquisition of the hylEfm plasmids may explain, at least in part, the recent successful emergence of some E. faecium strains as nosocomial pathogens.
Until the mid- to late 1980s, Enterococcus faecium was considered a “docile” organism; in fact, it was (and some strains still are) used in the food industry as cheese or fermentation starter cultures and as probiotics and was known to colonize many animal species (26). The situation has changed substantially in recent years, and isolates of E. faecium are now commonly recovered from infections seen in U.S. hospitals. Indeed, in a recent report on health-care associated infections, E. faecium accounted for 5.6% of isolates from health-care associated infections; it was the third-most-common bacterial species isolated after coagulase-negative staphylococci and Staphylococcus aureus and was even more frequent than E. faecalis (12). Moreover, 93% of vancomycin-resistant enterococcal isolates from a recent survey of U.S. hospitals were E. faecium (9). The ability of E. faecium to disseminate in the hospital environment is thought to be due not only to the presence of antibiotic resistance genes but also to the acquisition of determinants which may increase the capacity of the organism to survive in, colonize, and/or infect patients in the clinical setting. However, in terms of virulence factors, E. faecium appears to lack certain important determinants, such as hemolysin, gelatinase, and the fsr locus, previously shown to play a role in Enterococcus faecalis infections (7). Aggregation substance, encoded by pheromone-responsive E. faecalis plasmids, has been found much less frequently in E. faecium (7). Previously, a gene designated hylEfm, which encodes a protein with homology to hyaluronidases, was postulated to be a potential virulence factor of clinical isolates of E. faecium since this gene was found to be more common in vancomycin-resistant E. faecium isolates than in vancomycin-susceptible isolates of the same species (21). Moreover, the hylEfm gene was not found in any of 186 fecal E. faecium isolates from community-based sources (feces of healthy human volunteers, animals, probiotics, and wastewater), whereas it was identified in 34% of 264 U.S. clinical isolates of E. faecium (21). The gene has also been associated with isolates belonging to the genogroup referred to as clonal cluster 17 (CC17), a group of genetically related isolates that account for the majority of E. faecium clinical isolates in hospitals worldwide (26). Also, a plasmid carrying the hylEfm gene has been previously shown to be an important factor for gastrointestinal colonization in a mouse model (23). In this work, we studied the transferability of the hylEfm gene along with antibiotic resistance genes from clinical strains and characterized the role of the hylEfm-containing plasmid in virulence in the mouse peritonitis model.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this work are shown in Table 1. In order to simplify the strain nomenclature, we will refer to the names listed in the second column of Table 1, which are different from the published strain names (Table 1, first column). These bacteria include the hylEfm-positive (hylEfm+) clinical isolates of E. faecium strain A [TX0016 (DO)] (http://www.hgsc.bcm.tmc.edu) (19) and strain C (C-68) (4), both isolated in the United States, and strain B (ERV-99), obtained in Colombia. The Colombian isolate B is a multidrug-resistant E. faecium isolate recovered from the peritoneal fluid of a patient with abdominal sepsis in February 2005. All clinical strains (A, B, and C, classified as sequence type 16 [ST-16], ST-18, and ST-16, respectively) belong to CC17 by multilocus sequence typing (MLST) (Table 1) (13). TX1330RF and D344SRF (a pbp5-deficient E. faecium laboratory strain) (22) are rifampin (rifampicin)- and fusidic acid-resistant derivatives generated from the parental strains by sequential selection on brain-heart infusion (BHI) agar containing inhibitory concentrations of each antibiotic. E. faecium TX1330RF-A1, TX1330RF-B1/-B2, and D344SRF-C6 (23) are derivatives of TX1330RF and D344SRF to which hylEfm from strains A, B, and C was transferred by mating (see below).
TABLE 1.
