The gram-positive bacterium Staphylococcus aureus is a leading cause of hospital- and community-associated infections (16, 85, 108). In the hospital, S. aureus is the most frequent cause of surgical, lower respiratory tract, and cardiovascular infections. Furthermore, it is the second most common cause of health care-associated pneumonia and bloodstream infections (108, 151, 152). Historically, β-lactam antibiotics have exhibited potent activity against S. aureus, which along with good safety profiles make them the agents of choice for the treatment of staphyloccocal infections. Of particular concern now is the growing prevalence of methicillin (meticillin)-resistant S. aureus (MRSA) in both hospital- and community-associated infections (24, 70, 133). The development of resistance to β-lactam antimicrobials, often concurrently with resistance to other antimicrobial agents, poses a great challenge to the prevention and treatment of S. aureus infections (7, 108). Staphylococci have two primary mechanisms for resistance to β-lactam antibiotics: the expression of an enzyme (the PC1 β-lactamase) capable of hydrolyzing the β-lactam ring, thus rendering the antibiotic inactive, and the acquisition of a gene encoding a modified penicillin-binding protein (PBP), known as PBP 2a, found in MRSA and coagulase-negative staphylococci. PBP 2a is intrinsically resistant to inhibition by β-lactams (59). PBP 2a remains active in the presence of concentrations of β-lactam antibiotics that inhibit most endogenous PBP enzymes, thus substituting for their functions in cell wall synthesis and allowing growth in the presence of the β-lactam inhibitors. This review briefly discusses the structure and synthesis of the S. aureus cell wall, the resistance to β-lactam antibiotics through the acquisition of PBP 2a, the evolution of MRSA, and the involvement of other protein factors in methicillin resistance. In addition, the characteristics of new β-lactam antibiotics that target PBP 2a are discussed, along with their role as important new entities in the antibacterial pipeline for the treatment of MRSA infections.
NATIVE STAPHYLOCOCCAL PBPs AND PBP 2A: CELL WALL SYNTHESIS AND β-LACTAM RESISTANCE
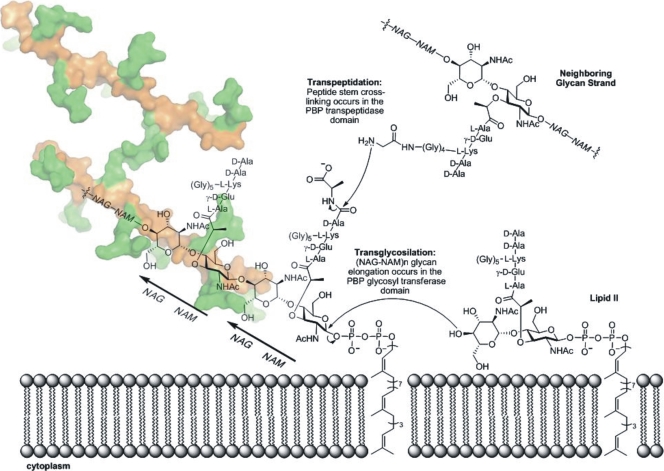
In gram-positive bacteria the cell wall, a key structural component, comprises the outermost layer of the cell, whereas in gram-negative bacteria the cell wall lies underneath an additional layer known as the outer membrane. At the molecular level, the cell wall is a meshwork of glycan (polysaccharide) chains interconnected by peptide cross-links, known as peptidoglycan (Fig. 1) (176). The full three-dimensional structure of the cell wall is unknown. Biosynthesis of the peptidoglycan is accomplished by the membrane-bound enzymes known as penicillin-binding proteins (PBPs) (161). Each bacterium contains several different types of PBPs that not only act as enzyme catalysts for peptidoglycan synthesis during cell growth but also function in cell septation and, in some species, sporulation. PBPs are localized to the extracellular surface of the cytoplasmic membrane via a membrane anchor. These biosynthetic proteins may catalyze both a glycosyltransferase activity (for the elongation of glycan strands) and a transpeptidase activity (for the interconnection of the glycan strands by peptide cross-linking) (Fig. 1) (68, 116, 173). While all PBPs have a transpeptidase domain, both glycosyltransferase and transpeptidase domains exist in certain bifunctional PBPs. β-Lactam antibiotics irreversibly acylate the catalytic serine within the transpeptidase active site of the biosynthetic PBPs and hence inhibit peptide cross-linking (59, 178). The loss of this catalytic activity impairs the ability of the bacterium to control the integrity of its cell wall. The loss of this control is ultimately bactericidal. Moreover, the cell wall also contributes to infectivity and pathogenicity, and the glycopeptides (muropeptides) released from the cell wall during cell growth (and cell death) activate the human immune system (30, 71, 72, 138, 155, 174).
FIG. 1.
Schematic representation of cell wall synthesis in S. aureus. The transpeptidase and transglycosylase reactions assemble the peptidoglycan on the surface of the cytoplasmic membrane. The three-dimensional structure of the peptidogycan was recently solved by our laboratory (120a), and it is depicted as a Connolly surface with the sugar backbone (NAG-NAM polymer) in orange and the peptides in green. Two strands of peptidoglycan are shown with cross-linking through a peptide bridge shared between the two. NHAc, N-acetyl group.
The structure of the glycan strands of the peptidoglycan consists of repeats of a β-1,4-linked N-acetyl-glucosamine-N-acetyl-muramic acid (NAG-NAM) disaccharide (Fig. 1). The NAM saccharide of this pair contains a pentapeptide stem (in S. aureus, NAM-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala), which forms a cross-link (by displacement of the terminal d-Ala of this stem, during the transpeptidation reaction) with the residue before the first d-Ala of the peptide stem of an adjacent glycan chain (Fig. 1) (59). There are some variations among bacteria in the characteristics of the cross-linking, and in some there is an interpeptide bridge (176). In S. aureus the cross-linking occurs to a pentaglycine pentapeptide extension attached to the ɛ-amine of the l-Lys of the peptide stem of an adjacent glycan strand (Fig. 1). Synthesis of the glycan strand occurs in the transglycosylase domain of a biosynthetic bifunctional PBP, and stem cross-linking occurs in the transpeptidase domain of the biosynthetic PBPs. In the bifunctional PBPs, such as PBP 2 of S. aureus (59), the transglycosylase and transpeptidase domains are spatially well separated. Carboxypeptidases, also members of the PBP family, remove the terminal d-Ala of the peptidoglycan peptide stems. This reaction moderates the degree of cross-linking of the peptides (66).
The structural similarity of β-lactam antibiotics to the acyl-d-Ala-d-Ala moiety of the peptidoglycan provides the mechanistic basis for the inactivation of the PBP transpeptidase (and carboxypeptidase) domain, according to the Tipper-Strominger hypothesis (93, 178). When the β-lactam is bound in the transpeptidase/carboxypeptidase PBP domains, the identical serine nucleophile used for catalytic transpeptidation attacks the carbonyl of the β-lactam ring, resulting in serine acylation. This β-lactam-derived acyl-enzyme complex, which is stable, in contrast to the transient acyl-enzyme complex derived from d-Ala-d-Ala, is incapable of transpeptidation and undergoes very slow hydrolysis to regenerate the free serine. This functionally irreversible acylation stops the transpeptidase activity of the biosynthetic PBPs and the carboxypeptidase activity of the PBPs involved in maturation of the cell wall structure. The basis for β-lactam resistance by PBP 2a-containing MRSA strains is the intrinsically lower reactivity of its transpeptidase domain to β-lactam acylation (58, 185). As PBP 2a lacks transglycosylase activity, cell wall biosynthesis in MRSA is completely dependent on the cooperative function of the PBP 2a transpeptidase with the transglycosylase domain of PBP 2 (141). Hence, when the transpeptidase activity of the native staphylococcal PBPs is abolished by β-lactam acylation, PBP 2a functions as a surrogate to maintain cell wall synthesis at β-lactam concentrations that inhibit β-lactam-sensitive PBPs (59).
The kinetic mechanism for the reaction of PBP 2a with β-lactams is identical to that for the transpeptidase/carboxypeptidase domains of all other PBPs. A noncovalent enzyme-β-lactam complex, characterized by an equilibrium dissociation constant, KD, is converted to the covalent acyl-enzyme form with a rate constant, k2 (58). Since the acyl-enzyme is incapable of transpeptidation, the only mechanism for regenerating free enzyme is slow hydrolytic deacylation (k3), releasing the ring-opened β-lactam as the product. For most PBPs (and for most β-lactams), k3 is manifested over a longer duration than cell viability in the presence of the β-lactam antibiotic. The KD values for PBP 2a are in the millimolar range (0.2 to 1.3 mM, depending on the β-lactam structure), which is much higher than those observed for PBP 2 (58, 112). Moreover, the acylation rate constant (k2) observed for PBP 2a is 3 to 4 orders of magnitude smaller than the acylation rate observed with β-lactam-sensitive PBPs (58). Once it is acylated, however, hydrolytic PBP 2a deacylation occurs with a small rate constant, similar to those for the other S. aureus PBPs. Therefore, deacylation does not determine β-lactam susceptibility.
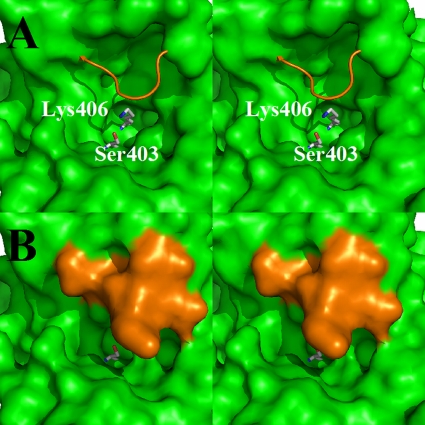
The basis for the resistance of PBP 2a to β-lactam acylation is a higher KD of the noncovalent β-lactam-PBP 2a complex and lower rates for serine acylation following binding. High-resolution crystal structures of PBP 2a and its acylated forms with different β-lactams suggest a structural basis for both observations (105). The higher KD is proposed to reflect the diminished accessibility of the β-lactams due to the presence of a loop that covers the active site (Fig. 2) (58), whereas inefficient acylation is thought to result from a suboptimal position for the nucleophilic serine (105). Circular dichroism analysis demonstrates a conformational change in PBP 2a upon acylation (57, 58). Experiments using muropeptide mimetics of the peptidoglycan show a greater accessibility of the PBP 2a active site to the β-lactam antibiotic (56). Binding of the peptidoglycan to an allosteric site in PBP 2a was proposed to facilitate a conformational change, thus displacing the peptide loop (Fig. 2) (56). The displacement of the loop enables access of both the substrate and the β-lactam to the active site (56). As such, it might be a critical step in the course of the physiological events involved in cell wall biosynthesis in MRSA.