E. faecium strains used in this work
| Published or laboratory strain nomenclature | Nomenclature used for this work | Relevant characteristicsa | Presence of hylEfm | Reference |
|---|---|---|---|---|
| TX0016 (DO) | A | Clinical isolate from the United States; Eryr Strr, ST-16 | Yes | 19 |
| ERV-99 | B | Clinical isolate from Colombia; Vanr Genr, ST-18 | Yes | This study |
| C-68 | C | Clinical isolate from the United States; Vanr, ST-16 | Yes | 4 |
| TX1330 | TX1330 | Susceptible fecal-colonizing strain from a healthy human volunteer | No | 19 |
| TX1330RF | TX1330RF | Derivative of TX1330; Fusr Rifr | No | This study |
| TX2158 | TX1330RF-A1 | Derivative of TX1330RF to which the hylEfm-containing plasmid was transferred by conjugation from strain A as donor; Eryr Strr | Yes | This study |
| TX6068 | TX1330RF-B1 | Transconjugant resulting from a mating expt between strain B (donor) and TX1330RF (recipient); Vanr Eryr Genr Strr | Yes | This study |
| TX6069 | TX1330RF-B2 | Transconjugant resulting from a mating expt between strain B (donor) and TX1330RF (recipient); Vanr Genr | Yes | This study |
| D344SRF | D344SRF | Laboratory strain with spontaneous loss of pbp5; Fusr Rifr | No | 23 |
| TC6 | D344SRF-C6 | Transconjugant resulting from a mating expt between strain C (donor) and D344SRF (recipient) | Yes | 24 |
Eryr, erythromycin resistance; Fusr, fusidic acid resistance; Genr, HLR to gentamicin; Rifr, rifampin resistance; Strr, HLR to streptomycin; Vanr, vancomycin resistance.
Filter matings, PFGE, and hybridizations.
The hylEfm gene had been previously transferred by filter mating from the clinical isolate E. faecium C to D344SRF (Table 1), resulting in the transconjugant D344SRF-C6, as described previously (23). Similarly, hylEfm was transferred from clinical strains A and B to TX1330RF by using the same mating approach (17), and transconjugants were selected on medium supplemented with some antibiotics whose resistance determinants are known to be transferable to determine if the resistance genes cotransfer with hylEfm. Transconjugants from the strain A and TX1330RF mating were selected on BHI agar supplemented with erythromycin (200 μg/ml)-fusidic acid (25 μg/ml) and streptomycin (2,000 μg/ml)-fusidic acid (25 μg/ml). Single colonies from each mating experiment were purified, grown on BHI agar (Difco Laboratories, Detroit, MI) supplemented with rifampin (100 μg/ml), and then screened by PCR with primers 5′-TTCTGAAACATGAACCATCA (forward) and 5′-TCAATATCATTTCCAGGACTAA (reverse) (nucleotides 398 to 674 of the hylEfm gene). Ten hylEfm+ colonies from each experiment were further characterized by pulsed-field gel electrophoresis (PFGE) (SmaI) and hybridizations following a methodology described before (16) and using a probe generated with the hylEfm primers. Transconjugants from the mating of strains B and TX1330RF were selected on BHI agar supplemented with vancomycin (128 μg/ml)-fusidic acid (25 μg/ml) and gentamicin (1,000 μg/ml)-fusidic acid (25 μg/ml). Purified colonies from each mating were then grown on rifampin (100 μg/ml). Screening by PCR, PFGE (SmaI), and hylEfm hybridizations was then performed as described above. SmaI digests of total DNA from the clinical strains A, B, and C (donors) and corresponding selected transconjugant strains (TX1330RF-A1, TX1330RF-B1 and TX1330RF-B2, and D344SRF-C6) were subsequently transferred to a nylon membrane and hybridizations performed with probes for erm(B), aac(6′)-aph(2") (25), ant(6)-I (25), and vanA (10), targeting genes involved in resistance to macrolides and high-level resistance (HLR) to gentamicin, streptomycin, and vancomycin, respectively. The erm(B) and ant(6)-I probes were generated using DNA from strain A as template, whereas total DNA from strain B was used to obtain the aac(6′)-aph(2") and vanA probes.
Plasmid extractions, S1 nuclease/I-CeuI digestions, and hybridizations.
In the current work, plasmids from the D344SRF-C6 transconjugant (23) containing the hylEfm gene were extracted by cesium chloride gradient centrifugation (8), and the digested (EcoRI) fragments were transferred to a nylon membrane and hybridized with the hylEfm probe. In order to confirm that hylEfm was present in plasmids in the donor and transconjugant strains, an S1 nuclease assay was performed by following the protocol of Barton et al. (3). This method allows the detection and estimation of the size of large bacterial plasmids in the presence of genomic DNA using PFGE and has been validated in gram-positive and gram-negative organisms (3). Briefly, agarose gel plugs containing bacterial cells were incubated for 45 min with 0.01 U of Aspergillus oryzae S1 nuclease (Sigma-Aldrich, St. Louis, MO) in 200 μl of 50 mM NaCl, 30 mM sodium acetate (pH 4.5), and 5 mM ZnSO4. Digested plugs were subjected to PFGE using the same methodology described before (16). I-CeuI is an enzyme that recognizes specific sequences found only in the 23S rRNA genes (14) and, thus, digests only bacterial chromosomes and can be used to differentiate the presence of extrachromosomal elements (e.g., plasmids) using PFGE and hybridizations (6). I-CeuI digests of bacterial strains were performed in agarose plugs, followed by separation by PFGE as described previously (6). Subsequently, S1 and I-CeuI digests were transferred to a nylon membrane and hybridized with a labeled hylEfm probe (both S1 nuclease and I-CeuI digests) and with a probe targeting the 23S rRNA genes obtained from the genome sequence of strain A (http://www.hgsc.bcm.tmc.edu) (19) (I-CeuI digests only).
Antimicrobial susceptibility testing.
The antimicrobial susceptibility profiles of selected strains were determined by the agar dilution method for ampicillin, vancomycin, and erythromycin; additionally, evaluation of the presence of HLR to gentamicin and streptomycin was also performed. These susceptibility tests were carried out following the Clinical and Laboratory Standards Institute (CLSI) guidelines (5).
Mouse peritonitis model.
Female (4- to 6-week-old), outbred ICR mice (Harlan Sprague Dawley, Houston) were used as previously described (24). Groups of 10 mice per inoculum (ranging from 1 × 108 to 4 × 109 CFU/ml) were included in each experiment. Strains of E. faecium for inoculation were grown on BHI agar (Difco Laboratories, Detroit, MI) and subsequently grown in BHI broth for 24 h at 37°C. Mice were injected intraperitoneally with 1 ml of a solution containing one of the E. faecium strains in 50% suspended sterile rat fecal extract (24) and monitored every 3 to 6 h for 120 h. Experiments were repeated at least twice. Comparison of the survival curves at similar inocula was performed using a log-rank test with Prism for Windows (GraphPad Software version 4.00); a P value of <0.05 was considered significant.
RESULTS
Conjugative cotransfer of the hylEfm-containing plasmid and antibiotic resistance genes.
Since CC17 clinical strains A (ST-16) and B (ST-18) harbor several antibiotic resistance genes (15), we thought it possible that hylEfm and some of these genes might be cotransferred by conjugation. Table 2 shows the MICs of parent and transconjugant strains, which indicate that resistance to erythromycin and vancomycin and HLR to streptomycin and gentamicin were transferred to the recipient strains. The conjugation efficiencies for resistance to erythromycin and HLR to streptomycin in the TX1330RF mating with strain A were 1.7 × 10−5 and 6.25 × 10−5 transconjugants per donor, respectively, and 7 out of 10 colonies in each mating experiment harbored hylEfm. On the other hand, among the selected hylEfm+ transconjugants from the TX1330RF mating with clinical strain B, the following two phenotypes were evident: (i) resistance to vancomycin and erythromycin and HLR to both gentamicin and streptomycin represented by the transconjugant strain TX1330RF-B1 and (ii) resistance only to vancomycin and HLR to gentamicin (transconjugant TX1330RF-B2) (Table 2). The conjugation efficiencies for vancomycin resistance and HLR to gentamicin were 5.4 × 10−5 and 5.5 × 10−5 transconjugants per donor, respectively. All selected vancomycin- and gentamicin-resistant transconjugants from the strain B-TX1330RF mating carried the hylEfm gene upon screening by PCR (10/10).