FIG. 2.
(A) Stereo view of the active site of PBP 2a of S. aureus depicted as a Connolly solvent-accessible surface (green). The side chains of the active-site serine and lysine are shown as capped sticks and colored by atom types (carbon in gray, oxygen in red, and nitrogen in blue). The backbone of the loop that caps the active site is shown as an orange wire. (B) Stereo view of the active site from the same perspective shown in panel A, except the loop is now shown as a solvent-accessible surface for both the backbone and the side chain functionalities. The presence of the loop blocks the active-site access for molecules the size of typical β-lactam antibiotics.
mecA GENE AS A DETERMINANT OF RESISTANCE TO β-LACTAMS
A compelling question is the origin of the gene encoding the PBP 2a enzyme in MRSA. The gene encoding PBP 2a, known as mecA, is localized in a unique segment of DNA referred to as the staphylococcal chromosome cassette (SCC) (10, 45). The SCC is a large mobile element integrated into the S. aureus chromosome at a site-specific location (attBscc) near the origin of replication (102). The mobility of SCCmec is in part due to the presence of fully functional recombinases of the invertase/resolvase family, encoded by the ccr gene complex (87). The SCC has its own evolutionary history and is capable of delivering a variety of different resistance or virulence determinants to S. aureus beyond mecA. On the basis of the genetic organization in the vicinity of the mecA gene, three SCC classes have been described: class A contains the complete mecA regulon (mecI-mecR1-mecA), while classes B and C contain the mecA regulatory genes disrupted by insertion sequences (48, 88). SCCmec types are defined by combining the class of the mec gene with the ccr allotype (ccrAB allotypes 1 to 4 or ccrC) (31, 48). The SCCmec element may also encode additional functions, including antibiotic resistance determinants that may contribute to the survival or pathogenic potential of the bacteria (31, 49, 81, 113, 124, 166). De Lencastre et al. (45) and Deurenberg and Stobberingh (48) discuss in detail the SCCmec core structure and definitions. Resistance to some antibiotics by S. aureus may also be plasmid mediated (67, 92), as is the case for plasmids that confer resistance to the glycopeptide vancomycin (179).
Transcription of the blaZ β-lactamase gene and the PBP 2a gene in MRSA is controlled by the BlaR1-BlaI-BlaZ and MecR1-MecI-MecA regulatory systems, respectively. The two systems are remarkably similar in structure and function (35, 65, 79, 104) but retain distinct identities. For example, they exhibit corepression but not coinduction (119, 156). BlaI and MecI are DNA-binding proteins that repress gene transcription by binding to the operator region of the operon. The regulation of the mec system is believed to parallel the better-characterized bla gene regulatory system. BlaR1 and MecR1 are β-lactam antibiotic sensor/signal transducer integral membrane proteins. When the bacterium is exposed to a β-lactam, a serine residue in the extracellular sensor domain of BlaR1 and MecR1 undergoes irreversible acylation by the β-lactam. This results in a conformational change in the sensor domain that is proposed to transmit the signal (i.e., the presence of a β-lactam) to the cytoplasmic domain of BlaR1/MecR1 (23, 69, 167). The molecular details of the signal transmission events are unknown. One conjecture is that the activated cytoplasmic domain has proteolytic activity and that it proteolyzes BlaI (either directly or through a surrogate protease) (187). Proteolysis disrupts the BlaI repressor dimer, leading to derepression of the operon (187). Despite the profound similarities at the genetic level, there is a significant temporal difference in the expression of the two systems. The induction of the bla system, which results in blaZ transcription, occurs within minutes, whereas the induction of mecA takes several hours (119, 156).
The earliest MRSA isolates are believed to have acquired the mecA and mecR1-mecI sequences en bloc from an unknown bacterial donor (78). These isolates would likely have appeared to be phenotypically methicillin susceptible because of the strong MecI repression of mecA transcription and the failure of MecR1 to mediate the induction of resistance. Many clinical MRSA isolates have specific deletions of mecI (which confer constitutive mecA expression), mutations in the presumed mecI operator, or point mutations in the mecI gene (78, 165). All of these events confer methicillin resistance. In some strains with intact mecI genes, MecI-mediated repression appears to be dysfunctional, which results in heterotypic resistance expression independent of mecA transcriptional regulation (130). This might reflect the presence or absence of additional regulatory cofactors not yet identified.
PBP 2a is the most abundant PBP in the MRSA cell (146); however, the abundance of PBP 2a does not fully correlate with the level of resistance (130, 139, 153, 157). Some MRSA strains present heterogeneous methicillin resistance, in which the majority of the cells are resistant to relatively low concentrations of methicillin, while a much smaller proportion are able to grow at higher antibiotic concentrations.
EVOLUTION OF METHICILLIN RESISTANCE IN S. AUREUS
Staphylococcus sciuri is a frequent inhabitant of the skin of rodents, primitive mammals, and domestic animals (45, 164). Recent work has identified a mecA homologous gene in S. sciuri (designated pbpD) as the genetic determinant of an 84-kDa PBP (PBP 4) (188). The majority of S. sciuri isolates are fully susceptible to β-lactam antibiotics, in spite of the presence of the mecA homologue (37, 38). In some unusual S. sciuri strains isolated from humans, however, upregulation of the function of the promoter of this gene by mutational change of the promoter sequence provides broad-spectrum β-lactam resistance (39). A similar upregulation phenomenon was observed in S. sciuri strains selected in vitro for methicillin resistance (182). In addition, transfer of the upregulated S. sciuri mecA homologue to a fully β-lactam-susceptible S. aureus strain resulted in a substantial increase in the oxacillin MIC of the S. aureus recipient. A recent study demonstrated that transduction of methicillin-susceptible S. aureus (MSSA) strain COL-S with the S. sciuri gene coding for PBP 4 resulted in strains with broad-spectrum, high-level, and homogeneous resistance to structurally different β-lactams (5). The protein product of the S. sciuri mecA homologue has properties similar to those of PBP 2a: a low affinity for β-lactam antibiotics, an allosteric site for binding of peptidoglycan precursors, and a sheltered active site requiring a conformational change for substrate and β-lactam access (57, 188). Evolution of the complex mecA-based resistance mechanism has been proposed to have occurred in S. sciuri under the selective pressure of penicillin β-lactams (47), which began to be used extensively as prophylactic agents in veterinary medicine in 1949 (45, 149). An upregulated version of the native S. sciuri mecA homologue may have emerged from colonizers of the skin of domestic animals, under the selective pressure of the prophylactic use of penicillin (45). The first instances of mecA-mediated methicillin resistance were identified in clinical isolates within 1 to 2 years after the introduction of the first penicillinase-resistant β-lactam into clinical use (45), a time frame that appears to be too short for the evolution of mecA-mediated resistance mechanisms. The mecA gene was most likely incorporated into an SCC prior to its transfer into S. aureus by a mechanism that remains to be elucidated. One possible mechanism for SCC transfer is by transduction by one of the many staphylococcal phages (45). Only a small number of clonal lineages dominate the global population structure of MRSA, so the concept has developed that MRSA emerged on a few occasions after penicillinase-stable β-lactam antibiotics were introduced into clinical practice, followed by the intercontinental spread of individual clones (40, 48, 52, 80).
Recent experiments suggest that many S. aureus genetic backgrounds have a barrier against maintaining and expressing a plasmid-borne mecA gene (90). Bypassing this barrier appears to be the property of a select group of S. aureus genetic backgrounds. It has been hypothesized that this barrier may explain the relatively low number of MRSA lineages identified in epidemiological studies (90). Alternatively, it is possible that methicillin resistance has arisen on multiple independent occasions through the acquisition of SCCmec elements by MSSA strains and that MRSA clones appear to be ubiquitous only because they cannot be distinguished by currently used typing procedures (multilocus sequence typing and differentiation of surface protein genes) (133). The most common sequence variations in bacterial genomes are point mutations, which result in single-nucleotide polymorphisms (SNPs) (133). Genome-wide SNPs can reveal the details of bacterial evolutionary history with great precision (133). Nubel et al. (133) have investigated the evolutionary history of an MRSA clone (sequence type 5) by mutation discovery at 108 loci (46 kb) within a global collection of 135 isolates. The analysis of the SNPs provided strong evidence that the geographical spread of MRSA over long distances and across cultural borders is a rare event compared with the frequency with which the SCC island has been imported.
PBP 2A: THE SOLE DETERMINANT OF METHICILLIN RESISTANCE?
Community-associated MRSA (CA-MRSA) strains have a different genetic background, coinciding with increased virulence in animal models, compared with the genetic background of hospital-associated MRSA (HA-MRSA) strains (6, 10, 42, 50, 51, 134, 143, 144). Recent cases of infections caused by CA-MRSA in healthy individuals who have no known risk factors for the acquisition of MRSA have raised great concern. A recent study suggests that CA-MRSA will become the dominant MRSA strain in hospitals and health care facilities (41). CA-MRSA is primarily associated with skin and soft tissue infections (abscesses, cellulitis, and furunculosis); but severe cases of CA-MRSA infection associated with septic shock, bacteremia, endocarditis, and necrotizing pneumonia have also been reported (19, 26, 111, 121, 186).