TABLE 2.
MICs of E. faecium strains
| Strain | Presence of hylEfm | MIC (μg/ml)a
|
HLRb
|
|||
|---|---|---|---|---|---|---|
| AMP | VAN | ERY | GEN | STR | ||
| A | Yes | 4 | 1 | >512 | − | + |
| TX1330RF | No | 1 | 0.5 | 0.5 | − | − |
| TX1330RF-A1 | Yes | 1 | 0.5 | >512 | − | + |
| B | Yes | 256 | 512 | >512 | + | + |
| TX1330RF-B1 | Yes | 1 | 128 | >512 | + | + |
| TX1330RF-B2 | Yes | 1 | 256 | 0.25 | + | − |
| C | Yes | 128 | 256 | >512 | + | + |
| D344SRF | No | <0.25 | 2 | 512 | − | + |
| D344SRF-C6 | Yes | <0.25 | 2 | >512 | − | + |
AMP, ampicillin; VAN, vancomycin; ERY, erythromycin.
GEN, gentamicin; STR, streptomycin; + or −, strain possesses or does not possess HLR, respectively.
The hylEfm gene is carried on large transferable plasmids in clinical strains of E. faecium.
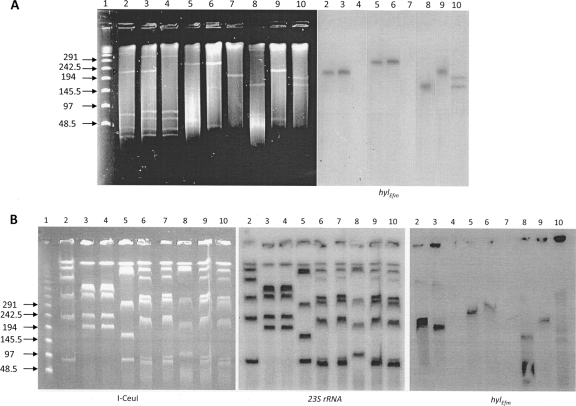
Previously, hylEfm was found to be on a large transferable plasmid in strain C (23), a finding that was confirmed here by cesium chloride plasmid extractions from strain D344SRF-C6 (a transconjugant resulting from the mating of the clinical strain C and the laboratory strain D344SRF) (23) and hybridization with a hylEfm probe (data not shown). Therefore, we sought to investigate whether hylEfm was carried similarly on plasmids in other clinical strains of E. faecium belonging to CC17 and unrelated epidemiologically to the C strain. Thus, we performed S1 nuclease digests with PFGE and hybridizations with a hylEfm probe using the three E. faecium clinical strains and corresponding transconjugants (Fig. 1). This method is very useful since large plasmids appear as linearized bands in the presence of a faint genomic DNA background, which allows the detection and estimation of the size of large bacterial plasmids using PFGE (Fig. 1A). The results showed that plasmid bands hybridizing with hylEfm are present in all clinical strains and transconjugants but are absent in the recipients (D344SRF and TX1330RF) (Fig. 1A), confirming that hylEfm readily transfers as part of large plasmids from CC17 E. faecium strains. The plasmids appeared to differ in size in the clinical strains, with the largest observed in clinical strain A (running between 242.5 and 291 kb) and the smallest in strain B (between 145 and 194 kb). Interestingly, transfer of the hylEfm-containing plasmid from strains A and C resulted in identical plasmid bands in the transconjugants; however, conjugative plasmid transfer from clinical strain B resulted in the following two events: (i) a transconjugant (TX1330RF-B1) carrying a hylEfm-containing plasmid band that was larger than that of the donor (Fig. 1A, lane 9) and (ii) a second transconjugant (TX1330RF-B2) which appeared to carry two hylEfm-containing plasmids, one similar in size to that of the donor strain and an additional one of ca. 194 kb (Fig. 1A, lane 10). The extrachromosomal location of hylEfm was confirmed by I-CeuI digestion, PFGE, and hybridization with a hylEfm probe (Fig. 1B). I-CeuI recognizes a sequence in the genes encoding the 23S rRNA, usually six copies for E. faecium; thus, hybridization with a probe targeting the 23S rRNA results in six chromosomal bands in each strain (Fig. 1B, center panel). Conversely, hybridization with a hylEfm probe resulted in bands that appeared as smears and did not correspond to the chromosomal bands (Fig. 1B, left panel), suggesting the presence of different forms of the plasmid.
FIG. 1.
Results of S1 nuclease and I-CeuI digests, PFGE, and hybridizations. (A) S1 digestion of total DNA of E. faecium strains was followed by PFGE (left panel) and hybridization with hylEfm (right panel). Lane 1, lambda ladder (molecular sizes in kilobases are shown to the left); lanes 2, strain C; lanes 3, transconjugant D344SRF-C6; lanes 4, D344SRF; lanes 5, clinical strain A; lanes 6, transconjugant TX1330RF-A1 (using strain A as donor); lanes 7, TX1330RF (recipient); lanes 8, clinical strain B; lanes 9, transconjugant TX1330RF-B1 (using strain B as donor); lanes 10, transconjugant TX1330RF-B2 (second transconjugant using clinical strain B as donor). Plasmid bands are shown as linearized fragments on the gel; the white arrowheads indicate the plasmid bands hybridizing with hylEfm (right panel). (B) I-CeuI digestion of total DNA was followed by PFGE (left panel) and hybridization with 23S rRNA (center panel) and hylEfm (right panel) probes. Lane 1, lambda ladder (molecular sizes in kilobases are shown to the left); lanes 2, strain C; lanes 3, transconjugant D344SRF-C6; lanes 4, D344SRF; lanes 5, clinical strain A; lanes 6, transconjugant TX1330RF-A1; lanes 7, TX1330RF; lanes 8, clinical strain B; lanes 9, transconjugant TX1330RF-B1; lanes 10, second transconjugant, TX1330RF-B2. The figure is a composite of gels and hybridizations from different experiments.
Vancomycin resistance genes are physically linked to hylEfm in the plasmids.
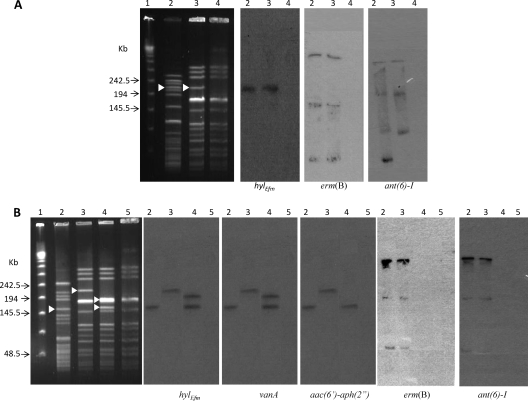
The results for cotransfer of hylEfm and antibiotic resistance determinants led to the hypothesis that some antibiotic resistance markers might be present on the same mobilizable elements as the hylEfm gene. Thus, we generated probes for genes commonly associated with resistance to vancomycin (vanA) and erythromycin [erm(B)] and HLR to gentamicin [aac(6′)-aph(2")] and streptomycin [ant(6′)-I] in E. faecium and hybridized SmaI-digested total DNA of donor strains and transconjugants after PFGE. In clinical strain A and its transconjugant (TX1330RF-A1), hybridizations with erm(B) and ant(6′)-I yielded several bands of different molecular weights that were clearly different from those hybridizing with the hylEfm probe (Fig. 2A), suggesting that erm(B) and ant(6′)-I were likely to be on different genetic elements than those carrying hylEfm. Conversely, the patterns of hybridization of hylEfm were different in the two selected transconjugants from the mating of clinical strain B (donor) and TX1330RF (TX1330RF-B1 and TX1330RF-B2); the two transconjugants also differed slightly in their SmaI PFGE pattern (Fig. 2B). In TX1330RF-B2, two bands yielding a hylEfm signal were observed (one of which was at the same level as the one hybridizing with the hylEfm probe in the donor strain, B) (Fig. 2B, lane 4), whereas a single band of a higher molecular weight was identified in the TX1330RF-B1 transconjugant (Fig. 2B, lane 3), confirming the results obtained with S1 nuclease digestion (see above). Interestingly, hybridization patterns identical to that of hylEfm were observed for the vanA gene in clinical strain B and its transconjugants (Fig. 2B); the aac(6′)-aph(2") probe also hybridized to the same bands as hylEfm did, with the exception of the transconjugant TX1330RF-B2, in which only one band yielding a signal (instead of two) was detected (Fig. 2B). The results suggest that vancomycin resistance genes cotransfer with the hylEfm-containing plasmid and that the gene clusters are likely to be physically linked.