CA-MRSA strains that are positive for mecA and PBP 2a but that appear to be phenotypically oxacillin susceptible have been reported (55, 82, 83, 86). Although mecA expression is normally a prerequisite for methicillin resistance, mecA expression alone does not appear to be sufficient to guarantee phenotypic methicillin resistance, which suggests the existence of additional molecular targets that could be associated with the susceptibility to oxacillin in certain strains (83). Recent publications suggest the involvement of genes other than the known effectors of methicillin resistance in CA-MRSA. The vraS/vraR two-component regulatory system is required for oxacillin resistance in CA-MRSA (9, 18, 61). The study of vraS mutants of strains that lacked the regulatory sequences mecI and mecR1 showed that mecA transcription was increased but that the abundance of PBP 2a was similar to that in the wild type. In addition, pbp2 transcription decreased but overexpression of the pbp2 operon did not restore resistance. These results indicate that the vraS/vraR regulatory system might have an important role in the CA-MRSA response to β-lactams and suggests the importance of regulatory effects on genes other than these known effectors of methicillin resistance. In a recent study, deletion of the gene encoding PBP 4 in two common CA-MRSA isolates, USA300 and MW2 (USA400), resulted in a 16-fold reduction in oxacillin and nafcillin resistance in these particular stains (120). However, the same deletion introduced into HA-MRSA strains had little or no effect on oxacillin MICs (120). In CA-MRSA MW2, the loss of pbp4 diminished the induced expression of pbp2 upon exposure to the cell wall-active antibiotics oxacillin and vancomycin, while the transcript level of mecA was not altered (120). The decrease in PBP 2 expression correlated with a significant decrease in peptidoglycan cross-linking. These studies suggest that PBP 4 could represent a significant target for the discovery of agents effective against CA-MRSA. Although PBP 4 has a low affinity for most β-lactams, cefoxitin, a semisynthetic β-lactam, efficiently acylates PBP 4. As predicted from these observations, cefoxitin is synergistic with oxacillin in killing CA-MRSA strains, including clinical isolates (120).
A recent study described the selection and characterization of S. aureus strains resistant to ceftobiprole (8). Ceftobiprole is an anti-MRSA cephalosporin currently under development. Ceftobiprole-resistant mutants were selected from strains with and without the mecA gene, which was located on a plasmid. Resistant mutants selected from the control strain that lacked the mecA gene displayed even higher levels of resistance compared with the levels of resistance of mutants selected from strains that carried the mecA gene. The highest levels of resistance came from mutations in chromosomal genes, which have not yet been identified. Sequence analysis of the pbp genes in these resistant mutants identified two amino acid substitutions in PBP 4, although the contribution of these mutations to ceftobiprole resistance is not known. As mentioned before, PBP 4 inactivation in HA-MRSA strains has minimal effects on methicillin resistance (89, 120). Hence, the full scope of PBP 4 involvement in β-lactam resistance is unknown.
Unidentified chromosomal loci were previously implicated in oxacillin resistance in clinical S. aureus isolates lacking mecA (169). Many other studies have demonstrated the participation of additional factors in the expression of high levels of homogeneous methicillin resistance in MRSA (11-13, 25, 44, 46, 95-99, 114, 126, 181). A number of auxiliary genes influencing methicillin resistance have either proven or probable roles in cell wall synthesis. These include proteins (murE, femA, and femV) involved in the recruitment of PBPs to the septum (the site of cell wall synthesis) and autolysins that control cell wall turnover (12-14, 46, 142, 148). Others are involved in the regulation of the global stress response (e.g., the σB gene) and in metabolism (e.g., protein kinase and ABC transporter genes).
Among the identified methicillin resistance factors, fmtA is part of the cell wall stimulon, a group of genes that is commonly induced upon treatment with cell wall-active antibiotics, including the β-lactams (101, 117, 118, 172). Inactivation of the fmtA gene is associated with reduced peptidoglycan cross-linking and methicillin resistance (97). Hence, the protein FmtA, whose function is unknown, has been hypothesized to participate in peptidoglycan biosynthesis under antibiotic-induced cell wall stress conditions. β-Lactams acylate a serine residue of FmtA but at a very low rate, even compared with that of PBP 2a (54). FmtA is the only native PBP of S. aureus intrinsically resistant to inactivation by β-lactams. In the presence of β-lactams, FmtA may cooperate with PBP 2a for functional cross-linking of the peptidoglycan (54). Additional roles for FmtA are suggested by a study of biofilm formation-defective S. aureus mutants (170). Mutants with transposon insertions in fmtA showed the undetectable formation of cell wall teichoic acid and defective biofilm formation. Hence, drugs targeting FmtA might provide a potential solution for two major problems confronting the clinical management of staphylococcal infections: biofilm formation and methicillin resistance.
CURRENT CLINICAL TREATMENTS FOR MRSA INFECTIONS
The difficulty in identifying new compounds with suitable antibacterial activity is one of the major problems faced in the fight against resistant organisms (140, 180). Natural product (NP)-derived drugs have played a pivotal role in anti-infective drug development. Most antibacterial drugs have been developed from modifications of NP scaffolds. Antibacterial NPs have evolved to penetrate bacteria and to interact with specific protein targets. A microorganism producing these compounds, either for the purpose of defense or for intercellular communication, has an evolutionary advantage (21). Penicillin, the first class of the β-lactam antibiotics discovered, gave rise to successive groups of related β-lactams for the treatment of bacterial infections. With the development of resistance, however, many of these β-lactams are ineffective against a significant proportion of S. aureus clinical strains (73).
The glycopeptides belong to a different class of NP-derived antibiotics effective against gram-positive organisms. Vancomycin and teicoplanin are the preeminent members of this class of antibiotics (108), and vancomycin is the antibiotic most commonly used to treat MRSA infections. The inhibition of cell wall biosynthesis by the glycopeptides results from their stable noncovalent binding to the d-Ala-d-Ala termini of peptidoglycan precursors. Some MRSA strains have evolved resistance to vancomycin (94, 159). Linezolid, quinupristin-dalfopristin, daptomycin, and tigecycline represent newer agents for the treatment of S. aureus infections, including those caused by non-vancomycin-susceptible MRSA. Linezolid (Zyvox; Pfizer, New York, NY) is a synthetic oxazolidinone antimicrobial agent that blocks the formation of protein synthesis initiation complexes. Quinupristin-dalfopristin (Synercid; DSM Pharmaceuticals Inc., Greenville, NC) is a mixture of semisynthetic streptogramin derivatives that bind to different sites of the 50S ribosomal subunit, resulting in the irreversible inhibition of bacterial protein synthesis. Daptomycin (Cubicin; Cubist Pharmaceuticals, Inc., Lexington, MA) is a cyclic lipopeptide that forms a Ca2+ complex in the bacterial cytoplasmic membrane, causing the loss of the transmembrane electrical potential gradient (2). Inhibition of protein, RNA, DNA, peptidoglycan, lipoteichoic acid, and lipid biosynthesis are also observed, although it is not yet clear if these are consequences of the loss of the transmembrane electrical potential gradient or the independent effects of daptomycin. Tigecycline (Tygacil; Wyeth Pharmaceuticals Inc., Philadelphia, PA), a glycylcycline, is a new tetracycline that inhibits protein translation in bacteria. Several recent reviews present an excellent overview of the mechanisms of action of these antibiotics as well as their clinical efficacies (1, 2, 28, 33, 74, 108, 122, 137, 177).
The remaining portion of this review focuses on four new β-lactam antibiotics, ceftobiprole, ceftaroline, ME1036, and PZ-601, which inhibit PBP 2a, the main protein involved in S. aureus resistance to β-lactams. In view of the great threat that MRSA represents, investigation of multiple alternative treatments aimed at different targets, such as protein synthesis, DNA or RNA synthesis, and the cytoplasmic membrane, is warranted. Several new glycopeptides (dalbavancin, oritavancin, telavancin), oxazolidinones (RX-1741, TR-701), streptogramins (flopristin/linopristin), and tetracyclines (PTK 0796), as well as other new antibacterial agents (e.g., friulimicin and iclaprim), are also being developed for the treatment of MRSA infections (1, 17, 74, 137, 145). Furthermore, nontraditional anti-infection targets, such as inhibitors of resistance mechanisms, virulence factors, and other proteins involved in cell wall synthesis, are also emerging as alternative strategies (27, 53, 75, 103, 106, 109, 134, 180). Peptidoglycan glycosyltransferases are essential enzymes, even for antibiotic-resistant bacteria such as MRSA. The structures of S. aureus PBP 2 unliganded and complexed with the substrate analog moenomycin gave insight into the key interactions required for enzyme inhibition (110). This approach was used to design an inhibitor of S. aureus PBP 2 (184). The development of high-throughput peptidoglycan glycosyltransferase assays (29) will facilitate further inhibitor discovery.
The overwhelming capacity of bacteria to develop resistance to antibiotics demands a constant effort to identify new antibiotics against both traditional and new targets. A renaissance in NP discovery, based on an expanding effort to explore nontraditional sources of microbial biodiversity and molecular tools, is revealing new chemical classes with the potential to prime the antibiotic pipeline (33, 177, 180).
β-LACTAM ANTIBIOTICS THAT INHIBIT PBP 2a: NEW WEAPONS IN THE PIPELINE
Four β-lactam antibiotics (Fig. 3) are currently being evaluated for the treatment of MRSA-associated infections by targeted inhibition of PBP 2a transpeptidase activity. In a review published in 2005, Guignard et al. (73) described 16 novel anti-MRSA β-lactam antibiotics, only 1 of which (ceftobiprole) remains in clinical development. In a later review, Page reported on a new cephalosporin (ceftaroline) also active against MRSA (135). In the last 3 years, two additional β-lactam antibiotics, ME1036 and PZ-601, have been added to the anti-MRSA pipeline (1, 21, 22, 91).
FIG. 3.
Chemical structures of the β-lactam antibiotics that are in advanced stages of evaluation for the treatment of MRSA infections. The chemical groups that are cleaved upon activation of the ceftobiprole medocaril and ceftaroline fosamil prodrugs are highlighted in gray.
Ceftobiprole medocaril.
Ceftobiprole medocaril (BAL-5788), a water-soluble cephalosporin prodrug, belongs to a new class of cephem antibiotics with activity against a wide range of gram-positive organisms, including MRSA and penicillin-resistant Streptococcus pneumoniae (PRSP), and gram-negative pathogenic bacteria (3, 20, 136). Ceftobiprole medocaril is being developed by Johnson & Johnson Pharmaceutical Research & Development, L.L.C., (Raritan, NJ) and Basilea Pharmaceutica Ltd. (Basel, Switzerland) and is currently under regulatory review in the United States for the treatment of complicated skin and skin structure infections (cSSSIs). Ceftobiprole is marketed in Canada and approved in Switzerland for the treatment of cSSSIs, including diabetic foot infections.