FIG. 2.
Results of PFGE (SmaI digestion; leftmost panels) and hybridizations with hylEfm, erm(B), ant(6)-I, vanA, and aac(6′)-aph(2") with E. faecium strains. (A) Lane 1, lambda ladder; lanes 2, clinical strain A (TX16), lanes 3, transconjugant TX1330RF-A1 to which the hylEfm gene from strain A was transferred; lanes 4, TX1330RF. (B) Lane 1, lambda ladder; lanes 2, clinical strain B; lanes 3, TX1330RF-B1 (transconjugant of TX1330RF to which the hylEfm gene, resistance to vancomycin and erythromycin, and HLR to gentamicin and streptomycin from clinical strain B were transferred); lanes 4, TX1330RF-B2 (second transconjugant of TX1330RF, to which the hylEfm gene, resistance to erythromycin, and HLR to streptomycin from clinical strain B were transferred); lanes 5, TX1330RF. White arrowheads indicate the bands to which the hylEfm probe hybridized. The figure was created using blots from different experiments.
The hylEfm-containing plasmid increases virulence in experimental peritonitis.
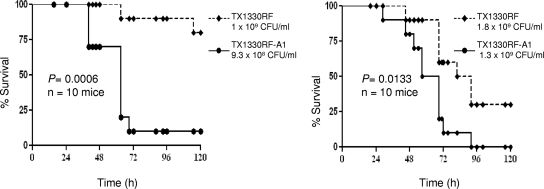
The hylEfm+ TX1330RF transconjugant (TX1330RF-A1) was compared to its parental strain (lacking the hylEfm plasmid) in the mouse peritonitis model (24). Acquisition of the hylEfm-containing plasmid by the transconjugant resulted in significantly decreased survival of mice injected with different inocula and in experiments performed on different days (P = 0.0006 and P = 0.0133, respectively) (Fig. 3). The increased lethality was maintained even when a slightly higher inoculum was used for the parental strain TX1330RF (Fig. 3), indicating that acquisition of the hylEfm-containing plasmid from E. faecium TX16 confers increased virulence in vivo in the mouse peritonitis model.
FIG. 3.
Survival of mice after intraperitoneal inoculation with E. faecium strains. Survival curves using strains TX1330RF and TX1330RF-A1 (TX1330RF transconjugant to which the hylEfm-containing plasmid was transferred) in the mouse peritonitis model (10 mice per group) at two different inocula in independent experiments are shown.