Rapid cleavage of ceftobiprole medocaril in plasma produces the active drug, ceftobiprole (127). Ceftobiprole inactivates all four S. aureus PBPs and PBP 2a, as indicated by competition assays against a fluorescent β-lactam. The 50% inhibitory concentration (IC50s) for the acylation of S. aureus PBPs 1, 2, 3, and 4 by ceftobiprole are 0.1, 0.5, 0.05, and 1 μg/ml, respectively, while the IC50 for the acylation of PBP 2a is 0.9 μg/ml (43). The greater activity of ceftobiprole against MRSA (MIC = 2 μg/ml) compared with the activities of other older cephalosporins (ceftriaxone, MIC > 64 μg/ml; ceftazidime, MIC > 128 μg/ml) correlates with the increased affinity of ceftobiprole for PBP 2a and native PBPs (32, 43). Ceftobiprole is relatively stable toward class C β-lactamases and demonstrates a low propensity to induce these enzymes (135). It does not select for stably derepressed mutants in strains producing these enzymes. Ceftobiprole is resistant to hydrolysis by the common staphylococcal PC1 β-lactamase, the class A TEM-1 β-lactamase, and the class C AmpC β-lactamase but is labile to hydrolysis by class B, class D, and class A extended-spectrum β-lactamases (147).
In some in vitro resistance development studies, the MICs for ceftobiprole did not increase in highly oxacillin-resistant subpopulations from individual MRSA strains (15, 76). However, in a more recent study, passage in high-volume broth cultures containing subinhibitory concentrations selected ceftobiprole-resistant mutants from strains carrying either wild-type mecA or mutant mecA (8). A plasmid with mutated mecA isolated from one of these ceftobiprole-resistant strains conferred resistance to a sensitive S. aureus strain, and the loss of this mecA plasmid restored susceptibility. Modeling studies suggested that some of the mutations might alter ceftobiprole binding, whereas other mutations may influence PBP 2a interactions with other proteins (8).
Ceftobiprole medocaril, the prodrug, is converted rapidly and almost completely by type A esterases to active ceftobiprole (127). Ceftobiprole has a steady-state volume of distribution of 18.4 liters and binds minimally (16%) to plasma proteins (127). Ceftobiprole undergoes minimal hepatic metabolism and is eliminated primarily through renal excretion (127). In a single- and multidose study using intravenous infusion over 30 min, ceftobiprole exhibited linear pharmacokinetics across the dose range of 125 to 1,000 mg (162, 163). Steady-state drug concentrations were attained on the first day of dosing, and there was no appreciable accumulation when it was administered three times or twice daily in subjects with normal renal function (127). The pharmacodynamics of ceftobiprole are similar in males and females (127). In patients with moderate to severe renal impairment, dose adjustments should be based on creatinine clearance (127). Hence, the pharmacokinetics and pharmacodynamics of ceftobiprole describe a drug that should be appropriate for the early empirical hospital treatment of patients with infections (127).
One phase 3 clinical trial for community-acquired pneumonia has been completed (1), and two phase 3 trials for cSSSIs (125, 131, 132) are currently ongoing in the United States. Ceftobiprole monotherapy was as effective as vancomycin plus ceftazidime for the treatment of patients with a broad range of cSSSIs and infections due to gram-positive bacteria (including MRSA) and gram-negative bacteria (131). In another phase 3 study, the cure rates for patients with MRSA infections were 92% (56/61) with ceftobiprole treatment and 90% (54/60) with vancomycin treatment (132). Ceftobiprole monotherapy was as effective as vancomycin monotherapy (132) or vancomycin plus ceftazidime (131).
Ceftaroline fosamil.
Ceftaroline fosamil (PPI-0903; formerly known as TAK-599), a water-soluble N-phosphono-type cephalosporin prodrug, is a member of a new class of cephem antibiotics having antibacterial activity against a wide range of species, including the resistant gram-positive pathogens MRSA and multidrug-resistant Streptococcus pneumoniae, as well as common gram-negative pathogenic bacteria. Ceftaroline fosamil, discovered by Takeda Chemical Industries (Osaka, Japan), is currently being developed by Forest Laboratories (New York, NY) for the treatment of infections, including cSSSIs and community-acquired pneumonia.
Ceftaroline, the active form of ceftaroline fosamil, is a potent inhibitor of PBP 2a of MRSA (IC50 = 0.16 to 0.18 μg/ml) (123, 175), which translates into a high level of inhibitory activity (MIC for MRSA = 0.25 to 0.5 μg/ml) (123, 158). As stated earlier, the crystal structure of PBP 2a reveals a closed active site (105) that presumably requires allosteric interactions (such as with the peptidoglycan) to trigger a conformational change to open the active site. Ceftaroline appears to be capable of triggering this conformational change (175). Ceftaroline binds with degrees of high affinity to MSSA PBPs 1, 2, and 3 (IC50s = 0.1, 0.034, and 0.049 μg/ml, respectively) and with less affinity to PBP 4 (IC50 > 1 μg/ml) (123). Ceftaroline has potent in vitro bactericidal activity against vancomycin-intermediate S. aureus (VISA; MIC90 = 1 μg/ml), vancomycin-resistant S. aureus (MIC = 0.12 to 1 μg/ml), and non-daptomycin-susceptible S. aureus (MIC = 0.5 to 1 μg/ml) isolates (160). Against CA-MRSA, ceftaroline has activity (MIC90 = 1 μg/ml) equal to the activities of vancomycin and daptomycin and superior to the activities of clindamycin, linezolid, trimethoprim-sulfamethoxazole, and ceftriaxone. In a rabbit model of endocarditis, ceftaroline exhibited bactericidal activity in vivo against resistant S. aureus strains superior to that of either linezolid or vancomycin (84). Ceftaroline was also more effective than linezolid and vancomycin against a heterogeneous glycopeptide-intermediate S. aureus strain (84).
MSSA, MRSA, CA-MRSA, VISA, penicillin-susceptible S. pneumoniae, PRSP, and β-lactamase-negative ampicillin-resistant Haemophilus influenzae did not develop detectable resistance to ceftaroline (77). Vancomycin-sensitive enterococci and vancomycin-resistant enterococci developed resistance to four times the MIC during 10 serial passages. Overall, ceftaroline demonstrated a low propensity for resistance development among the pathogens tested (77, 128). In addition, resistance among the members of the Enterobacteriaceae family due to class A β-lactamases was reversed by clavulanate (128).
Ceftaroline fosamil, the prodrug, undergoes rapid conversion by plasma phosphatases to active ceftaroline. The maximum plasma concentrations of ceftaroline following multiple intravenous doses (600 mg administered over 1 h every 12 h for 14 days) were 19.0 μg/ml for the first dose and 21.0 μg/ml for the last dose (62). Ceftaroline has a volume of distribution of 0.37 liters/kg (28.3 liters), <20% protein binding, and a serum half-life of 2.6 h (64). No accumulation of ceftaroline is observed with multiple doses. Ceftaroline is eliminated primarily through renal excretion. Dose adjustment is recommended for patients with moderate renal impairment (creatinine clearance, 30 to 50 ml/min) but is not needed for patients with mild renal impairment (63). In a single- and multidose study that used the intramuscular injection of ceftaroline fosamil with healthy adults, excellent bioavailability, predictable pharmacokinetics, a sufficient time with drug levels above the MIC, a low volume of injection, and an acceptable tolerability profile were observed (150). These results indicate that for outpatients intramuscular injection may provide an alternative to intravenous dosing.
To date, two phase 3 clinical trials with patients with cSSSIs have been completed, and two phase 3 clinical trials with patients with community-acquired pneumonia are currently ongoing in United States. In a randomized, double-blinded study of the efficacy and safety of ceftaroline versus those of vancomycin plus aztreonam in patients with cSSSIs, ceftaroline monotherapy (intravenous) was as effective and well tolerated as vancomycin plus aztreonam combination therapy for the treatment of patients infected with both gram-positive and gram-negative pathogens (36). The clinical cure rates were similar for ceftaroline and vancomycin plus aztreonam in clinically evaluable subjects (91% and 93%, respectively). The clinical cure rate for MRSA infections was 95% for both ceftaroline and vancomycin plus aztreonam. Microbiological success was similar for ceftaroline and vancomycin plus aztreonam overall (92% and 93%, respectively) and for MRSA in particular (95% and 92%, respectively). In conclusion, ceftaroline monotherapy was as effective as vancomycin plus aztreonam (36).
ME1036.
ME1036 (CP5609; developed by Meiji Seika, licensed by Forest) is a broad-spectrum carbapenem that binds with a high affinity to PBP 2a of MRSA (IC50 = 0.13 to 0.73 μg/ml) (100, 175) and that exhibits potent in vitro inhibitory activity against MRSA (158). The inhibitory effect of ME1036 on the enzymatic activity of PBP 2a also appears to coincide with facilitated opening of the active site by allosteric interactions (175).
ME1036 has activity against MRSA and multidrug-resistant streptococci, in addition to broad-spectrum activity against organisms that include extended-spectrum β-lactamase-producing Enterobacteriaceae and common anaerobes (100). In a rat model of S. aureus endocarditis, ME1036 at 100 mg/kg of body weight with cilastin (an inhibitor of dehydropeptidase) at 50 mg/kg (both administered twice daily) was effective in decreasing the bacterial densities in the three assayed tissues (cardiac valve vegetations, kidneys, and spleen) and in preventing relapse over 3 days following treatment (183). Real-time bioluminescent imaging demonstrated a greater reduction in the cardiac bioluminescence signal with ME1036 therapy than with vancomycin (120 mg/kg twice daily) and daptomycin (10 mg/kg once daily) therapy (183). Likewise, the efficacy of ME1036 in a rabbit model of endocarditis was superior to that of vancomycin (129). The in vivo pharmacokinetic activity of ME1036 in a murine thigh infection model against multiple bacteria (including MRSA) showed that the correlations of the time with drug levels above the MIC and efficacy were similar to those of other carbapenems (4).
A series of 1β-methyl carbapenems having structural similarity to ME1036 showed potent activities against MRSA and PRSP, as well as against the gram-negative organism ampicillin-resistant, β-lactamase-negative Haemophilus influenzae (115).
PZ-601 (Razupenem).