DISCUSSION
To explain the changing trends in the epidemiology of E. faecium, several studies have evaluated the possible association of “epidemic” E. faecium isolates with putative virulence genes. In Europe, the association of a gene called espfm (a homolog of the E. faecalis esp gene) with isolates from CC17 has been reported (27); the espfm gene has been shown to be transferable by conjugation (20), and it has also been noted to be relevant for biofilm formation in vitro (11). Recently, an intact acm gene, encoding a cell wall-anchored collagen adhesin (the expression of which is enriched in CC17 isolates), was shown in vivo to play an important role in E. faecium pathogenesis in a rat model of endocarditis (18). However, the presence of this gene did not increase virulence in the mouse model of peritonitis (18), indicating that acm expression may not be as important in other E. faecium infections. In the current study, we present evidence for the first time that a plasmid plays a role in E. faecium pathogenesis in the mouse peritonitis model and that this plasmid contains the hylEfm gene and antibiotic resistance genes. We also show that the hylEfm-containing plasmids from CC17 clinical strains isolated in different parts of the world were transferable by conjugation, confirming that the acquisition of the hylEfm-containing plasmid may be a strategy to increase the in vivo virulence or survival ability of nosocomial E. faecium strains. Using a combination of S1 nuclease and I-CeuI digestions and PFGE (which have been shown to be accurate in the detection, linearization, and size estimation of large plasmids) (3), we determined that the hylEfm-containing plasmids are larger than 145 kb in all the clinical strains studied. The presence of such large hylEfm-containing plasmids indicates that many potential genes could participate and increase the pathogenic properties of E. faecium carrying this plasmid. Although hylEfm has been the focus of attention and has even been designated a “virulence determinant” in several recent surveys worldwide (9, 28), the exact role of hylEfm in pathogenesis has not been elucidated and our preliminary data indicate that this gene alone is not sufficient to explain all the virulence properties of the plasmid carrying it (not shown). In a previous study, the transfer of the hylEfm-containing plasmid to E. faecium strains also conferred an increased ability to colonize the gastrointestinal tract of mice which was independent of the expression of antibiotic resistance determinants (23). Therefore, it appears that E. faecium isolates (mostly from the CC17 genotype) have recruited important genes in a conjugative plasmid which appear to play an important role in both virulence and colonization.
Similarly, the acquisition of antimicrobial resistance genes in E. faecium poses immense therapeutic challenges for severe infections caused by these organisms, with resistance emerging to virtually all antibiotics used in clinical practice (1). In this work, we also show that acquisition and cotransfer (with hylEfm) of important antibiotic resistance determinants, such as those conferring glycopeptide resistance, among others, readily occur, and we can draw the following conclusions from the results of our mating experiments. (i) Antibiotic resistance genes also cotransferred with the hylEfm-containing plasmid at relatively high frequencies. (ii) Vancomycin resistance genes, as well as aac(6′)-aph(2") in transconjugant TX1330RF-B1, were physically linked to hylEfm and are likely to be in the same plasmid. (iii) erm(B) and ant(6′)-I appeared to be present in different genetic elements (likely on a different plasmid which does not have a SmaI restriction site) which, nonetheless, also cotransfer with hylEfm, albeit at a lower rate of frequency. (iv) Gene rearrangements of the hylEfm and vanA regions appeared to occur during the conjugation process, leading to gene duplications and/or deletions; this may have resulted from the movement of transposable elements with which hylEfm (21) and the vanA gene cluster are known to be associated (2). The latter point is illustrated by the results of the mating experiment using the Colombian clinical strain B as donor. As opposed to the mating experiments with clinical strains A and C which resulted in transconjugants harboring hylEfm-containing plasmids of the same size (Fig. 1), conjugative transfer of the hylEfm-containing plasmid from the B strain yielded plasmids of different sizes. Furthermore, one of the recipients ended up having two distinct hylEfm- and vanA-containing plasmids. These findings support the hypothesis that movement and duplication of transposable elements containing hylEfm and vancomycin-resistance genes to other plasmids can readily occur during conjugative transfer in E. faecium clinical strains. Our results illustrate the striking plasticity and adaptation of enterococci in the hospital environment to become a successful pathogen; in a single genetic unit, these organisms are capable of recruiting and transferring genes that could help with outcompeting other bacteria in the gut in the presence of antibiotics and at the same time increase their ability to produce disease. The increasing prevalence of this multidrug-resistant CC17 E. faecium containing a “virulence” plasmid in the United States and Europe is a worrisome prospect.
In summary, our work indicates that large transferable plasmids containing the hylEfm gene cotransferred with antibiotic resistance markers and that the acquisition of one of these plasmids was associated with enhanced virulence of a commensal strain of E. faecium. Thus, this single genetic unit appears to enhance the colonizing, survival, and virulence properties of members of the CC17 E. faecium genogroup.