PZ-601 (formerly known as SMP-601; licensed from Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan) is a new carbapenem currently being developed by Protez Pharmaceuticals (now Novartis) that has demonstrated a high degree of potency against MRSA. Against clinical isolates of MRSA from Japan, PZ-601 has shown MICs in the range of 0.25 to 4 μg/ml (MIC90 = 2 μg/ml) and minimum bactericidal concentrations (MBCs) in the range of 0.5 to 4 μg/ml (MBC/MIC ratio = 1 to 4) (60). The bactericidal activity against MRSA at concentrations 1 to 16 times the MIC was better than the activities of vancomycin and linezolid. The in vivo 50% effective doses of PZ-601, vancomycin, and linezolid in a mouse model of systemic infection with MRSA were 4.7, 6.0, and 7.4 mg/kg, respectively (60). For 6/10 strains of MRSA, after 18 to 49 days of passage in the presence of sub-MICs of the drug in vitro, the PZ-601 MICs increased from 0.016 to 2 μg/ml to 0.125 to 8 μg/ml (34). The postantibiotic effects of PZ-601 against S. aureus were equivalent to those of vancomycin and linezolid (171). PZ-601 has in vivo efficacy against VISA and is currently in phase 2 clinical trials for the treatment of cSSSIs (1). In a study carried out to determine the safety and multiple-dose pharmacokinetics of PZ-601 in healthy male volunteers, PZ-601 did not cause any serious adverse events (107).
CONCLUSIONS
The growing prevalence of MRSA in hospital- and community-associated infections is of great clinical concern. The development of resistance to vancomycin and teicoplanin (94, 108, 159) further underscores the urgent need to develop innovative antibiotics active against panresistant organisms. Recently, several new antibiotics have been approved for use to help meet this challenge. However, the development of resistance following the introduction of these compounds in the clinical setting is likely (154, 168). Despite the fact that there is no evidence that the need for novel therapeutics to treat drug-resistant infections will be met in the foreseeable future (17), discernible progress is being made in the development of antibiotics active against MRSA. The development of new β-lactam antibiotics that inhibit PBP 2a is significant progress and offers a glimmer of hope for the future treatment of MRSA infections, including those caused by vancomycin-resistant and -intermediate strains.
Acknowledgments
Leticia I. Llarrull is a Pew Latin American Fellow in the Biomedical Sciences, supported by The Pew Charitable Trusts.
The opinions expressed are those of the authors and do not necessarily reflect the views of The Pew Charitable Trusts.
The authors acknowledge the financial support of this work by the NIH.
Biography
 Leticia I. Llarrull received her B.S. degree in biotechnology in 2001 from the National University of Rosario, Argentina. She obtained her Ph.D. in biological sciences at the same institution in 2007 under the supervision of Prof. Alejandro Vila, characterizing the catalytic mechanism of the metallo-β-lactamase BcII. She is currently a Pew Latin American Fellow in the Biomedical Sciences in the laboratory of Shahriar Mobashery. Her particular interest is the elucidation of the mechanisms of resistance of pathogenic bacteria to the β-lactam antibiotics.
Leticia I. Llarrull received her B.S. degree in biotechnology in 2001 from the National University of Rosario, Argentina. She obtained her Ph.D. in biological sciences at the same institution in 2007 under the supervision of Prof. Alejandro Vila, characterizing the catalytic mechanism of the metallo-β-lactamase BcII. She is currently a Pew Latin American Fellow in the Biomedical Sciences in the laboratory of Shahriar Mobashery. Her particular interest is the elucidation of the mechanisms of resistance of pathogenic bacteria to the β-lactam antibiotics.
 Jed F. Fisher had the extraordinary fortune to learn chemistry from a series of outstanding mentors: Bill Fowler (SUNY Stony Brook), Chris Walsh (MIT), and Jeremy Knowles (Harvard). His subsequent independent career focused on drug mechanisms (University of Minnesota) and drug discovery (The Upjohn Company). In 2005, he joined the University of Notre Dame with a renewed focus on the challenges of antimicrobial mechanisms and resistance, working in collaboration with his friend Shahriar Mobashery.
Jed F. Fisher had the extraordinary fortune to learn chemistry from a series of outstanding mentors: Bill Fowler (SUNY Stony Brook), Chris Walsh (MIT), and Jeremy Knowles (Harvard). His subsequent independent career focused on drug mechanisms (University of Minnesota) and drug discovery (The Upjohn Company). In 2005, he joined the University of Notre Dame with a renewed focus on the challenges of antimicrobial mechanisms and resistance, working in collaboration with his friend Shahriar Mobashery.
 Shahriar Mobashery received his undergraduate (1981) and doctoral (1985) degrees from the University of Southern California and the University of Chicago, respectively. Subsequent to postdoctoral research (1986 to 1988) at the Rockefeller University, he joined the faculty at Wayne State University in 1989. Mobashery and his research group relocated to the University of Notre Dame in 2003, where he holds the position of Navari Family Professor of Life Sciences in the Department of Chemistry and Biochemistry. A major focus of research in Mobashery's lab for the past 2 decades has been studies of antibiotic resistance mechanisms.
Shahriar Mobashery received his undergraduate (1981) and doctoral (1985) degrees from the University of Southern California and the University of Chicago, respectively. Subsequent to postdoctoral research (1986 to 1988) at the Rockefeller University, he joined the faculty at Wayne State University in 1989. Mobashery and his research group relocated to the University of Notre Dame in 2003, where he holds the position of Navari Family Professor of Life Sciences in the Department of Chemistry and Biochemistry. A major focus of research in Mobashery's lab for the past 2 decades has been studies of antibiotic resistance mechanisms.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Abbanat, D., B. Morrow, and K. Bush. 2008. New agents in development for the treatment of bacterial infections. Curr. Opin. Pharmacol. 8:582-592. [DOI] [PubMed] [Google Scholar]
- 2.Aksoy, D. Y., and S. Unal. 2008. New antimicrobial agents for the treatment of gram-positive bacterial infections. Clin. Microbiol. Infect. 14:411-420. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, S. D., and J. G. Gums. 2008. Ceftobiprole: an extended-spectrum anti-methicillin-resistant Staphylococcus aureus cephalosporin. Ann. Pharmacother. 42:806-816. [DOI] [PubMed] [Google Scholar]
- 4.Andes, D., and W. Craig. 2008. In vivo pharmacodynamic activity of carbapenem ME1036 in a murine thigh infection model, poster A-032. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 5.Antignac, A., and A. Tomasz. 2009. Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob. Agents Chemother. 53:435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appelbaum, P. C. 2007. Microbiology of antibiotic resistance in Staphylococcus aureus. Clin. Infect. Dis. 45(Suppl. 3):S165-S170. [DOI] [PubMed] [Google Scholar]
- 7.Arias, C. A., and B. E. Murray. 2009. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N. Engl. J. Med. 360:439-443. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee, R., M. Gretes, L. Basuino, N. Strynadka, and H. F. Chambers. 2008. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2089-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. [DOI] [PubMed] [Google Scholar]
- 10.Ben Zakour, N. L., C. M. Guinane, and J. R. Fitzgerald. 2008. Pathogenomics of the staphylococci: insights into niche adaptation and the emergence of new virulent strains. FEMS Microbiol. Lett. 289:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Berger-Bachi, B. 1999. Genetic basis of methicillin resistance in Staphylococcus aureus. Cell. Mol. Life Sci. 56:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger-Bachi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165-171. [DOI] [PubMed] [Google Scholar]
- 13.Berger-Bachi, B., A. Strassle, J. E. Gustafson, and F. H. Kayser. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger-Bachi, B., and M. Tschierske. 1998. Role of fem factors in methicillin resistance. Drug Resist. Updat. 1:325-335. [DOI] [PubMed] [Google Scholar]
- 15.Bogdanovich, T., L. M. Ednie, S. Shapiro, and P. C. Appelbaum. 2005. Antistaphylococcal activity of ceftobiprole, a new broad-spectrum cephalosporin. Antimicrob. Agents Chemother. 49:4210-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boucher, H. W., and G. R. Corey. 2008. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl. 5):S344-S349. [DOI] [PubMed] [Google Scholar]
- 17.Boucher, H. W., G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg, and J. Bartlett. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1-12. [DOI] [PubMed] [Google Scholar]
- 18.Boyle-Vavra, S., S. Yin, and R. S. Daum. 2006. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262:163-171. [DOI] [PubMed] [Google Scholar]
- 19.Braaten, D. 2007. Bugs vs drugs. Nat. Med. 13:522-523. [DOI] [PubMed] [Google Scholar]
- 20.Bush, K., M. Heep, M. J. Macielag, and G. J. Noel. 2007. Anti-MRSA beta-lactams in development, with a focus on ceftobiprole: the first anti-MRSA beta-lactam to demonstrate clinical efficacy. Expert Opin. Investig. Drugs 16:419-429. [DOI] [PubMed] [Google Scholar]
- 21.Butler, M. S. 2008. Natural products to drugs: natural product-derived compounds in clinical trials. Nat. Prod. Rep. 25:475-516. [DOI] [PubMed] [Google Scholar]
- 22.Butler, M. S., and A. D. Buss. 2006. Natural products—the future scaffolds for novel antibiotics? Biochem. Pharmacol. 71:919-929. [DOI] [PubMed] [Google Scholar]
- 23.Cha, J., S. B. Vakulenko, and S. Mobashery. 2007. Characterization of the β-lactam antibiotic sensor domain of the MecR1 signal sensor/transducer protein from methicillin-resistant Staphylococcus aureus. Biochemistry 46:7822-7831. [DOI] [PubMed] [Google Scholar]
- 24.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers, H. F. 2005. Community-associated MRSA—resistance and virulence converge. N. Engl. J. Med. 352:1485-1487. [DOI] [PubMed] [Google Scholar]