Acknowledgments
C.A.A. is supported by NIH Pathway to Independence award K99/R00 AI72961 from the National Institute of Allergy and Infectious Diseases (NIAID). This work was also supported in part by NIH grant R56 AI042399 to B.E.M. from the Division of Microbiology and Infectious Diseases, NIAID. D.P. was partially funded by a graduate scholarship from The Instituto Colombiano para el Desarrollo de la Ciencia y Tecnología Francisco José de Caldas, Colciencias.
Footnotes
Published ahead of print on 10 August 2009.
REFERENCES
- 1.Arias, C. A., and B. E. Murray. 2008. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti-Infect. Ther. 6:637-655. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., and R. Quintiliani, Jr. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 4.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Coburn, P. S., A. S. Baghdayan, G. T. Dolan, and N. Shankar. 2007. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Mol. Microbiol. 63:530-544. [DOI] [PubMed] [Google Scholar]
- 7.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 8.Currier, T. C., and E. W. Nester. 1976. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal. Biochem. 76:431-441. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande, L. M., T. R. Fritsche, G. J. Moet, D. J. Biedenbach, and R. N. Jones. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 58:163-170. [DOI] [PubMed] [Google Scholar]
- 10.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heikens, E., M. J. Bonten, and R. J. Willems. 2007. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J. Bacteriol. 189:8233-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hidron, A. I., J. R. Edwards, J. Patel, T. C. Horan, D. M. Sievert, D. A. Pollock, and S. K. Fridkin. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996-1011. [DOI] [PubMed] [Google Scholar]
- 13.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malathum, K., T. M. Coque, K. V. Singh, and B. E. Murray. 1999. In vitro activities of two ketolides, HMR 3647 and HMR 3004, against gram-positive bacteria. Antimicrob. Agents Chemother. 43:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nallapareddy, S. R., K. V. Singh, and B. E. Murray. 2006. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl. Environ. Microbiol. 72:334-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallapareddy, S. R., K. V. Singh, and B. E. Murray. 2008. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect. Immun. 76:4120-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nallapareddy, S. R., G. M. Weinstock, and B. E. Murray. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 47:1733-1747. [DOI] [PubMed] [Google Scholar]
- 20.Oancea, C., I. Klare, W. Witte, and G. Werner. 2004. Conjugative transfer of the virulence gene, esp, among isolates of Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 54:232-235. [DOI] [PubMed] [Google Scholar]
- 21.Rice, L. B., L. Carias, S. Rudin, C. Vael, H. Goossens, C. Konstabel, I. Klare, S. R. Nallapareddy, W. Huang, and B. E. Murray. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508-512. [DOI] [PubMed] [Google Scholar]
- 22.Rice, L. B., L. L. Carias, S. Marshall, S. D. Rudin, and R. Hutton-Thomas. 2005. Tn5386, a novel Tn916-like mobile element in Enterococcus faecium D344R that interacts with Tn916 to yield a large genomic deletion. J. Bacteriol. 187:6668-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice, L. B., V. Lakticova, L. L. Carias, S. Rudin, R. Hutton, and S. H. Marshall. 2009. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J. Infect. Dis. 199:342-349. [DOI] [PubMed] [Google Scholar]
- 24.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 25.Swenson, J. M., M. J. Ferraro, D. F. Sahm, N. C. Clark, D. H. Culver, F. C. Tenover, et al. 1995. Multilaboratory evaluation of screening methods for detection of high-level aminoglycoside resistance in enterococci. J. Clin. Microbiol. 33:3008-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Top, J., R. Willems, and M. Bonten. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297-308. [DOI] [PubMed] [Google Scholar]
- 27.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 28.Worth, L. J., M. A. Slavin, V. Vankerckhoven, H. Goossens, E. A. Grabsch, and K. A. Thursky. 2008. Virulence determinants in vancomycin-resistant Enterococcus faecium vanB: clonal distribution, prevalence and significance of esp and hyl in Australian patients with haematological disorders. J. Hosp. Infect. 68:137-144. [DOI] [PubMed] [Google Scholar]