- 27.Chambers, H. F. 2009. Pathogenesis of staphylococcal infection: a manner of expression. J. Infect. Dis. 199:291-293. [DOI] [PubMed] [Google Scholar]
- 28.Chambers, H. F., and S. S. Hegde. 2007. Combating the growing problem of methicillin-resistant Staphylococcus aureus: do the newer antibiotics represent a better alternative to vancomycin? Expert. Rev. Anti-Infect. Ther. 5:333-335. [DOI] [PubMed] [Google Scholar]
- 29.Cheng, T. J., M. T. Sung, H. Y. Liao, Y. F. Chang, C. W. Chen, C. Y. Huang, L. Y. Chou, Y. D. Wu, Y. H. Chen, Y. S. Cheng, C. H. Wong, C. Ma, and W. C. Cheng. 2008. Domain requirement of moenomycin binding to bifunctional transglycosylases and development of high-throughput discovery of antibiotics. Proc. Natl. Acad. Sci. USA 105:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho, S., Q. Wang, C. P. Swaminathan, D. Hesek, M. Lee, G. J. Boons, S. Mobashery, and R. A. Mariuzza. 2007. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 104:8761-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung, M., A. Antignac, C. Kim, and A. Tomasz. 2008. Comparative study of the susceptibilities of major epidemic clones of methicillin-resistant Staphylococcus aureus to oxacillin and to the new broad-spectrum cephalosporin ceftobiprole. Antimicrob. Agents Chemother. 52:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clardy, J., M. A. Fischbach, and C. T. Walsh. 2006. New antibiotics from bacterial natural products. Nat. Biotechnol. 24:1541-1550. [DOI] [PubMed] [Google Scholar]
- 34.Clark, C., K. Kosowska-Shick, P. McGhee, and P. C. Appelbaum. 2008. Capability of PZ-601 (SMP-601) to select for resistant mutants of vancomycin-susceptible and non-susceptible MRSA, poster C1-195. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 35.Clarke, S. R., and K. G. Dyke. 2001. Studies of the operator region of the Staphylococcus aureus beta-lactamase operon. J. Antimicrob. Chemother. 47:377-389. [DOI] [PubMed] [Google Scholar]
- 36.Corey, G. R., M. Wilcox, G. H. Talbot, T. Baculik, and D. Thye. 2008. CANVAS-1: randomized, double-blinded, phase 3 study (P903-06) of the efficacy and safety of ceftaroline vs. vancomycin plus aztreonam in complicated skin and skin structure infections (cSSSI), poster L-1515a. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 37.Couto, I., H. de Lencastre, E. Severina, W. Kloos, J. A. Webster, R. J. Hubner, I. S. Sanches, and A. Tomasz. 1996. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 2:377-391. [DOI] [PubMed] [Google Scholar]
- 38.Couto, I., I. S. Sanches, R. Sa-Leao, and H. de Lencastre. 2000. Molecular characterization of Staphylococcus sciuri strains isolated from humans. J. Clin. Microbiol. 38:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couto, I., S. W. Wu, A. Tomasz, and H. de Lencastre. 2003. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J. Bacteriol. 185:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Agata, E. M., G. F. Webb, M. A. Horn, R. C. J. Moellering, and S. Ruan. 2009. Modeling the invasion of community-acquired methicillin-resistant Staphylococcus aureus into hospitals. Clin. Infect. Dis. 48:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David, M. Z., D. Glikman, S. E. Crawford, J. Peng, K. J. King, M. A. Hostetler, S. Boyle-Vavra, and R. S. Daum. 2008. What is community-associated methicillin-resistant Staphylococcus aureus? J. Infect. Dis. 197:1235-1243. [DOI] [PubMed] [Google Scholar]
- 43.Davies, T. A., M. G. Page, W. Shang, T. Andrew, M. Kania, and K. Bush. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:2621-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lencastre, H., B. L. de Jonge, P. R. Matthews, and A. Tomasz. 1994. Molecular aspects of methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 33:7-24. [DOI] [PubMed] [Google Scholar]
- 45.de Lencastre, H., D. Oliveira, and A. Tomasz. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 10:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Lencastre, H., and A. Tomasz. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Lencastre, H., and A. Tomasz. 2007. Multiple stages in the evolution of the methicillin resistant Staphylococcus aureus, p. 333-346. In F. Baquero, C. Nombela, G. H. Cassell, and J. A. Gutierrez-Fuentes (ed.), Evolutionary biology of bacterial and fungal pathogens. ASM Press, Washington, DC.
- 48.Deurenberg, R. H., and E. E. Stobberingh. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8:747-763. [DOI] [PubMed] [Google Scholar]
- 49.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 50.Diep, B. A., and M. Otto. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diep, B. A., A. M. Palazzolo-Ballance, P. Tattevin, L. Basuino, K. R. Braughton, A. R. Whitney, L. Chen, B. N. Kreiswirth, M. Otto, F. R. DeLeo, and H. F. Chambers. 2008. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One 3:e3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escaich, S. 2008. Antivirulence as a new antibacterial approach for chemotherapy. Curr. Opin. Chem. Biol. 12:400-408. [DOI] [PubMed] [Google Scholar]
- 54.Fan, X., Y. Liu, D. Smith, L. Konermann, K. W. Siu, and D. Golemi-Kotra. 2007. Diversity of penicillin-binding proteins. Resistance factor FmtA of Staphylococcus aureus. J. Biol. Chem. 282:35143-35152. [DOI] [PubMed] [Google Scholar]
- 55.Forbes, B. A., K. Bombicino, K. Plata, A. Cuirolo, D. Webber, C. L. Bender, and A. E. Rosato. 2008. Unusual form of oxacillin resistance in methicillin-resistant Staphylococcus aureus clinical strains. Diagn. Microbiol. Infect. Dis. 61:387-395. [DOI] [PubMed] [Google Scholar]
- 56.Fuda, C., D. Hesek, M. Lee, K. Morio, T. Nowak, and S. Mobashery. 2005. Activation for catalysis of penicillin-binding protein 2a from methicillin-resistant Staphylococcus aureus by bacterial cell wall. J. Am. Chem. Soc. 127:2056-2057. [DOI] [PubMed] [Google Scholar]
- 57.Fuda, C., M. Suvorov, Q. Shi, D. Hesek, M. Lee, and S. Mobashery. 2007. Shared functional attributes between the mecA gene product of Staphylococcus sciuri and penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. Biochemistry 46:8050-8057. [DOI] [PubMed] [Google Scholar]
- 58.Fuda, C., M. Suvorov, S. B. Vakulenko, and S. Mobashery. 2004. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279:40802-40806. [DOI] [PubMed] [Google Scholar]
- 59.Fuda, C. C. S., J. F. Fisher, and S. Mobashery. 2005. Beta-lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell. Mol. Life Sci. 62:2617-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujimoto, K., T. Uda, and K. Kanazawa. 2008. In vitro and in vivo efficacy of SMP-601 (SMP; PZ-601), a novel anti-MRSA carbapenem, against clinical isolates of MRSA in Japan, poster F1-360. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 61.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge, Y., R. Redman, L. Floren, S. Liao, and M. Wikler. 2006. The pharmacokinetics (PK) and safety of ceftaroline (PPI-0903) in healthy subjects receiving multiple-dose intravenous (IV) infusions, poster A-1937. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 63.Ge, Y., D. Thye, S. Liao, and G. H. Talbot. 2006. Pharmacokinetics (PK) of ceftaroline (PPI0903) in subjects with mild or moderate renal impairment (RI), poster A-1939. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 64.Ge, Y., S. Liao, and G. H. Talbot. 2007. Population pharmacokinetics (PK) analysis of ceftaroline (CPT) in volunteers and patients with complicated skin and skin structure infection (cSSSI), poster A-34. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 65.Geronimus, L. H., and S. Cohen. 1957. Induction of staphylococcal penicillinase. J. Bacteriol. 73:28-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghosh, A. S., C. Chowdhury, and D. E. Nelson. 2008. Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 16:309-317. [DOI] [PubMed] [Google Scholar]
- 67.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goffin, C., and J. M. Ghuysen. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66:702-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Golemi-Kotra, D., J. Y. Cha, S. O. Meroueh, S. B. Vakulenko, and S. Mobashery. 2003. Resistance to β-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J. Biol. Chem. 278:18419-18425. [DOI] [PubMed] [Google Scholar]
- 70.Grundmann, H., M. Aires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874-885. [DOI] [PubMed] [Google Scholar]
- 71.Guan, R., and R. A. Mariuzza. 2007. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 15:127-134. [DOI] [PubMed] [Google Scholar]
- 72.Guan, R., A. Roychowdhury, B. Ember, S. Kumar, G. J. Boons, and R. A. Mariuzza. 2004. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 101:17168-17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guignard, B., J. M. Entenza, and P. Moreillon. 2005. Beta-lactams against methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 5:479-489. [DOI] [PubMed] [Google Scholar]
- 74.Hancock, R. E. 2005. Mechanisms of action of newer antibiotics for gram-positive pathogens. Lancet Infect. Dis. 5:209-218. [DOI] [PubMed] [Google Scholar]
- 75.Haydon, D. J., N. R. Stokes, R. Ure, G. Galbraith, J. M. Bennett, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington, and L. G. Czaplewski. 2008. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321:1673-1675. [DOI] [PubMed] [Google Scholar]
- 76.Heller, S., E. Marrer, M. G. P. Page, S. Shapiro, and L. Thenoz. 2004. Development of endogenous resistance by staphylococci to BAL9141 and comparators, abstr. P675. Clin. Microbiol. Infect. 10:163.14759242 [Google Scholar]
- 77.Hinshaw, R. R., R. D. Schaadt, B. Murray, D. Stapert, D. Biek, Y. Ge, G. E. Zurenko, and D. Shinabarger. 2008. Spontaneous mutation frequency and serial passage resistance development studies with ceftaroline (CPT), poster C1-185. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 78.Hiramatsu, K. 1995. Molecular evolution of MRSA. Microbiol. Immunol. 39:531-543. [DOI] [PubMed] [Google Scholar]
- 79.Hiramatsu, K., K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133-136. [DOI] [PubMed] [Google Scholar]
- 80.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 81.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hososaka, Y., H. Hanaki, H. Endo, Y. Suzuki, Z. Nagasawa, Y. Otsuka, T. Nakae, and K. Sunakawa. 2007. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: a new type of MRSA. J. Infect. Chemother. 13:79-86. [DOI] [PubMed] [Google Scholar]
- 83.Ikonomidis, A., G. Michail, A. Vasdeki, M. Labrou, V. Karavasilis, C. Stathopoulos, A. N. Maniatis, and S. Pournaras. 2008. In vitro and in vivo evaluations of oxacillin efficiency against mecA-positive oxacillin-susceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3905-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jacqueline, C., J. Caillon, V. Le Mabecque, A. F. Miegeville, A. Hamel, D. Bugnon, J. Y. Ge, and G. Potel. 2007. In vivo efficacy of ceftaroline (PPI-0903), a new broad-spectrum cephalosporin, compared with linezolid and vancomycin against methicillin-resistant and vancomycin-intermediate Staphylococcus aureus in a rabbit endocarditis model. Antimicrob. Agents Chemother. 51:3397-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones, R. N. 2003. Global epidemiology of antimicrobial resistance among community-acquired and nosocomial pathogens: a five-year summary from the SENTRY Antimicrobial Surveillance Program (1997-2001). Semin. Respir. Crit. Care. Med. 24:121-134. [DOI] [PubMed] [Google Scholar]
- 86.Kampf, G., S. Adena, H. Ruden, and K. Weist. 2003. Inducibility and potential role of mecA-gene-positive oxacillin-susceptible Staphylococcus aureus from colonized healthcare workers as a source for nosocomial infections. J. Hosp. Infect. 54:124-129. [DOI] [PubMed] [Google Scholar]
- 87.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katayama, Y., H. Z. Zhang, and H. F. Chambers. 2003. Effect of disruption of Staphylococcus aureus PBP4 gene on resistance to beta-lactam antibiotics. Microb. Drug Resist. 9:329-336. [DOI] [PubMed] [Google Scholar]
- 90.Katayama, Y., H. Z. Zhang, D. Hong, and H. F. Chambers. 2003. Jumping the barrier to beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 185:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kattan, J. N., M. V. Villegas, and J. P. Quinn. 2008. New developments in carbapenems. Clin. Microbiol. Infect. 14:1102-1111. [DOI] [PubMed] [Google Scholar]
- 92.Khan, S. A., and R. P. Novick. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10:251-259. [DOI] [PubMed] [Google Scholar]
- 93.Koch, A. L. 2003. Bacterial wall as target for attack: past, present, and future research. Clin. Microbiol. Rev. 16:673-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kollef, M. H. 2007. Limitations of vancomycin in the management of resistant staphylococcal infections. Clin. Infect. Dis. 45(Suppl. 3):S191-S195. [DOI] [PubMed] [Google Scholar]
- 95.Komatsuzawa, H., G. H. Choi, T. Fujiwara, Y. Huang, K. Ohta, M. Sugai, and H. Suginaka. 2000. Identification of a fmtA-like gene that has similarity to other PBPs and beta-lactamases in Staphylococcus aureus. FEMS Microbiol. Lett. 188:35-39. [DOI] [PubMed] [Google Scholar]
- 96.Komatsuzawa, H., K. Ohta, T. Fujiwara, G. H. Choi, H. Labischinski, and M. Sugai. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 203:49-54. [DOI] [PubMed] [Google Scholar]
- 97.Komatsuzawa, H., K. Ohta, H. Labischinski, M. Sugai, and H. Suginaka. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Komatsuzawa, H., K. Ohta, M. Sugai, T. Fujiwara, P. Glanzmann, B. Berger-Bachi, and H. Suginaka. 2000. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 45:421-431. [DOI] [PubMed] [Google Scholar]
- 99.Komatsuzawa, H., M. Sugai, K. Ohta, T. Fujiwara, S. Nakashima, J. Suzuki, C. Y. Lee, and H. Suginaka. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2355-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kurazono, M., T. Ida, K. Yamada, Y. Hirai, T. Maruyama, E. Shitara, and M. Yonezawa. 2004. In vitro activities of ME1036 (CP5609), a novel parenteral carbapenem, against methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 48:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 102.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 103.Lappchen, T., V. A. Pinas, A. F. Hartog, G. J. Koomen, C. Schaffner-Barbero, J. M. Andreu, D. Trambaiolo, J. Lowe, A. Juhem, A. V. Popov, and T. den Blaauwen. 2008. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem. Biol. 15:189-199. [DOI] [PubMed] [Google Scholar]
- 104.Lewis, R. A., and K. G. Dyke. 2000. MecI represses synthesis from the beta-lactamase operon of Staphylococcus aureus. J. Antimicrob. Chemother. 45:139-144. [DOI] [PubMed] [Google Scholar]
- 105.Lim, D., and N. C. Strynadka. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870-876. [DOI] [PubMed] [Google Scholar]
- 106.Liu, C. I., G. Y. Liu, Y. Song, F. Yin, M. E. Hensler, W. Y. Jeng, V. Nizet, A. H. Wang, and E. Oldfield. 2008. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319:1391-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lo, T. S., J. M. Welch, A. M. Alonto, and E. A. Vicaldo-Alonto. 2008. A review of the carbapenems in clinical use and clinical trials. Recent Patents Anti-Infect. Drug Disc. 3:123-131. [DOI] [PubMed] [Google Scholar]
- 108.Loffler, C. A., and C. Macdougall. 2007. Update on prevalence and treatment of methicillin-resistant Staphylococcus aureus infections. Expert Rev. Anti-Infect. Ther. 5:961-981. [DOI] [PubMed] [Google Scholar]
- 109.Loughman, J. A., S. A. Fritz, G. A. Storch, and D. A. Hunstad. 2009. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lovering, A. L., L. H. de Castro, D. Lim, and N. C. Strynadka. 2007. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402-1405. [DOI] [PubMed] [Google Scholar]
- 111.Lowy, F. D. 2007. Secrets of a superbug. Nat. Med. 13:1418-1420. [DOI] [PubMed] [Google Scholar]
- 112.Lu, W. P., Y. Sun, M. D. Bauer, S. Paule, P. M. Koenigs, and W. G. Kraft. 1999. Penicillin-binding protein 2a from methicillin-resistant Staphylococcus aureus: kinetic characterization of its interactions with beta-lactams using electrospray mass spectrometry. Biochemistry 38:6537-6546. [DOI] [PubMed] [Google Scholar]
- 113.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184:3623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maki, H., T. Yamaguchi, and K. Murakami. 1994. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J. Bacteriol. 176:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maruyama, T., Y. Kano, Y. Yamamoto, M. Kurazono, K. Iwamatsu, K. Atsumi, and E. Shitara. 2007. Synthesis and SAR study of novel 7-(pyridinium-3-yl)-carbonyl imidazo[5,1-b]thiazol-2-yl carbapenems. Bioorg. Med. Chem. 15:392-402. [DOI] [PubMed] [Google Scholar]
- 116.Massova, I., and S. Mobashery. 1998. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob. Agents Chemother. 42:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McAleese, F., S. W. Wu, K. Sieradzki, P. Dunman, E. Murphy, S. Projan, and A. Tomasz. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate- S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McCallum, N., G. Spehar, M. Bischoff, and B. Berger-Bachi. 2006. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim. Biophys. Acta 1760:1475-1481. [DOI] [PubMed] [Google Scholar]
- 119.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Memmi, G., S. R. Filipe, M. G. Pinho, Z. Fu, and A. Cheung. 2008. Staphylococcus aureus PBP 4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120a.Meroueh, S. O., K. Z. Bencze, D. Hesek, M. Lee, J. F. Fisher, T. L. Stemmler, and S. Mobashery. 2006. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA 103:4404-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Millar, B. C., B. D. Prendergast, and J. E. Moore. 2008. Community-associated MRSA (CA-MRSA): an emerging pathogen in infective endocarditis. J. Antimicrob. Chemother. 61:1-7. [DOI] [PubMed] [Google Scholar]
- 122.Moellering, R. C. 2008. Current treatment options for community-acquired methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46:1032-1037. [DOI] [PubMed] [Google Scholar]
- 123.Moisan, H., M. Pruneau, and F. Malouin. 2008. Binding of ceftaroline (CPT) to penicillin-binding proteins (PBPs) of Streptococcus pneumoniae (SPN) and methicillin-resistant Staphylococcus aureus (MRSA), poster C1-183. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 124.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moreillon, P. 2008. New and emerging treatment of Staphylococcus aureus infections in the hospital setting. Clin. Microbiol. Infect. 14(Suppl. 3):32-41. [DOI] [PubMed] [Google Scholar]
- 126.Murakami, K., and A. Tomasz. 1989. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 171:874-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murthy, B., and A. Schmitt-Hoffmann. 2008. Pharmacokinetics and pharmacodynamics of ceftobiprole, an anti-MRSA cephalosporin with broad-spectrum activity. Clin. Pharmacokinet. 47:21-33. [DOI] [PubMed] [Google Scholar]
- 128.Mushtaq, S., M. Warner, Y. Ge, K. Kaniga, and D. M. Livermore. 2007. In vitro activity of ceftaroline (PPI-0903M, T-91825) against bacteria with defined resistance mechanisms and phenotypes. J. Antimicrob. Chemother. 60:300-311. [DOI] [PubMed] [Google Scholar]
- 129.Nagura, J., K. Kijima, M. Kurazono, S. Takahata, T. Sugano, Y. Tanaka, Y. Hirai, K. Yamada, Y. Takayama, E. Shitara, and M. Yonezawa. 2005. Therapeutic effect of ME1036 on endocarditis experimentally induced by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3526-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Noel, G. J., K. Bush, P. Bagchi, J. Ianus, and R. S. Strauss. 2008. A randomized, double-blind trial comparing ceftobiprole medocaril with vancomycin plus ceftazidime for the treatment of patients with complicated skin and skin-structure infections. Clin. Infect. Dis. 46:647-655. [DOI] [PubMed] [Google Scholar]
- 132.Noel, G. J., R. S. Strauss, K. Amsler, M. Heep, R. Pypstra, and J. S. Solomkin. 2008. Results of a double-blind, randomized trial of ceftobiprole treatment of complicated skin and skin structure infections caused by gram-positive bacteria. Antimicrob. Agents Chemother. 52:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nubel, U., P. Roumagnac, M. Feldkamp, J. H. Song, K. S. Ko, Y. C. Huang, G. Coombs, M. Ip, H. Westh, R. Skov, M. J. Struelens, R. V. Goering, B. Strommenger, A. Weller, W. Witte, and M. Achtman. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 105:14130-14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nygaard, T. K., F. R. DeLeo, and J. M. Voyich. 2008. Community-associated methicillin-resistant Staphylococcus aureus skin infections: advances toward identifying the key virulence factors. Curr. Opin. Infect. Dis. 21:147-152. [DOI] [PubMed] [Google Scholar]
- 135.Page, M. G. 2006. Anti-MRSA beta-lactams in development. Curr. Opin. Pharmacol. 6:480-485. [DOI] [PubMed] [Google Scholar]
- 136.Page, M. G. 2007. Emerging cephalosporins. Expert Opin. Emerg. Drugs 12:511-524. [DOI] [PubMed] [Google Scholar]
- 137.Pan, A., S. Lorenzotti, and A. Zoncada. 2008. Registered and investigational drugs for the treatment of methicillin-resistant Staphylococcus aureus infection. Recent Patents Anti-Infect. Drug Disc. 3:10-33. [DOI] [PubMed] [Google Scholar]
- 138.Park, J. W., C. H. Kim, J. H. Kim, B. R. Je, K. B. Roh, S. J. Kim, H. H. Lee, J. H. Ryu, J. H. Lim, B. H. Oh, W. J. Lee, N. C. Ha, and B. L. Lee. 2007. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. USA 104:6602-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Parvez, M. A., H. Shibata, T. Nakano, S. Niimi, N. Fujii, N. Arakaki, and T. Higuti. 2008. No relationship exists between PBP 2a amounts expressed in different MRSA strains obtained clinically and their beta-lactam MIC values. J. Med. Investig. 55:246-253. [DOI] [PubMed] [Google Scholar]
- 140.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 141.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 98:10886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pinho, M. G., and J. Errington. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871-881. [DOI] [PubMed] [Google Scholar]
- 143.Popovich, K. J., B. Hota, and R. A. Weinstein. 2008. Treatment of community-associated methicillin-resistant Staphylococcus aureus. Curr. Infect. Dis. Rep. 10:411-420. [DOI] [PubMed] [Google Scholar]
- 144.Popovich, K. J., R. A. Weinstein, and B. Hota. 2008. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin. Infect. Dis. 46:787-794. [DOI] [PubMed] [Google Scholar]
- 145.Projan, S. J., and P. A. Bradford. 2007. Late stage antibacterial drugs in the clinical pipeline. Curr. Opin. Microbiol. 10:441-446. [DOI] [PubMed] [Google Scholar]
- 146.Pucci, M. J., and T. J. Dougherty. 2002. Direct quantitation of the numbers of individual penicillin-binding proteins per cell in Staphylococcus aureus. J. Bacteriol. 184:588-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Queenan, A. M., W. Shang, M. Kania, M. G. Page, and K. Bush. 2007. Interactions of ceftobiprole with beta-lactamases from molecular classes A to D. Antimicrob. Agents Chemother. 51:3089-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Raju, S., R. Kelmani Chandrakanth, and S. A. Patil. 2007. High-level oxacillin and gentamycin resistance with reduced susceptibility to vancomycin in Staphylococcus aureus-carrying mecA and femA gene complex. Curr. Microbiol. 54:429-434. [DOI] [PubMed] [Google Scholar]
- 149.Rasmussen, F. 2007. Discovery, isolation, production and introduction of penicillin for veterinary use in Denmark during World War II. Vet. History 13:339-353. [Google Scholar]
- 150.Riccobene, T., E. Fang, and D. Thye. 2008. A single- and multiple-dose study to determine the safety, tolerability, and pharmacokinetics (PK) of ceftaroline (CPT) administered by intramuscular (IM) injection to healthy subjects, poster A-1888. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 151.Richards, M. J., J. R. Edwards, D. H. Culver, R. P. Gaynes, et al. 1999. Nosocomial infections in pediatric intensive care units in the United States. Pediatrics 103:e39. [DOI] [PubMed] [Google Scholar]
- 152.Richards, M. J., J. R. Edwards, D. H. Culver, R. P. Gaynes, et al. 1999. Nosocomial infections in medical intensive care units in the United States. Crit. Care Med. 27:887-892. [DOI] [PubMed] [Google Scholar]
- 153.Rohrer, S., and B. Berger-Bachi. 2003. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob. Agents Chemother. 47:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ross, J. E., T. R. Fritsche, H. S. Sader, and R. N. Jones. 2007. Oxazolidinone susceptibility patterns for 2005: International Report from the Zyvox Annual Appraisal of Potency and Spectrum Study. Int. J. Antimicrob. Agents 29:295-301. [DOI] [PubMed] [Google Scholar]
- 155.Royet, J., and R. Dziarski. 2007. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 5:264-277. [DOI] [PubMed] [Google Scholar]
- 156.Ryffel, C., F. H. Kayser, and B. Berger-Bachi. 1992. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 36:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ryffel, C., A. Strassle, F. H. Kayser, and B. Berger-Bachi. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:724-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sader, H. S., T. R. Fritsche, and R. N. Jones. 2008. Antimicrobial activities of ceftaroline and ME1036 tested against clinical strains of community-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52:1153-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sakoulas, G., and R. C. Moellering. 2008. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin. Infect. Dis. 46(Suppl. 5):S360-S367. [DOI] [PubMed] [Google Scholar]
- 160.Saravolatz, L. D., J. Pawlak, and L. Johnson. 2008. In vitro activity of ceftaroline against CA-MRSA, VISA, VRSA and daptomycin-non-susceptible Staphylococcus aureus (DNSSA), poster C1-162. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 161.Sauvage, E., F. Kerff, M. Terrak, J. A. Ayala, and P. Charlier. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234-258. [DOI] [PubMed] [Google Scholar]
- 162.Schmitt-Hoffmann, A., L. Nyman, B. Roos, M. Schleimer, J. Sauer, N. Nashed, T. Brown, A. Man, and E. Weidekamm. 2004. Multiple-dose pharmacokinetics and safety of a novel broad-spectrum cephalosporin (BAL5788) in healthy volunteers. Antimicrob. Agents Chemother. 48:2576-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schmitt-Hoffmann, A., B. Roos, M. Schleimer, J. Sauer, A. Man, N. Nashed, T. Brown, A. Perez, E. Weidekamm, and P. Kovacs. 2004. Single-dose pharmacokinetics and safety of a novel broad-spectrum cephalosporin (BAL5788) in healthy volunteers. Antimicrob. Agents Chemother. 48:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stepanovic, S., V. Dimitrijevic, D. Vukovic, I. Dakic, B. Savic, and M. Svabic-Vlahovic. 2001. Staphylococcus sciuri as a part of skin, nasal and oral flora in healthy dogs. Vet. Microbiol. 82:177-185. [DOI] [PubMed] [Google Scholar]
- 165.Suzuki, E., K. Kuwahara-Arai, J. F. Richardson, and K. Hiramatsu. 1993. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob. Agents Chemother. 37:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Thumanu, K., J. Cha, J. F. Fisher, R. Perrins, S. Mobashery, and C. Wharton. 2006. Discrete steps in sensing of beta-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc. Natl. Acad. Sci. USA 103:10630-10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Toh, S. M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tomasz, A., H. B. Drugeon, H. M. de Lencastre, D. Jabes, L. McDougall, and J. Bille. 1989. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob. Agents Chemother. 33:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tu Quoc, P. H., P. Genevaux, M. Pajunen, H. Savilahti, C. Georgopoulos, J. Schrenzel, and W. L. Kelley. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 75:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Uda, T., K. Takemoto, K. Fujimoto, K. Eguchi, and K. Kanazawa. 2008. Post-antibiotic effect (PAE) of SMP-601 (PZ-601) against Staphylococcus aureus including MRSA and VISA, poster F1-361. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 172.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 173.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 174.van Heijenoort, J., and L. Gutmann. 2000. Correlation between the structure of the bacterial peptidoglycan monomer unit, the specificity of transpeptidation, and susceptibility to beta-lactams. Proc. Natl. Acad. Sci. USA 97:5028-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Villegas-Estrada, A., M. Lee, D. Hesek, S. B. Vakulenko, and S. Mobashery. 2008. Co-opting the cell wall in fighting methicillin-resistant Staphylococcus aureus: potent inhibition of PBP 2a by two anti-MRSA beta-lactam antibiotics. J. Am. Chem. Soc. 130:9212-9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149-167. [DOI] [PubMed] [Google Scholar]
- 177.von Nussbaum, F., M. Brands, B. Hinzen, S. Weigand, and D. Habich. 2006. Antibacterial natural products in medicinal chemistry—exodus or revival? Angew. Chem. Int. Ed. Engl. 45:5072-5129. [DOI] [PubMed] [Google Scholar]
- 178.Waxman, D. J., and J. L. Strominger. 1983. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 52:825-869. [DOI] [PubMed] [Google Scholar]
- 179.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 180.Wright, G. D., and A. D. Sutherland. 2007. New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 13:260-267. [DOI] [PubMed] [Google Scholar]
- 181.Wu, S., H. de Lencastre, A. Sali, and A. Tomasz. 1996. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb. Drug Resist. 2:277-286. [DOI] [PubMed] [Google Scholar]
- 182.Wu, S. W., H. de Lencastre, and A. Tomasz. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 183:2417-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xiong, Y. Q., Y. Li, W. Abdel Hady, D. Biek, Y. Ge, and A. S. Bayer. 2008. Real-time evaluation of a new carbapenem, ME1036, in a rat Staphylococcus aureus endocarditis (IE) model using in vivo bioluminescent imaging, poster F1-343. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents. Chemother. (ICAAC)/Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 184.Yuan, Y., S. Fuse, B. Ostash, P. Sliz, D. Kahne, and S. Walker. 2008. Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design. ACS Chem. Biol. 3:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zapun, A., C. Contreras-Martel, and T. Vernet. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361-385. [DOI] [PubMed] [Google Scholar]
- 186.Zetola, N., J. S. Francis, E. L. Nuermberger, and W. R. Bishai. 2005. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275-286. [DOI] [PubMed] [Google Scholar]
- 187.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]
- 188.Zhou, Y., A. Antignac, S. W. Wu, and A. Tomasz. 2008. Penicillin-binding proteins and cell wall composition in beta-lactam-sensitive and -resistant strains of Staphylococcus sciuri. J. Bacteriol. 190:508-514. [DOI] [PMC free article] [PubMed] [Google Scholar]